Abstract
Motivation: Microsatellite instability (MSI) is an important indicator of larger genome instability and has been linked to many genetic diseases, including Lynch syndrome. MSI status is also an independent prognostic factor for favorable survival in multiple cancer types, such as colorectal and endometrial. It also informs the choice of chemotherapeutic agents. However, the current PCR–electrophoresis-based detection procedure is laborious and time-consuming, often requiring visual inspection to categorize samples. We developed MSIsensor, a C++ program for automatically detecting somatic microsatellite changes. It computes length distributions of microsatellites per site in paired tumor and normal sequence data, subsequently using these to statistically compare observed distributions in both samples. Comprehensive testing indicates MSIsensor is an efficient and effective tool for deriving MSI status from standard tumor-normal paired sequence data.
Availability and implementation: https://github.com/ding-lab/msisensor
Contact: kye@genome.wustl.edu or lding@genome.wustl.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Microsatellites are repetitive sequences with unit length varying from 1–6 bp. During DNA replication, those repetitive sequences may expand or shrink due to strand slippage (Kawai et al., 1998). Those errors are often recognized and repaired by the DNA mismatch repair machinery, which includes genes such as MLH1 and MSH2 (Prolla et al., 1994). However, when mismatch repair enzymes are defective, replication errors will evade repair and will persist in somatic cells, as occurs in patients with hereditary non-polyposis colorectal cancer, also known as Lynch syndrome (Vasen et al., 1999).
Levels of microsatellite instability (MSI) are traditionally determined experimentally. The sizes of microsatellite marker sets in tumor DNA are compared via electrophoresis with corresponding DNA isolated from a normal tissue sample of the same patient. The most widely used set of markers was recommended by a National Cancer Institute consensus group and consists of either five or seven repeat markers (Vasen et al., 1999). Samples are normally classified as microsatellite instability high (MSI-H), microsatellite instability low (MSI-L) and microsatellite stable (MSS).
Although the experimental approach is considered the gold standard, its detection procedure is expensive and its scope is limited to only a small subset of microsatellites. Conversely, paired tumor-normal genome sequencing allows for comprehensive investigation of MSI sites simultaneously and will likely become a routine part of diagnosis and treatment procedures. Recently, it was reported that MSI status can be derived from RNA-seq data (Lu et al., 2013), but no stand-alone pipeline is provided in that study. Here, we describe MSIsensor, a software tool that quantifies MSI in paired tumor-normal genome sequencing data and reports somatic status of corresponding microsatellite sites in the human genome.
2 METHODS
2.1 Approach
2.1.1 Cataloging of sites in the reference genome
MSIsensor starts with scanning a given reference genome for the locations of homopolymers and microsatellites. By default, it records homopolymers of at least 5-bp length and microsatellites of maximum repeat unit length 5 from the reference genome. The location and flanking sequences of each site are saved to a loci file for the subsequent analysis. This file can be used for additional samples as long as the reference remains unchanged.
2.1.2 Generation of null and candidate distributions for hypothesis testing
For each microsatellite site examined in 2.1.1, all read pairs having at least one read mapped within 2 kb of the site are extracted from both tumor and normal Binary Sequence Alignment/Map format (BAM) files. The histogram of read counts for a set of k-mer alleles is then constructed in a two-step process. First, a dictionary is created by concatenating the flanking sequences to all possible repeat lengths in the range 0 to L-10, where L is read length. Then, observed instances for each entry are tallied over the set of sequencing reads associated with this site. For example, when a homopolymer (10 As, ‘A’ as the repeat unit) has flanking sequences ACGAT and CCGAC, the process will tally k-mers of ACGATCCGAC (repeat length 0), ACGATACCGAC (repeat length 1), ACGATAACCGAC (repeat length 2), ACGATAAACCGAC (repeat length 3) and so forth. Tallies are recorded separately in both tumor and normal data. This process furnishes candidate and null distributions for tumor and normal, respectively, for each site.
2.1.3 Goodness-of-fit test for somatic calling
A standard χ2 test is performed at each site having at least 20 reads in both the tumor and normal samples to assess the goodness-of-fit between their respective k-mer distributions (Sokal and Rohlf, 2012). A site is tagged as somatic if distributions are significantly different, as quantified by standard multiple testing correction of χ2 P-values (Benjamini and Hochberg, 1995). MSIsensor specifies a default false discovery rate (FDR) threshold of 0.05, but this is user configurable.
2.1.4 Quantification of MSI
For each sample, we note the total number of sites with sufficient data (at least 20 spanning reads in both normal and tumor) and also the number of somatic sites. The percentage of somatic sites is the score for MSI.
2.2 Test dataset
A cohort of 242 endometrial exome-sequenced cases was used to test the performance of MSIsensor. Of these, 85 had been designated as MSI based on experimental measurement (The Cancer Genome Atlas Research Network, 2013).
3 RESULTS
3.1 Runtime and memory
MSIsensor is implemented in C++ and was applied to The Cancer Genome Atlas (TCGA) endometrial exome sequence data from 242 tumor-normal pairs. Each pair requires roughly 660 MB of memory and 30 min of compute time on a Dell PowerEdge™ Blade with Ubuntu OS and with Intel® Xeon® CPU E5420 2.50 GHz with four threads for parallel computing.
3.2 Correlation with experimental measure
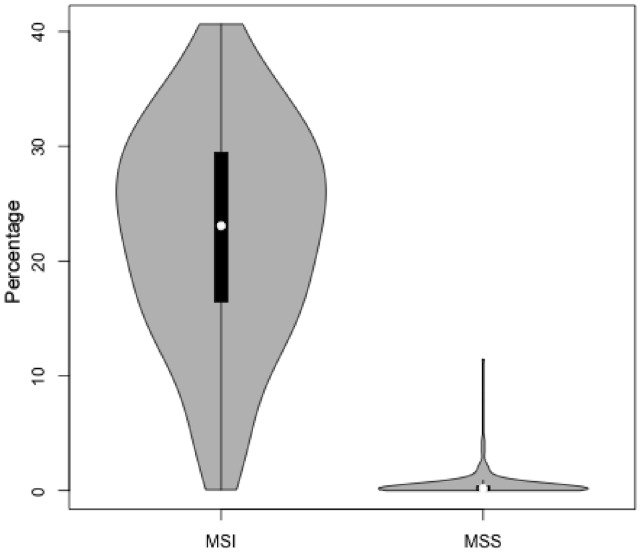
As shown in Figure 1, MSIsensor’s score correlates well with the experimental measurements of MSI in 242 endometrial tumor-normal pairs (Pearson coefficient 0.79). We consider one sample as MSI if both five and seven markers indicate positive. Otherwise it is MSS. Among 71 MSI samples, 70 have an MSI score >3.5. In addition, 165 of 168 MSS samples have a score <3.5. Of those three special cases, one is classified as MSI-H by five marks and MSI-L by seven markers, whereas the other two are MSI-L by both sets of markers (Supplementary Table S1).
Fig. 1.
MSIsensor differentiates MSI (microsatellite instable) samples from MSS (microsatellite stable) ones. The x-axis is clinical classification of MSI and MSS. The y-axis is the percentage of microsatellite sites with a somatic indel
4 CONCLUSION
MSIsensor is an efficient and effective software tool for deriving MSI status from tumor-normal paired genome sequencing data. We will apply it to TCGA data for MSI screening and anticipate a wider usage of this tool in cancer clinical sequencing.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Cyriac Kandoth for helpful discussions and comments.
Funding: National Human Genome Research Institute (U01HG006517) and National Cancer Institute (R01CA180006) to L.D.
Conflict of Interest: none declared.
REFERENCES
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995;57:289–300. [Google Scholar]
- The Cancer Genome Atlas Research Network. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai A, et al. Prognostic factors for patients with sarcomas of the pelvic bones. Cancer. 1998;82:851–859. [PubMed] [Google Scholar]
- Lu Y, et al. A novel approach for characterizing microsatellite instability in cancer cells. PLoS One. 2013;8:e63056. doi: 10.1371/journal.pone.0063056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prolla TA, et al. MLH1, PMS1, and MSH2 interactions during the initiation of DNA mismatch repair in yeast. Science. 1994;265:1091–1093. doi: 10.1126/science.8066446. [DOI] [PubMed] [Google Scholar]
- Sokal RR, Rohlf FJ. Biometry: The Principles and Practice of Statistics in Biological Research. New York: W.H. Freeman and Co; 2012. [Google Scholar]
- Vasen HF, et al. New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC. Gastroenterology. 1999;116:1453–1456. doi: 10.1016/s0016-5085(99)70510-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.