How homologous chromosomes recognize each other during meiosis remains a fundamental question in chromosome biology. Ishiguro et al. now show that homolog recognition in spermatocytes starts in early meiotic prophase. Intriguingly, meiosis-specific cohesin plays an essential role in homolog recognition far prior to DNA double-strand break formation. The findings suggest the possibility that homolog recognition is achieved primarily by searching for homology in the chromosome architecture as defined by meiosis-specific cohesin rather than the DNA sequence itself.
Keywords: homolog pairing, cohesin, DSB, bouquet
Abstract
During meiosis, homologous chromosome (homolog) pairing is promoted by several layers of regulation that include dynamic chromosome movement and meiotic recombination. However, the way in which homologs recognize each other remains a fundamental issue in chromosome biology. Here, we show that homolog recognition or association initiates upon entry into meiotic prophase before axis assembly and double-strand break (DSB) formation. This homolog association develops into tight pairing only during or after axis formation. Intriguingly, the ability to recognize homologs is retained in Sun1 knockout spermatocytes, in which telomere-directed chromosome movement is abolished, and this is the case even in Spo11 knockout spermatocytes, in which DSB-dependent DNA homology search is absent. Disruption of meiosis-specific cohesin RAD21L precludes the initial association of homologs as well as the subsequent pairing in spermatocytes. These findings suggest the intriguing possibility that homolog recognition is achieved primarily by searching for homology in the chromosome architecture as defined by meiosis-specific cohesin rather than in the DNA sequence itself.
Meiosis generates haploid gametes from a diploid parental cell. This process is initiated by the pairing of homologous chromosomes (homologs) and subsequent double-strand break (DSB)-mediated recombination (Neale and Keeney 2006; Baudat and de Massy 2007). A number of mechanisms are involved in chromosome pairing and alignment. The first step is the attachment of telomeres (or pairing centers in Caenorhabditis elegans) to the nuclear envelope (NE) (Scherthan 2001; Hiraoka and Dernburg 2009). The following telomere-led nuclear movement and polarized chromosome arrangement bouquet facilitate chromosome alignment and homolog pairing/synapsis (Zickler and Kleckner 1999; Scherthan 2001; Page and Hawley 2004). The physical recognition of homologs might be driven by DSB-dependent recombination machinery that involves a homology search on the basis of DNA sequence. However, since repetitive elements comprise 30%–50% of mammalian genomes, wide-range homology search, rather than regional DNA sequence identity, might be important to avoid nonallelic pairing and recombination in meiosis (Zickler and Kleckner 1999; Page and Hawley 2004; Sasaki et al. 2010). In fact, in some organisms undergoing recombinationless meiosis, homologs are properly paired and even synapsed in a DSB-independent manner (Dernburg et al. 1998; McKim et al. 1998). Also, in fungi (yeasts and Sordaria), while a tight association is established by DSB-dependent recombination and subsequent synapsis, DSB-independent mechanisms seem to promote the juxtaposition of homologs (Weiner and Kleckner 1994; Peoples et al. 2002; Storlazzi et al. 2003; Bhuiyan and Schmekel 2004; Ding et al. 2004; Peoples-Holst and Burgess 2005). A recent study in mice reported that a DSB-independent mechanism promotes homolog pairing during premeiotic S phase prior to DSB formation (Boateng et al. 2013). The molecular mechanism of DSB-independent homolog recognition remains a mystery, although the aggregation of heterochromatin, noncoding RNA, the SPO11 protein, or sequence-specific DNA-binding proteins such as transcription factors are suggested to contribute to this process (Page and Hawley 2004; Barzel and Kupiec 2008; Dombecki et al. 2011; Ding et al. 2012; Boateng et al. 2013).
The cohesin complex in meiosis differs from that in mitosis, in which the α-kleisin subunit SCC1/RAD21 is largely replaced by a meiotic counterpart, REC8 (Watanabe 2004; Nasmyth and Haering 2005). Meiotic cohesin is crucial not only for sister chromatid cohesion but also to act as a structural basis for axial element (AE) formation and synaptonemal complex (SC) assembly during prophase I (Klein et al. 1999; Zickler and Kleckner 1999; Prieto et al. 2001; Page and Hawley 2004; Novak et al. 2008; Llano et al. 2012). Recent studies in mice have identified a novel meiosis-specific α-kleisin subunit of cohesin, RAD21L (Herran et al. 2011; Ishiguro et al. 2011; Lee and Hirano 2011), in addition to the previously identified REC8 (Bannister et al. 2004; Xu et al. 2005). RAD21L and REC8 form distinct cohesin complexes and distribute uniquely along each chromosome but identically between homologs during early meiotic prophase I, whereas the mitotic α-kleisin subunit RAD21 appears only transiently at the later pachytene stage in meiosis (Ishiguro et al. 2011; Lee and Hirano 2011). Although RAD21L and REC8 may play predominant roles in meiotic prophase, as their knockout spermatocytes show zygotene-like arrest, the precise functions of these meiosis-specific cohesin complexes remain largely elusive.
In this study, we demonstrate that Spo11 knockout spermatocytes retain the ability to promote homolog pairing, indicating that DSB-independent homolog pairing operates prior to or independently of DSB-dependent homology search. Furthermore, we show that the disruption of Rad21L in spermatocytes causes a primary defect in the process of meiotic homolog pairing, resulting in arrest at the bouquet stage with aberrant synapsis. Our study highlights previously unknown properties of the atypical meiotic cohesin RAD21L in DSB-independent homolog pairing and the bouquet exit checkpoint.
Results
Homolog pairing initiates in early leptotene
To examine homolog pairing in mouse spermatocytes, we performed fluorescent in situ hybridization (FISH) assays using structurally preserved nuclei (∼10-µm diameter) and probes that detect specific DNA sequences in the mid-arm regions of chromosomes 8 and 3 and the subtelomeric region of chromosome 18 (Fig. 1A). In wild-type spermatocytes, homolog pairing was undetectable in both the arm and subtelomeric regions during premeiotic S phase, which is identified as EdU-positive, patchy SYCP3 immunostaining and relatively spherical nuclei larger than those of somatic cells (Fig. 1B). However, the overall distance between FISH signals in both the arm and subtelomeric regions decreased from the early to late leptotene stages, the period during which AEs are assembled with accompanying DMC1 and RAD51 foci, but the SC remains absent (Fig. 1A; Supplemental Fig. 1). The overall distance between FISH signals decreased further during the zygotene stage, when most chromosomes are paired but not completely synapsed. Homologs are mostly paired in the pachytene stage, during which AEs are totally synapsed by SC assembly (Fig. 1A,B). We confirmed that this pairing is significant by showing that FISH probes hybridized to different chromosomes do not pair (Supplemental Fig. 2A). These results are consistent with the previous observations (Scherthan et al. 1996) but partly contradictory to a recent study that reports the appearance of a transient pairing peak at late premeiotic S phase (see the Discussion; Boateng et al. 2013). Under our experimental conditions, the distance between paired FISH signals decreases to <1.35 µm in 45%∼60% of zygotene spermatocytes and in >95% of pachytene spermatocytes (Fig. 1B). Therefore, we define this distance between FISH signals as “paired” throughout the assays. By this criterion, pairing at the late leptotene stage reaches 30%∼40% in both the arm and subtelomeric regions in wild-type spermatocytes.
Figure 1.
Homologs recognize each other during early leptotene stage. (A, left) Structurally preserved nuclei from wild-type (WT) spermatocytes (4–8 wk old) immunostained with the indicated antibodies or EdU were subjected to FISH with a point probe (Chr. 3). (Right) The schematic illustrates three-dimensional (3D) measurement of distance between FISH signals and relative positions of the point probes. (B) Homolog pairing was examined using the point probes to detect the mid-region on chromosome 3, the mid-region on Chr. 8, or the subtelomeric region on Chr. 18. (Bottom) The distances between two probe signals are represented in a scatter plot with medians. The dashed line at 1.35 μm indicates the threshold for pairing. P-values (Mann-Whitney t-test) are shown. (*) P < 0.05; (**) P < 0.01. (Top) The homolog pairing ratio is shown in the graph. (Pre-mei. S) Premeiotic S; (e. Lep.) early leptotene; (l. Lep.) late leptotene; (Zyg.) zygotene; (Pachy.) pachytene. (C) Structurally preserved nuclei from wild-type spermatocytes (4 wk old) were stained as indicated and labeled by FISH with a Chr. 8 painting probe (green). (Left) Representative images are shown with four single Z-sections of Chr. 8 FISH signals. The schematic model illustrates the relative positioning of homolog chromosome territories. (Right) The proximity of chromosome territories of homologs was classified as separated, juxtaposed, or fused and is shown in the graph. (*) P < 0.05 (Pearson’s χ2 test). Bars, 5 μm. (D) Schematic illustration of the relative positioning of FISH point probes (green stars) and chromosome territories (gray) of homologs in meiotic prophase spermatocytes. AE and lateral element (LE) are shown (dark-gray bar).
To delineate the initial homolog association, we next performed FISH chromosome painting, a method to monitor chromosome territory. FISH chromosome painting using preserved nuclei of wild-type spermatocytes revealed that chromosome 8 homologs occupy spatially limited territories during meiotic prophase, as previously reported in human spermatogonia (Fig. 1C; Scherthan et al. 1996), which is similar to the chromosome territories observed in interphase somatic cells (Cremer and Cremer 2010). Although most homologs are separated from each other during premeiotic S phase, some homologs start to locate in close proximity to one another (juxtaposed) at the early leptotene stage (Fig. 1C) despite the fact that they have not yet developed chromosome axes and accompanying compaction (Fig. 1A,B). The homolog association develops into a single rod-shaped entity (fusion) from the leptotene to zygotene stages, when most chromosomal sites are paired (Fig. 1C). These results suggest that meiosis-specific homolog association starts presumably through interaction of the surface of the homologous chromosome territory in the early leptotene stage, where DNA replication is completed, but chromosome axes defined by SYCP3 staining are not yet formed.
We reason that homolog interaction is not always detected by paired point FISH signals (cutoff value of 1.35 µm), especially when chromosomes are associated but not paired at the specific site, and that the proximity of homologs can be measured more quantitatively by the overall distance between point FISH signals or its median (Fig. 1D). We therefore used this value throughout the study to inspect homolog association.
Homolog association in Sun1 knockout
A crucial event promoting homolog synapsis is telomere-mediated chromosome movement and the bouquet configuration of chromosomes, a process that depends on the NE proteins SUN/KASH and is disrupted in Sun1 knockout mice (Ding et al. 2007). We examined the effect of Sun1 knockout on the process of homolog pairing. FISH assays using point probes reveal that Sun1 knockout spermatocytes also retain pairing ability, albeit less than that of wild-type spermatocytes (Fig. 2A). This result is contradictory to the previous observation that homolog pairing is abolished in Sun1 knockout spermatocytes (see the Discussion; Boateng et al. 2013). Furthermore, FISH chromosome painting assays indicate that the initial chromosome association at the early leptotene stage is delayed in Sun1 knockout spermatocytes (Fig. 2B). These results indicate that homolog association and pairing progress slowly in Sun1 knockout spermatocytes, implying that active chromosome movement is not essential for homolog recognition, although it is required to facilitate and ensure pairing.
Figure 2.
Homolog pairing occurs independently of SUN1-mediated chromosome movement. (A) Structurally preserved nuclei from Sun1 knockout (KO) spermatocytes (4–8 wk old) immunostained with the indicated antibodies were subjected to FISH using the point probes to detect the mid-region on Chr. 8 (left) or the subtelomeric region on Chr. 18 (middle). (Bottom right) The distances between two probe signals are represented in a scatter plot with medians. P-values (Mann-Whitney t-test) are shown. (*) P < 0.05; (**) P < 0.01. (Top right) The homolog pairing ratio is shown in the graph. (B, left) Structurally preserved nuclei from Sun1 knockout spermatocytes (4 wk old) were stained as indicated and labeled by FISH with a Chr. 8 painting probe. (Right) The proximity of the chromosome territories of homologs was classified as separated, juxtaposed, or fused and is shown in the graph. P-values are shown (Pearson’s χ2 test). Bars, 5 μm.
Homolog association in Spo11 knockout
To examine whether DSB or its dependent DNA search is the primary determinant of homolog recognition, we analyzed Spo11 knockout mice in which DSB formation is abolished (Supplemental Fig. 1). Spo11 knockout spermatocytes form cohesin axes and AEs but fail to undergo proper synapsis, thus arresting at the late leptotene or zygotene-like stage (Fig. 3A; Baudat et al. 2000; Romanienko and Camerini-Otero 2000). Strikingly, in Spo11 knockout spermatocytes, FISH analysis using point probes detected homolog association and pairing at all tested chromosomal regions, although less than in wild-type spermatocytes (Fig. 3A). We confirmed that this pairing is significant by showing that FISH probes hybridized to different chromosomes do not pair (Supplemental Fig. 2B).
Figure 3.
Homolog pairing occurs independently of DSB. (A, top) Structurally preserved nuclei from Spo11 knockout (KO) spermatocytes (4–8 wk old) immunostained with the indicated antibodies were subjected to FISH with the Chr. 8 point probe. Homolog pairing was examined using the point probes to detect the mid-region on Chr. 3, the mid-region on Chr. 8, or the subtelomeric region on Chr. 18. (Bottom) The distances between two probe signals are represented in a scatter plot with medians. P-values (Mann-Whitney t-test) are shown. (*) P < 0.05; (**) P < 0.01. (B, left) Structurally preserved nuclei from Spo11 knockout spermatocytes (4 wk old) were stained as indicated and labeled by FISH with a Chr. 8 painting probe. (Right) The proximity of the chromosome territories of homologs was classified and is shown in the graph. (*) P < 0.05 (Pearson’s χ2 test). Note homologous synapsis in zygotene-like spermatocyte. (C) Mildly spread nuclei from wild-type (WT) and Spo11 knockout spermatocytes (4 wk old) were analyzed by FISH with a Chr. 8 painting probe (green). Images from a Z-section are shown. (Panels i–v) Enlarged images are shown at the bottom. Arrows indicate aligned or paired homologs. Bars, 5 μm.
To delineate Spo11-independent homolog pairing, we performed FISH chromosome painting. As in wild-type spermatocytes, homologs start to associate at the early leptotene stage in Spo11 knockout spermatocytes (Fig. 3B). Zooming in on the chromosome axes (SYCP3) reveals that the axes are synapsed during the zygotene stage only in wild-type spermatocytes (∼42%; n = 59), while the homolog synapsis is hardly detected in Spo11 knockout spermatocytes, at least in the early zygotene stage (Fig. 3C). (Note that Spo11 knockout spermatocytes produce aberrant nonhomologous synapsis in the late zygotene-like stage [Baudat et al. 2000; Romanienko and Camerini-Otero 2000].)
These results indicate that homolog recognition is retained even in the absence of SPO11, although SPO11-dependent events may also facilitate homolog pairing.
Cohesin and chromosome architecture
The foregoing results suggest that although meiotic homolog pairing indeed requires DSB or chromosome movement, another mechanism of homolog recognition still operates in their absence because disruption of either pathway does not remove the pairing property of meiotic chromosomes (Figs. 2, 3). Considering that the early leptotene stage is the period when homolog association initiates, we envisaged that some proteins that are already assembled on chromatin during this stage might be involved in homolog recognition. Because cohesin might be such a candidate, we decided to explore the function of meiotic cohesin by newly making a Rad21L knockout (Supplemental Fig. 3) and combining it with the Rec8(mei8) mutant (hereafter referred to as Rec8 knockout) (Bannister et al. 2004) and other meiotic mutants.
We first examined the meiotic chromosome structures of Rad21L knockout, Rec8 knockout, and Rec8/Rad21L double-knockout spermatocytes. Chromosome spread assays indicated that both Rad21L knockout and Rec8 knockout spermatocytes form AEs, presumably on the chromosome axis where the core cohesin subunit SMC3 locates, while they arrest at the leptotene/zygotene stage with aberrant SC formation (Fig. 4A,B; Supplemental Fig. 3J). Although SC assembly occurs solely between sisters in Rec8 knockout spermatocytes as reported (Bannister et al. 2004; Xu et al. 2005), Rad21L knockout spermatocytes assemble SC at both intersisters and interchromosomes (Fig. 4B; Llano et al. 2012). In contrast to the results using mice with a single mutation, Rec8/Rad21L double-knockout mice (male and female) did not form either cohesin axes or chromosome axes, including SYCP3 (Fig. 4A; Supplemental Fig. 3J,K), suggesting that RAD21L and REC8 redundantly function in forming the cohesin axis, a base for the assembly of the AE (Fig. 7, below; Llano et al. 2012). Immunogold electron microscopic analyses revealed that cohesin complexes localize on the innermost sides of the two lateral elements in wild-type bivalents (Fig. 4C), reminiscent of a study in Drosophila (Anderson et al. 2005). Furthermore, immunoprecipitation assays using spermatocyte extracts indicated that both REC8 and RAD21L precipitate the transverse filament protein SYCP1 of the SC but little of the central element protein SYCE1 (Fig. 4D). Thus, the assembly of meiotic chromosome axis structures (both AEs and SC) is largely mediated by REC8 and RAD21L in a redundant manner.
Figure 4.

RAD21L and REC8 are required redundantly for meiotic chromosome axis assembly. (A) SYCP3 and SMC3 signals were examined by immunostaining surface spread nuclei from the indicated mouse spermatocytes (4- to 8-wk-old males). (B) SYCP3 and SYCP1 signals were examined by immunostaining surface spread nuclei from the indicated mouse spermatocytes. (Bottom right) An enlarged image of aberrant SC and intersister SC from Rad21L knockout (KO) is shown. The arrow indicates interchromosome SC, where two different chromosomes are overlapped with SYCP1 staining. The arrowhead indicates intersister SC. (Right) The number of chromosomes showing intersister or interchromosome synapsis in Rec8 knockout and Rad21L knockout are shown in the scatter plot with the medians. (C) Post-embedding immunogold electron microscopy analysis of wild-type (WT) spermatocytes demonstrates that RAD21L and REC8 are located in the innermost part of the lateral element (LE) of the SC. A central element (CE) protein, SYCE1, was similarly detected. Red circles indicate the immunogold particles. Bar, 200 nm. A histogram for the location of SYCE1, RAD21L, and REC8 is shown on the bottom (see also Supplemental Fig. 5). (D) RAD21L and REC8 antibodies were immunoprecipitated (IP) from spermatocyte extracts (Input) and analyzed by immunoblot using the indicated antibodies (IP). Note that RAD21L and REC8 cohesin complexes coprecipitate with SYCP1 (transverse element) but not SYCE1 (central element). Bars (except C), 5 μm.
Figure 7.
Schematic models of homolog pairing in mouse spermatocytes. Cohesin axes are developed into AEs during leptotene, whereas DSB formation promotes SC assembly in zygotene. Some homologs already locate in close proximity during early leptotene even though AE is not yet developed. Pairing in late leptotene is mediated presumably through chromatin loops depending on meiotic cohesin complexes. The DSB-dependent recombination process drives DNA homology search and establishes pairing, further promoting synapsis between homologs.
RAD21L is an atypical cohesin
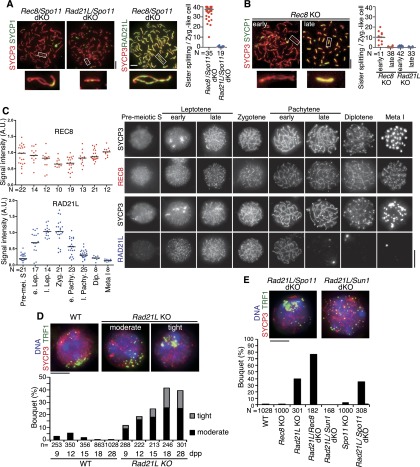
Rec8 knockout and Rad21L knockout spermatocytes show an accumulation of DMC1 and RAD51 foci (DSB markers) during zygotene-like arrest (Supplemental Fig. 1; Llano et al. 2012), suggesting that some recombination process that includes DSB formation is initiated, at least in part. When Spo11 was further depleted from Rec8 knockout or Rad21L knockout mice, SC formation was largely abolished (cf. Figs. 4B and 5A), suggesting that the intersister SC in both Rec8 and Rad21L mutants is promoted by DSBs and, conversely, that the residual SC formation in Spo11 mutants depends on the kleisins. We further noticed that Rec8/Spo11 double-knockout spermatocytes, but not Rad21L/Spo11 double-knockout spermatocytes, exhibit AE splitting in the zygotene-like stage, although RAD21L is fully loaded onto chromatin (Fig. 5A). Accordingly, even in Rec8 knockout spermatocytes, regional splitting of AEs is observed at the early zygotene stage but is suppressed in later stages (Fig. 5B), when DSB markers accumulate (Supplemental Fig. 1). Given that sister chromatid cohesion in Rec8 knockout spermatocytes is mediated primarily by RAD21L (Supplemental Fig. 4), the foregoing results suggest that RAD21L-mediated cohesion might be established largely depending on DSB formation in the leptotene stage rather than by coupling with premeiotic DNA replication. Because the SC assembled in Rec8 knockout spermatocytes seems intact (Supplemental Fig. 5), we infer that the “cohesion” of AEs mediated by RAD21L and SPO11 might be achieved mainly by a zipper-like assembly of SC rather than canonical sister chromatid cohesion. In excellent agreement with these results, RAD21L appears on the chromosomes mostly after DNA replication and culminates at the leptotene/zygotene stage, while REC8 fully localizes along chromosomes before DNA replication and persists until metaphase I (Fig. 5C; Lee and Hirano 2011). These results reveal the functional divergence of the two meiosis-specific α-kleisin subunits in mice, stressing the unique role of RAD21L during the leptotene/zygotene stage.
Figure 5.
RAD21L is an atypical cohesin. (A) SYCP3 and SYCP1 (or RAD21L) signals were examined in the indicated mice (4 wk old) by immunostaining surface spread nuclei of zygotene-like spermatocytes; enlarged images of a univalent are also shown on the bottom. (Right) Univalents with separated axes were measured and are represented as scatter plots with medians. (B, left) SYCP3 and SYCP1 signals were examined in Rec8 knockout (KO) zygotene-like spermatocytes as in A. (Right) Univalents with separated axes were measured and are represented as scatter plots with medians. (C) Immunolocalization of REC8 (top right) and RAD21L (bottom right) were examined in structurally preserved nuclei (with addition of 0.1% [v/v] Triton X-100) from premeiotic S to metaphase I stages in wild type (WT) (4-wk-old male). (Left) Relative intensities of immunofluorescent signals of REC8 and RAD21L were quantified and are represented as scatter plots with medians. (e. Pachy.) Early pachytene; (l. Pachy.) late pachytene; (Dip.) diplotene; (Meta I) metaphase I. (D, top) The indicated spermatocyte nuclei were immunostained with antibodies against SYCP3 (red), TRF1 (green), and SYCP1. Telomere clustering in wild-type and Rad21L knockout was scored at 9, 12, 15, 18, and 28 d post-partum (dpp). Telomere distribution in bouquet was classified into tight and moderate clustering. (Bottom) The frequency of bouquet stage spermatocytes is shown (see also Supplemental Fig. 6). (E) Rad21L/Spo11 double-knockout (dKO) and Rad21L/Sun1 double-knockout spermatocyte nuclei were immunostained as in D. The frequency of bouquet stage spermatocytes is shown for wild-type and the indicated mutant spermatocytes (4 wk old). Bars, 5 μm.
Curiously, immunostaining of the telomere protein TRF1 revealed that Rad21L knockout spermatocytes often arrest in the zygotene-like stage while exhibiting telomere clustering along the NE (bouquet configuration of chromosomes) (Supplemental Fig. 6). Furthermore, chronological analyses of spermatocytes from juvenile mice revealed that although wild-type spermatocytes show transient telomere clustering along the NE at ∼12 d post-partum (dpp), Rad21L knockout spermatocytes accumulate nuclei at the bouquet stage thereafter (Fig. 5D). This arrest is suppressed by Sun1 knockout (Fig. 5E), suggesting that chromosome movement mediated by SUN1 is indeed acting and is the reason of the bouquet configuration in Rad21L knockout spermatocytes. Notably, bouquet arrest is not abolished in Rad21L/Spo11 double knockout (Fig. 5E), implying that this arrest does not originate from aberrant recombination or DSB-mediated checkpoint activation. Although Rec8 knockout spermatocytes do not accumulate bouquet nuclei, Rad21L/Rec8 double-knockout spermatocytes show enhanced bouquet arrest (Fig. 5E), implying that REC8 only partly substitutes for RAD21L in bouquet exit. These results underscore the unique role of RAD21L.
RAD21L is required for DSB-independent homolog recognition
Given that cohesin-deficient spermatocytes proceed into a zygotene-like stage, we examined homolog pairing in these mutants by FISH analysis (Fig. 6; Supplemental Fig. 7). Despite the absence of synapsis between homologs in Rec8 knockout spermatocytes (Fig. 4B), a significant population of spermatocytes (28%∼35%) exhibits homolog pairing in the chromosome mid- and subtelomeric regions (Fig. 6A; Supplemental Fig. 7). These results are consistent with previous cytological observations (Bannister et al. 2004; Xu et al. 2005). Strikingly, however, homolog association in the chromosome arm regions is largely impaired in Rad21L knockout spermatocytes (Fig. 6B) regardless of the accumulation of bouquet nuclei (Fig. 5D). Although some association at the subtelomeric region can be detected in Rad21L knockout spermatocytes, this might be an indirect effect of telomere clustering because Rad21L/Sun1 double-knockout spermatocytes show abolishment of this superficial pairing as well as bouquet arrest (Figs. 5E, 6B). The suppression is specific because Sun1 knockout spermatocytes retain the pairing ability (Fig. 2A). Moreover, we found that Rec8/Spo11 double-knockout spermatocytes show significant homolog association, while Rad21L/Spo11 double-knockout spermatocytes do not (Fig. 6C). Crucially, FISH chromosome painting assays also demonstrate that homolog association during the leptotene stage initiates in Rec8 knockout spermatocytes while it rarely does in Rad21L knockout spermatocytes (Fig. 6D). Although some residual association of homologs can be detected in Rad21L knockout spermatocytes, this might be also an indirect effect of bouquet arrest because it is largely abolished in Rad21L/Sun1 double-knockout spermatocytes. Overall, these results suggest that RAD21L plays a prominent role in DSB-independent homolog association and pairing, while REC8 may play a minor role in this process.
Figure 6.

RAD21L is uniquely required for homolog recognition. (A) Homolog pairing was examined in structurally preserved nuclei from Rec8 knockout (KO) spermatocytes (4–8 wk old) by FISH using the point probes detecting the indicated chromosome sites. (Bottom) The distances between two probe signals are represented in a scatter plot with medians. (Top) The homolog pairing ratio is shown in the graph. (B) Homolog pairing was examined in structurally preserved nuclei from Rad21L knockout and Rad21L/Sun1 double-knockout (dKO) spermatocytes using the indicated point probes as in A. (C) Homolog pairing was examined in structurally preserved nuclei from Spo11 knockout, Rec8/Spo11 double-knockout, and Rad21L/Spo11 double-knockout spermatocytes using the point probe to detect the mid-region on Chr. 8 as in A. Note that the same data set of Spo11 knockout (Fig. 3A) is shown for reference. (Right) Examples of surface spread nuclei of wild-type (WT) and Rec8/Spo11 double-knockout spermatocytes immunostained by SYCP3 are shown. P-value (Mann-Whitney t-test) is shown. (*) P < 0.05. (D) Structurally preserved nuclei from Rec8 knockout, Rad21L knockout, and Rad21L/Sun1 double-knockout spermatocytes (3–4 wk old) were stained as indicated and labeled by FISH with a Chr. 8 painting probe. The proximity of the chromosome territories of homologs was classified into separated, juxtaposed, or fused and is shown in the graph. (*) P < 0.05 (Pearson’s χ2 test). Bars, 5 μm.
Discussion
Because of the multiple requirements of meiotic cohesin for overall chromosome structure, it remains elusive whether the observed pairing defects in cohesin mutants are direct or indirect consequences of cohesin loss. In mice, two meiotic α-kleisin subunits, REC8 and RAD21L, act redundantly in the assembly of the AE and SC, which are largely retained in both Rec8 knockout and Rad21L knockout mice but abolished in double-knockout mice. Our study reveals that homolog recognition is abolished in Rad21L knockout but largely retained in Rec8 knockout spermatocyte chromosomes. Since SMC1β knockout spermatocytes also show a partial defect in axis–loop structure but still progress to the pachytene stage with normal homolog pairing/synapsis (Revenkova et al. 2004; Novak et al. 2008), our results highlight a specific role for the α-kleisin subunit in homolog pairing/synapsis. Analyses of pairing in several mouse mutants further illuminate a fundamental mechanism of homolog recognition that is mediated by some chromosome architecture defined by meiosis-specific cohesin.
Homolog recognition precedes axis formation and DSB-dependent tight pairing
Our FISH chromosome painting assays in mouse spermatocytes revealed that some homologs are already placed in close proximity at the early leptotene stage, when the chromosome axis is not yet assembled (Fig. 1C), although assays using point FISH probes did not detect this association (Fig. 1B). We reason that because homolog association might be initiated through limited interstitial interaction, associations of specific chromosome sites will rarely be detected if the chromosome axes are not developed (Fig. 1D). Consistent with the existence of homolog association from the early leptotene stage, when DMC1 and RAD51 foci have not yet formed, homolog association occurs in the absence of the SPO11 protein (Fig. 3). Nevertheless, because Spo11 knockout spermatocytes show aberrant synapsis even between nonhomologous chromosomes (Baudat et al. 2000; Romanienko and Camerini-Otero 2000), we assume that SPO11 plays a crucial role in stabilizing homolog pairing and promoting synapsis, most likely through a recombination process (Boateng et al. 2013). Notably, at the leptotene/zygotene stage, chromosome axes, which do not assemble the SC, are aligned at some distance (Fig. 3C). Based on these results, we propose a model in which homolog association first appears in chromatin that lacks an axial structure at the early leptotene stage and is then retained and developed during axis formation in the leptotene/zygotene stage; axis assembly may preserve loop interaction (Fig. 7).
A recent study has identified transient pairing in late premeiotic S phase, which does not depend on DSB but depends on SPO11 protein (Boateng et al. 2013). However, neither our point nor chromosome paint FISH assays detected significant homolog association in premeiotic S phase. The reason for this difference is unclear, although some experiments in the earlier study used FACS sorting, which we did not use in this study. Furthermore, the earlier study showed that homolog pairing during the leptotene to zygotene stages is absent in Spo11 knockout and Sun1 knockout spermatocytes (Boateng et al. 2013), although we detected homolog association in these knockout mice. This can be accounted for by the fact that the experimental criteria for pairing or association differ somewhat between the two studies; the earlier study considered a 1.00-µm distance between FISH probes as “paired,” whereas we used a cutoff value of 1.35 µm. Moreover, we monitored the overall distance between point FISH signals and the association of painted chromosomes, thus enabling immature pairing of homologs to be detected. These results are consistent with the notion that homolog pairing is initiated but not completed in Spo11 knockout and Sun1 knockout spermatocytes.
Telomere-driven chromosome movement is not essential for homolog recognition
In meiotic prophase, telomere-driven chromosome movement is mediated by the NE protein SUN1. Arguably, chromosome movement that may promote alignment is thought to be crucial for homolog pairing (Voet et al. 2003; Lee et al. 2012; Boateng et al. 2013; Lui et al. 2013); however, it has been reported that 22%∼25% of zygotene-arrested Sun1 knockout spermatocytes form synapses (Ding et al. 2007). Sun1 knockout spermatocytes lack overall chromosome movement (Shibuya et al. 2014) but retain the ability to undergo homolog pairing (Fig. 2), although the pairing is delayed as compared with wild-type spermatocytes (Fig. 1). We noted, however, that centromeric heterochromatin moves and gathers along the nuclear periphery during the early leptotene stage in Sun1 knockout spermatocytes, as in wild-type spermatocytes (Scherthan et al. 1996; data not shown), suggesting that such mild chromosome movement may contribute to initial homolog search independent of SUN1.
The pairing initiation site on chromosomes is also an arguable issue in meiosis. Although pairing or synapsis might initiate at different sites among organisms, there are conflicting reports in mice. Some reports suggest that the telomere is the initiation site for pairing/synapsis, while others suggest interstitial sites on the chromosome arms (Bisig et al. 2012; Qiao et al. 2012; Boateng et al. 2013). Our point probe FISH assays indicate that the timing of homolog pairing/synapsis is similar between the arm and subtelomeric regions in wild-type and Spo11 knockout spermatocytes (Figs. 1B, 3A), suggesting that homolog recognition occurs via multiple interstitial interactions along the entire chromosome length.
RAD21L is an atypical cohesin
Our results indicate that although REC8 persists on the chromatin throughout prophase, RAD21L appears mostly after DNA replication, peaks during the leptone/zygotene stage, and declines thereafter (Fig. 5C). Strikingly, in Rec8 knockout spermatocytes, sister chromatid cohesion is largely impaired during the leptotene stage but restored in the zygotene stage, most likely by RAD21L-dependent SC assembly between sisters (Fig. 5A). Therefore, we assume that RAD21L plays only a minor role in canonical sister chromatid cohesion but a major role in homolog pairing and synapsis. In this regard, RAD21L might be functionally homologous to Drosophila meiotic α-kleisin C(2)M, which also localizes on chromatin after meiotic DNA replication and is required for events associated with meiotic exchange but not for sister chromatid cohesion (Heidmann et al. 2004; Tanneti et al. 2011). Divergence of the meiotic α-kleisin subunit is also observed in C. elegans, in which three different types of α-kleisin, REC-8, COH-3, and COH4, act redundantly to promote AE and SC formation, although each may have additional specialized functions as well (Severson et al. 2009).
Bouquet exit checkpoint?
We found that Rad21L knockout spermatocytes show extensive arrest at the bouquet stage (Fig. 5D,E). Importantly, however, Rad21L/Sun1 double-knockout spermatocytes, in which RAD21L is absent and the bouquets are disrupted, still show a defect in homolog pairing, suggesting that the bouquet arrest is the consequence rather than the cause of defective homolog pairing in Rad21L knockout spermatocytes (Figs. 5E, 6B). Notably, bouquet stage arrest is rarely observed in mouse mutants that show meiotic prophase arrest, except for a mutant in the ATM-mediated DSB repair process (Liebe et al. 2006). Because deletion of Spo11 from Atm knockout suppresses bouquet arrest, the ATM kinase-mediated DNA damage response, which is associated with SPO11-dependent DSBs, might be required for the exit from the bouquet stage (Pandita et al. 1999; Fernandez-Capetillo et al. 2003; Liebe et al. 2006). Also, in Sordaria Spo11 mutants, the bouquet stage persists longer than in wild-type cells. Therefore, it has been proposed that the exit from the bouquet is mediated by a regulatory process that senses the progression of a recombination-related process (Storlazzi et al. 2003). In contrast, bouquet arrest in Rad21L knockout spermatocytes is not suppressed by Spo11 knockout in mice (Fig. 5E). In this regard, it is notable that budding yeast meiotic cells depleted for Rec8 (the sole meiotic cohesin in this organism) show pairing loss and bouquet arrest, and neither the depletion of Spo11 nor the ectopic expression of Scc1 (mitotic cohesin) suppresses this arrest (Trelles-Sticken et al. 2005; Conrad et al. 2007). Thus, studies in budding yeast and mice suggest the existence of a conserved “bouquet exit checkpoint” that might monitor chromosome pairing rather than DSB-dependent recombination. We do not formally exclude the possibility that meiotic cohesin may provide the chromosome structure necessary for bouquet release or that bouquet arrest may reflect an independent function of RAD21L at telomeres.
Meiotic cohesins are most likely the first proteins to organize meiotic chromosome axes and localize on the chromatin prior to axis formation (Fig. 7). Our results suggest that homolog recognition occurs at the early leptotene stage and develops into pairing toward the late leptotene stage. This pairing is totally dependent on the function of RAD21L but only partly on REC8. However, it should be noted that sexual dimorphism is observed in Rad21L knockout mice; spermatocytes progress to the zygotene-like stage, while oocytes progress into the pachytene-like stage, where many more homologs are synapsed (Supplemental Fig. 3). This implies that the contribution of REC8 and RAD21L to homolog pairing might be weighted differently between meiosis in male and female mice. Moreover, our data show that homolog recognition does not always require the SPO11 protein, which acts in DSB formation. Taken together, we propose that homolog recognition is mediated by a specific chromosome architecture defined by the meiosis-specific cohesin rather than a DSB-dependent DNA sequence search. Our study provides an important framework for disclosing the molecular mechanism of homolog recognition in meiosis.
Materials and methods
Animal experiments
Rec8(mei8) mutant (although the Rec8 allele has a point mutation, here it is referred to as Rec8 knockout), Spo11 knockout, and Sun1 knockout mice were reported earlier (Baudat et al. 2000; Bannister et al. 2004; Ding et al. 2007). All single- and double-knockout mice were congenic with the C57BL/6J background. Since Rad21L and Spo11 alleles are linked on the same chromosome 2, two males of Rad21L/Spo11 double knockout from a total of 302 offspring were identified after intercrossing of Rad21L+/−/Spo11+/− mice. Whenever possible, each knockout animal was compared with littermates or age-matched (4–8 wk) nonlittermates from the same colony unless otherwise described. Animal experiments were approved by the Institutional Animal Care and Use Committee (approval nos. 23001, 23013, 24001, and 25012).
Immunostaining of spermatocytes and fetal oocytes
Structurally preserved nuclei from spermatocytes were prepared as described (Liebe et al. 2004) with modification. Briefly, testicular cells were collected in PBS by mincing seminiferous tubules into small pieces with fine-tipped tweezers, clipping them with flat forceps, and then pipetting. After removal of tissue pieces, the cell suspension was filtered through a cell strainer (BD Falcon) to remove debris. The cell suspension (∼5 μL) was dropped onto a MAS-coated slide glass (Matsunami), fixed with 10 μL of 2% paraformaldehyde (PFA)/100 mM sucrose in PBS for 10 min followed by the addition of 1.5 μL of 1.25 M glycine/PBS, and then air-dried at room temperature. Immediately before they were completely air-dried, the slide glasses were washed with PBS containing 0.1% Triton-X100 or frozen in liquid nitrogen for longer storage at −80°C.
Surface-spread nuclei from spermatocytes and oocytes were prepared by the dry-down method as described (Peters et al. 1997). Mildly spread nuclei from spermatocytes and oocytes (with diameters less than twice that of the intact nuclei) were prepared as described (Ishiguro et al. 2011) with modification. Briefly, cells were suspended in PBS without hypotonic treatment, dropped onto a slide glass together with an equal volume of 2% PFA and 0.2% (v/v) Triton X-100 in PBS, and incubated at room temperature in humidified chamber. The sides were then air-dried and processed as the structurally preserved nuclei preparation.
Immunofluorescence staining was performed as described previously (Ishiguro et al. 2011). DNA was counterstained with VectaShield DAPI. Images were captured with DeltaVision and processed with DeltaVision SoftWorx software (Applied Precision). All of the images shown were Z-stacked.
FISH against immunostained nuclei
Immunostained samples of structurally preserved nuclei or mildly spread nuclei were fixed in 4% paraformaldehyde for 5 min, washed with PBS, and subjected to sequential dehydration through 70%, 80%, 90%, and 100% ethanol. Immunostained samples were denatured in 50% formamide and 2× SSC for 10 min at 72°C and sequentially dehydrated through 70%, 80%, 90%, and 100% ethanol. Hybridization was conducted with a fluorescence-labeled point probe in buffer containing 50% formamide, 2× SSC, 20% dextran sulfate and 0.1 μg/μL of mouse Cot-1 DNA (Invitrogen) and sealed with a cover glass for 12–16 h at 37°C. The slides were washed sequentially in 2× SSC for 1 min and 0.4× SSC/0.3% Tween20 solution for 2 min at room temperature and in 2× SSC for 1 min at room temperature. The mouse point probe chromosome 8-A2 (ID Labs) detected the mid-region of chromosome 8. The mouse point probe derived from BAC clone RP23-6I6 detected the mid-region of chromosome 3. The mouse point probe derived from BAC clone RP23-321L15 detected the subtelomeric loop region of chromosome 18 (Kauppi et al. 2011). To examine sister chromatid cohesion, the point probe chromosomes 3 was employed, since it gives discrete foci, unlike the point probe chromosome 8. For whole-chromosome painting in structurally preserved nuclei, immunostaining was first conducted as described above followed by hybridization with chromosome 8 painting probe (Tokyo Instruments), after which the slides were washed sequentially in 50% formamide/2× SSC solution for 2 min, 2× SSC for 2 min, and 0.4× SSC/0.3% Tween 20 solution for 2 min at room temperature and in 2× SSC for 1 min at room temperature. For telomere FISH in structurally preserved nuclei, immunostained slides were treated with 100 μg/mL RNase A for 30 min at 37°C in 2× SSC followed by dehydration. Hybridization was conducted with TelC (CCCTAA)3 PNA probe (Panagene) for 4 h at 37°C.
For every measurement, images were acquired with Z-sections encompassing the entire nuclei. The distance between two hybridization signals was measured for a pair of homologs using structurally preserved nuclei. A pair of points was assigned for which the maximum signals were identified from Z-sections. If the FISH signals on one homolog were split (doublet), a measurement point was assigned to the center of those signals. The distance between two FISH signals was measured by calculating the square root of X2 + Y2 + Z2 using DeltaVision SoftWorx software (Applied Precision). The FISH experiments using mildly spread nuclei were similarly assayed.
Although structurally preserved nuclei appear approximately spherical in shape with less variation in size compared with mildly spread nuclei, some show an irregular rather than spherical shape. Structurally preserved nuclei showing such extremely irregular shape and size were omitted from the measurement if the longer axis was <7 μm or >12 μm. Since mildly spread nuclei size shows variations between different preparations, the pairing distances were normalized to the approximate diameter. The pairing indices are represented in scatter plots with the median. In cohesion-defective Rad21L/Rec8 double-knockout spermatocytes, averages of three or six distance measurements of one pair out of three or four probe signals were plotted, respectively.
For the classification of FISH chromosome painting, the overlapping of homologs was assessed by inspecting hybridization signal masses on each single Z-section (Fig. 1C). Classification was done as follows. Juxtaposed: Hybridization signals of homologs overlap but are discernible in some sections. Fused: Hybridization signals of homologs overlap and are not discernible. Separated: Hybridization signal masses are separated in all sections.
The data sets were pooled from different slide glass samples prepared from two or three mice (2–8 wk old) for each knockout strain and wild-type, except that one male was used for Rad21L/Spo11 double-knockout spermatocytes (4 wk old). Statistical analysis was carried out with GraphPad Prism5. For comparison of independent data sets, the two-tailed nonparametric Mann-Whitney t-test was used.
Acknowledgments
We thank S. Keeney for the Spo11 knockout mice, and H. Cooke for the antibody. We also thank H. Scherthan, D. Camerini-Otero, N. Numata, N. Kudo, A. Sato, Y. Itoh, Y. Fujii, and Y. Kishi as well as the Gotoh laboratory for technical advice, and all members of the Watanabe laboratory for their support and discussion. This work was supported in part by a Japan Society for the Promotion of Science Research Fellowship to J.K. and H.S.; a Grant-in-Aid for Scientific Research on Priority Areas, a Grant-in-Aid for Scientific Research on Innovative Areas, and a Grant-in-Aid for Young Scientists to K.I.; the Swedish Research Council to A.H.-H. and C.H.; the Swedish Cancer Society to A.H.-H. and C.H.; and a Grant-in-Aid for Specially Promoted Research (to Y.W.) from MEXT, Japan.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.237313.113.
References
- Anderson LK, Royer SM, Page SL, McKim KS, Lai A, Lilly MA, Hawley RS 2005. Juxtaposition of C(2)M and the transverse filament protein C(3)G within the central region of Drosophila synaptonemal complex. Proc Natl Acad Sci 102: 4482–4487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister LA, Reinholdt LG, Munroe RJ, Schimenti JC 2004. Positional cloning and characterization of mouse mei8, a disrupted allelle of the meiotic cohesin Rec8. Genesis 40: 184–194 [DOI] [PubMed] [Google Scholar]
- Barzel A, Kupiec M 2008. Finding a match: How do homologous sequences get together for recombination? Nat Rev Genet 9: 27–37 [DOI] [PubMed] [Google Scholar]
- Baudat F, de Massy B 2007. Regulating double-stranded DNA break repair towards crossover or non-crossover during mammalian meiosis. Chromosome Res 15: 565–577 [DOI] [PubMed] [Google Scholar]
- Baudat F, Manova K, Yuen JP, Jasin M, Keeney S 2000. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol Cell 6: 989–998 [DOI] [PubMed] [Google Scholar]
- Bhuiyan H, Schmekel K 2004. Meiotic chromosome synapsis in yeast can occur without spo11-induced DNA double-strand breaks. Genetics 168: 775–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisig CG, Guiraldelli MF, Kouznetsova A, Scherthan H, Hoog C, Dawson DS, Pezza RJ 2012. Synaptonemal complex components persist at centromeres and are required for homologous centromere pairing in mouse spermatocytes. PLoS Genet 8: e1002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boateng KA, Bellani MA, Gregoretti IV, Pratto F, Camerini-Otero RD 2013. Homologous pairing preceding SPO11-mediated double-strand breaks in mice. Dev Cell 24: 196–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad MN, Lee CY, Wilkerson JL, Dresser ME 2007. MPS3 mediates meiotic bouquet formation in Saccharomyces cerevisiae. Proc Natl Acad Sci 104: 8863–8868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T, Cremer M 2010. Chromosome territories. Cold Spring Harb Perspect Biol 2: a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg AF, McDonald K, Moulder G, Barstead R, Dresser M, Villeneuve AM 1998. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94: 387–398 [DOI] [PubMed] [Google Scholar]
- Ding DQ, Yamamoto A, Haraguchi T, Hiraoka Y 2004. Dynamics of homologous chromosome pairing during meiotic prophase in fission yeast. Dev Cell 6: 329–341 [DOI] [PubMed] [Google Scholar]
- Ding X, Xu R, Yu J, Xu T, Zhuang Y, Han M 2007. SUN1 is required for telomere attachment to nuclear envelope and gametogenesis in mice. Dev Cell 12: 863–872 [DOI] [PubMed] [Google Scholar]
- Ding DQ, Okamasa K, Yamane M, Tsutsumi C, Haraguchi T, Yamamoto M, Hiraoka Y 2012. Meiosis-specific noncoding RNA mediates robust pairing of homologous chromosomes in meiosis. Science 336: 732–736 [DOI] [PubMed] [Google Scholar]
- Dombecki CR, Chiang AC, Kang HJ, Bilgir C, Stefanski NA, Neva BJ, Klerkx EP, Nabeshima K 2011. The chromodomain protein MRG-1 facilitates SC-independent homologous pairing during meiosis in Caenorhabditis elegans. Dev Cell 21: 1092–1103 [DOI] [PubMed] [Google Scholar]
- Fernandez-Capetillo O, Liebe B, Scherthan H, Nussenzweig A 2003. H2AX regulates meiotic telomere clustering. J Cell Biol 163: 15–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidmann D, Horn S, Heidmann S, Schleiffer A, Nasmyth K, Lehner CF 2004. The Drosophila meiotic kleisin C(2)M functions before the meiotic divisions. Chromosoma 113: 177–187 [DOI] [PubMed] [Google Scholar]
- Herran Y, Gutierrez-Caballero C, Sanchez-Martin M, Hernandez T, Viera A, Barbero JL, de Alava E, de Rooij DG, Suja JA, Llano E, et al. 2011. The cohesin subunit RAD21L functions in meiotic synapsis and exhibits sexual dimorphism in fertility. EMBO J 30: 3091–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka Y, Dernburg AF 2009. The SUN rises on meiotic chromosome dynamics. Dev Cell 17: 598–605 [DOI] [PubMed] [Google Scholar]
- Ishiguro K, Kim J, Fujiyama-Nakamura S, Kato S, Watanabe Y 2011. A new meiosis-specific cohesin complex implicated in the cohesin code for homologous pairing. EMBO Rep 12: 267–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppi L, Barchi M, Baudat F, Romanienko PJ, Keeney S, Jasin M 2011. Distinct properties of the XY pseudoautosomal region crucial for male meiosis. Science 331: 916–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Mahr P, Galova M, Buonomo SBC, Michaelis C, Nairz K, Nasmyth K 1999. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 98: 91–103 [DOI] [PubMed] [Google Scholar]
- Lee J, Hirano T 2011. RAD21L, a novel cohesin subunit implicated in linking homologous chromosomes in mammalian meiosis. J Cell Biol 192: 263–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CY, Conrad MN, Dresser ME 2012. Meiotic chromosome pairing is promoted by telomere-led chromosome movements independent of bouquet formation. PLoS Genet 8: e1002730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebe B, Alsheimer M, Hoog C, Benavente R, Scherthan H 2004. Telomere attachment, meiotic chromosome condensation, pairing, and bouquet stage duration are modified in spermatocytes lacking axial elements. Mol Biol Cell 15: 827–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebe B, Petukhova G, Barchi M, Bellani M, Braselmann H, Nakano T, Pandita TK, Jasin M, Fornace A, Meistrich ML, et al. 2006. Mutations that affect meiosis in male mice influence the dynamics of the mid-preleptotene and bouquet stages. Exp Cell Res 312: 3768–3781 [DOI] [PubMed] [Google Scholar]
- Llano E, Herran Y, Garcia-Tunon I, Gutierrez-Caballero C, de Alava E, Barbero JL, Schimenti J, de Rooij DG, Sanchez-Martin M, Pendas AM 2012. Meiotic cohesin complexes are essential for the formation of the axial element in mice. J Cell Biol 197: 877–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui DY, Cahoon CK, Burgess SM 2013. Multiple opposing constraints govern chromosome interactions during meiosis. PLoS Genet 9: e1003197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim KS, Green-Marroquin BL, Sekelsky JJ, Chin G, Steinberg C, Khodosh R, Hawley RS 1998. Meiotic synapsis in the absence of recombination. Science 279: 876–878 [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH 2005. The structure and function of SMC and kleisin complexes. Annu Rev Biochem 74: 595–648 [DOI] [PubMed] [Google Scholar]
- Neale MJ, Keeney S 2006. Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature 442: 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak I, Wang H, Revenkova E, Jessberger R, Scherthan H, Hoog C 2008. Cohesin Smc1β determines meiotic chromatin axis loop organization. J Cell Biol 180: 83–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page SL, Hawley RS 2004. The genetics and molecular biology of the synaptonemal complex. Annu Rev Cell Dev Biol 20: 525–558 [DOI] [PubMed] [Google Scholar]
- Pandita TK, Westphal CH, Anger M, Sawant SG, Geard CR, Pandita RK, Scherthan H 1999. Atm inactivation results in aberrant telomere clustering during meiotic prophase. Mol Cell Biol 19: 5096–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peoples TL, Dean E, Gonzalez O, Lambourne L, Burgess SM 2002. Close, stable homolog juxtaposition during meiosis in budding yeast is dependent on meiotic recombination, occurs independently of synapsis, and is distinct from DSB-independent pairing contacts. Genes Dev 16: 1682–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peoples-Holst TL, Burgess SM 2005. Multiple branches of the meiotic recombination pathway contribute independently to homolog pairing and stable juxtaposition during meiosis in budding yeast. Genes Dev 19: 863–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AH, Plug AW, van Vugt MJ, de Boer P 1997. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res 5: 66–68 [DOI] [PubMed] [Google Scholar]
- Prieto I, Suja JA, Pezzi N, Kremer L, Martínez AC, Rufas JS, Barbero JL 2001. Mammalian STAG3 is a cohesin specific to sister chromatid arms in meiosis I. Nat Cell Biol 3: 761–766 [DOI] [PubMed] [Google Scholar]
- Qiao H, Chen JK, Reynolds A, Hoog C, Paddy M, Hunter N 2012. Interplay between synaptonemal complex, homologous recombination, and centromeres during mammalian meiosis. PLoS Genet 8: e1002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revenkova E, Eijpe M, Heyting C, Hodges CA, Hunt PA, Liebe B, Scherthan H, Jessberger R 2004. Cohesin SMC1β is required for meiotic chromosome dynamics, sister chromatid cohesion and DNA recombination. Nat Cell Biol 6: 555–562 [DOI] [PubMed] [Google Scholar]
- Romanienko PJ, Camerini-Otero RD 2000. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol Cell 6: 975–987 [DOI] [PubMed] [Google Scholar]
- Sasaki M, Lange J, Keeney S 2010. Genome destabilization by homologous recombination in the germ line. Nat Rev Mol Cell Biol 11: 182–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherthan H 2001. A bouquet makes ends meet. Nat Rev Mol Cell Biol 2: 621–627 [DOI] [PubMed] [Google Scholar]
- Scherthan H, Weich S, Schwegler H, Heyting C, Harle M, Cremer T 1996. Centromere and telomere movements during early meiotic prophase of mouse and man are associated with the onset of chromosome pairing. J Cell Biol 134: 1109–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson AF, Ling L, van Zuylen V, Meyer BJ 2009. The axial element protein HTP-3 promotes cohesin loading and meiotic axis assembly in C. elegans to implement the meiotic program of chromosome segregation. Genes Dev 23: 1763–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya H, Ishiguro K, Watanabe Y 2014. The TRF1-binding protein TERB1 promotes chromosome movement and telomere rigidity in meiosis. Nat Cell Biol 16: 145–156 [DOI] [PubMed] [Google Scholar]
- Storlazzi A, Tesse S, Gargano S, James F, Kleckner N, Zickler D 2003. Meiotic double-strand breaks at the interface of chromosome movement, chromosome remodeling, and reductional division. Genes Dev 17: 2675–2687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanneti NS, Landy K, Joyce EF, McKim KS 2011. A pathway for synapsis initiation during zygotene in Drosophila oocytes. Curr Biol 21: 1852–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trelles-Sticken E, Adelfalk C, Loidl J, Scherthan H 2005. Meiotic telomere clustering requires actin for its formation and cohesin for its resolution. J Cell Biol 170: 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voet T, Liebe B, Labaere C, Marynen P, Scherthan H 2003. Telomere-independent homologue pairing and checkpoint escape of accessory ring chromosomes in male mouse meiosis. J Cell Biol 162: 795–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y 2004. Modifying sister chromatid cohesion for meiosis. J Cell Sci 117: 4017–4023 [DOI] [PubMed] [Google Scholar]
- Weiner BM, Kleckner N 1994. Chromosome pairing via multiple interstitial interactions before and during meiosis in yeast. Cell 77: 977–991 [DOI] [PubMed] [Google Scholar]
- Xu H, Beasley MD, Warren WD, van der Horst GT, McKay MJ 2005. Absence of mouse REC8 cohesin promotes synapsis of sister chromatids in meiosis. Dev Cell 8: 949–961 [DOI] [PubMed] [Google Scholar]
- Zickler D, Kleckner N 1999. Meiotic chromosomes: integrating structure and function. Annu Rev Genet 33: 603–754 [DOI] [PubMed] [Google Scholar]