Abstract
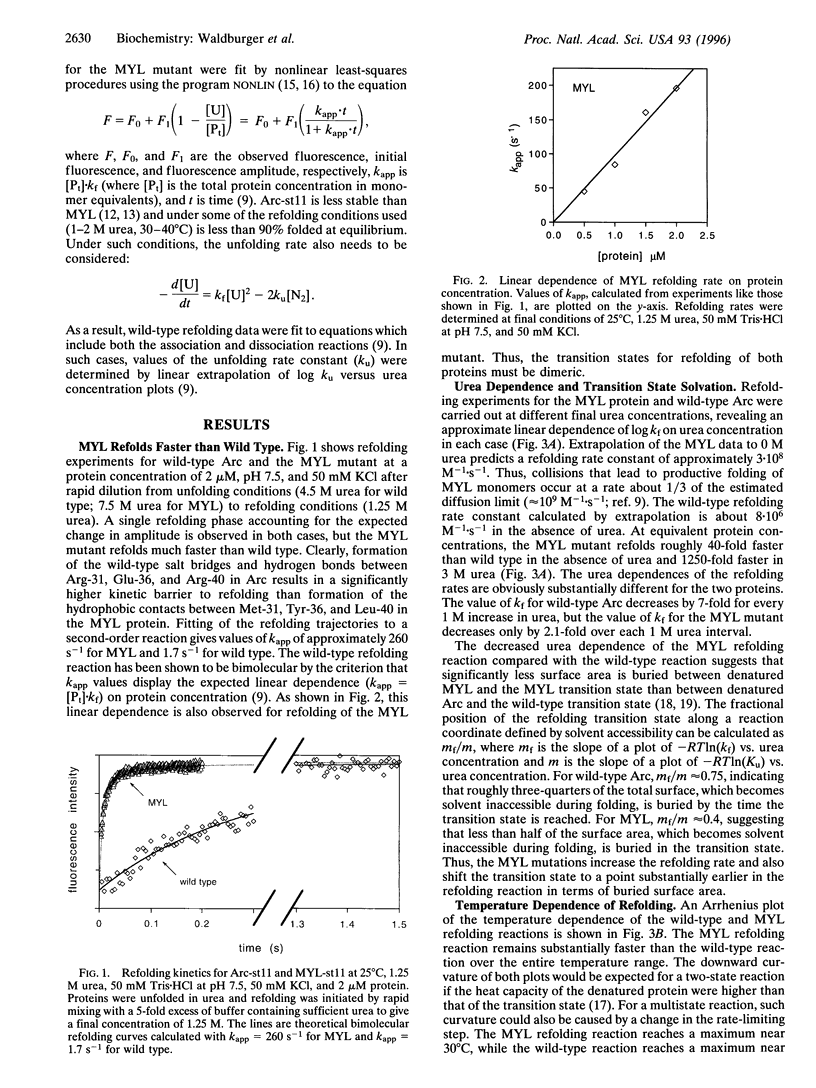
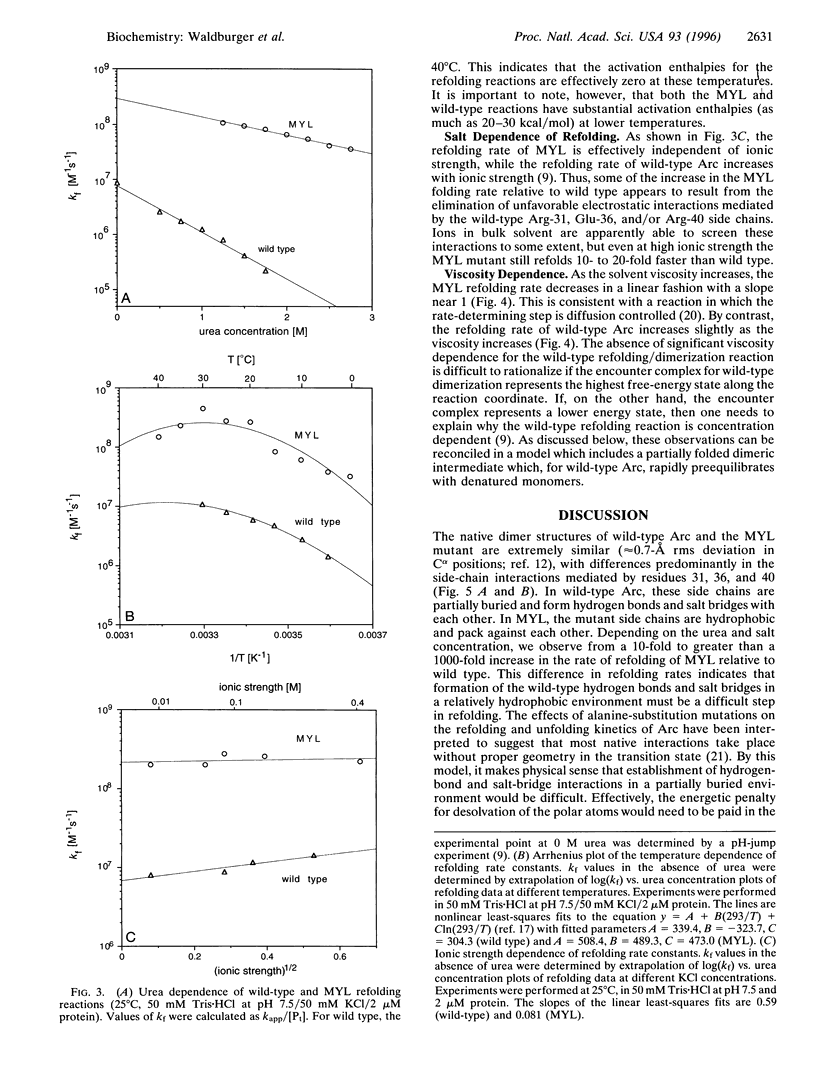
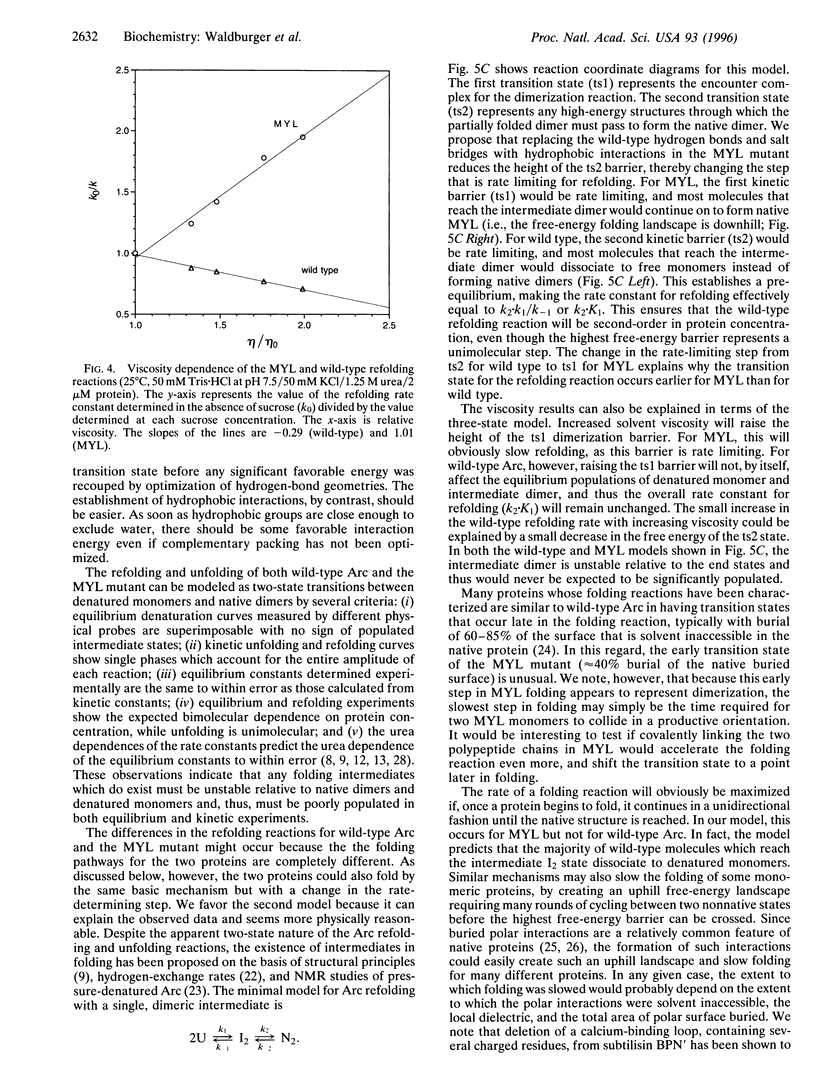
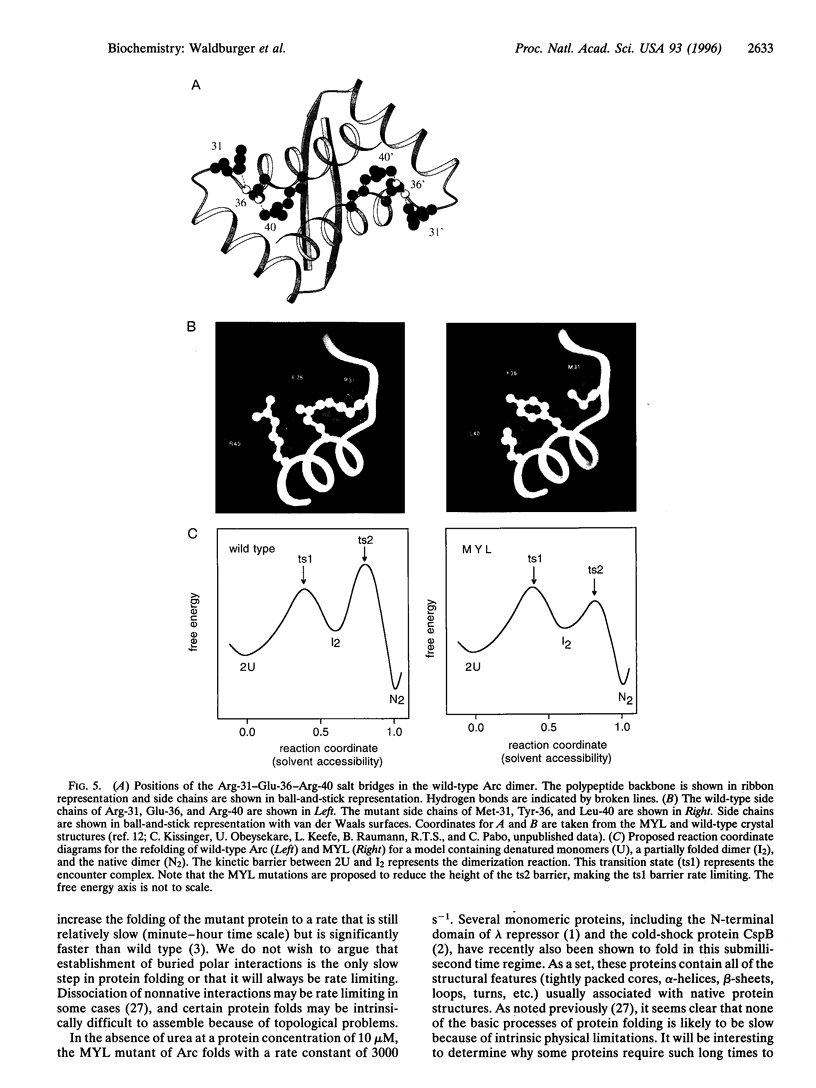
In the MYL mutant of the Arc repressor dimer, sets of partially buried salt-bridge and hydrogen-bond interactions mediated by Arg-31, Glu-36, and Arg-40 in each subunit are replaced by hydrophobic interactions between Met-31, Tyr-36, and Leu-40. The MYL refolding/dimerization reaction differs from that of wild type in being 10- to 1250-fold faster, having an earlier transition state, and depending upon viscosity but not ionic strength. Formation of the wild-type salt bridges in a hydrophobic environment clearly imposes a kinetic barrier to folding, which can be lowered by high salt concentrations. The changes in the position of the transition state and viscosity dependence can be explained if denatured monomers interact to form a partially folded dimeric intermediate, which then continues folding to form the native dimer. The second step is postulated to be rate limiting for wild type. Replacing the salt bridge with hydrophobic interactions lowers this barrier for MYL. This makes the first kinetic barrier rate limiting for MYL refolding and creates a downhill free-energy landscape in which most molecules which reach the intermediate state continue to form native dimers.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berg O. G., von Hippel P. H. Diffusion-controlled macromolecular interactions. Annu Rev Biophys Biophys Chem. 1985;14:131–160. doi: 10.1146/annurev.bb.14.060185.001023. [DOI] [PubMed] [Google Scholar]
- Bonvin A. M., Vis H., Breg J. N., Burgering M. J., Boelens R., Kaptein R. Nuclear magnetic resonance solution structure of the Arc repressor using relaxation matrix calculations. J Mol Biol. 1994 Feb 11;236(1):328–341. doi: 10.1006/jmbi.1994.1138. [DOI] [PubMed] [Google Scholar]
- Bowie J. U., Sauer R. T. Equilibrium dissociation and unfolding of the Arc repressor dimer. Biochemistry. 1989 Sep 5;28(18):7139–7143. doi: 10.1021/bi00444a001. [DOI] [PubMed] [Google Scholar]
- Breg J. N., van Opheusden J. H., Burgering M. J., Boelens R., Kaptein R. Structure of Arc repressor in solution: evidence for a family of beta-sheet DNA-binding proteins. Nature. 1990 Aug 9;346(6284):586–589. doi: 10.1038/346586a0. [DOI] [PubMed] [Google Scholar]
- Brown B. M., Milla M. E., Smith T. L., Sauer R. T. Scanning mutagenesis of the Arc repressor as a functional probe of operator recognition. Nat Struct Biol. 1994 Mar;1(3):164–168. doi: 10.1038/nsb0394-164. [DOI] [PubMed] [Google Scholar]
- Bryan P., Alexander P., Strausberg S., Schwarz F., Lan W., Gilliland G., Gallagher D. T. Energetics of folding subtilisin BPN'. Biochemistry. 1992 Jun 2;31(21):4937–4945. doi: 10.1021/bi00136a003. [DOI] [PubMed] [Google Scholar]
- Burgering M. J., Hald M., Boelens R., Breg J. N., Kaptein R. Hydrogen exchange studies of the Arc repressor: evidence for a monomeric folding intermediate. Biopolymers. 1995 Feb;35(2):217–226. doi: 10.1002/bip.360350210. [DOI] [PubMed] [Google Scholar]
- Chen B. L., Baase W. A., Schellman J. A. Low-temperature unfolding of a mutant of phage T4 lysozyme. 2. Kinetic investigations. Biochemistry. 1989 Jan 24;28(2):691–699. doi: 10.1021/bi00428a042. [DOI] [PubMed] [Google Scholar]
- Creighton T. E. Disulfide bonds as probes of protein folding pathways. Methods Enzymol. 1986;131:83–106. doi: 10.1016/0076-6879(86)31036-x. [DOI] [PubMed] [Google Scholar]
- Huang G. S., Oas T. G. Submillisecond folding of monomeric lambda repressor. Proc Natl Acad Sci U S A. 1995 Jul 18;92(15):6878–6882. doi: 10.1073/pnas.92.15.6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matouschek A., Fersht A. R. Application of physical organic chemistry to engineered mutants of proteins: Hammond postulate behavior in the transition state of protein folding. Proc Natl Acad Sci U S A. 1993 Aug 15;90(16):7814–7818. doi: 10.1073/pnas.90.16.7814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews C. R. Pathways of protein folding. Annu Rev Biochem. 1993;62:653–683. doi: 10.1146/annurev.bi.62.070193.003253. [DOI] [PubMed] [Google Scholar]
- Milla M. E., Brown B. M., Sauer R. T. P22 Arc repressor: enhanced expression of unstable mutants by addition of polar C-terminal sequences. Protein Sci. 1993 Dec;2(12):2198–2205. doi: 10.1002/pro.5560021219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milla M. E., Brown B. M., Waldburger C. D., Sauer R. T. P22 Arc repressor: transition state properties inferred from mutational effects on the rates of protein unfolding and refolding. Biochemistry. 1995 Oct 24;34(42):13914–13919. doi: 10.1021/bi00042a024. [DOI] [PubMed] [Google Scholar]
- Milla M. E., Sauer R. T. P22 Arc repressor: folding kinetics of a single-domain, dimeric protein. Biochemistry. 1994 Feb 8;33(5):1125–1133. doi: 10.1021/bi00171a011. [DOI] [PubMed] [Google Scholar]
- Peng X., Jonas J., Silva J. L. Molten-globule conformation of Arc repressor monomers determined by high-pressure 1H NMR spectroscopy. Proc Natl Acad Sci U S A. 1993 Mar 1;90(5):1776–1780. doi: 10.1073/pnas.90.5.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashin A. A., Honig B. On the environment of ionizable groups in globular proteins. J Mol Biol. 1984 Mar 15;173(4):515–521. doi: 10.1016/0022-2836(84)90394-2. [DOI] [PubMed] [Google Scholar]
- Raumann B. E., Rould M. A., Pabo C. O., Sauer R. T. DNA recognition by beta-sheets in the Arc repressor-operator crystal structure. Nature. 1994 Feb 24;367(6465):754–757. doi: 10.1038/367754a0. [DOI] [PubMed] [Google Scholar]
- Rozwarski D. A., Gronenborn A. M., Clore G. M., Bazan J. F., Bohm A., Wlodawer A., Hatada M., Karplus P. A. Structural comparisons among the short-chain helical cytokines. Structure. 1994 Mar 15;2(3):159–173. doi: 10.1016/s0969-2126(00)00018-6. [DOI] [PubMed] [Google Scholar]
- Schindler T., Herrler M., Marahiel M. A., Schmid F. X. Extremely rapid protein folding in the absence of intermediates. Nat Struct Biol. 1995 Aug;2(8):663–673. doi: 10.1038/nsb0895-663. [DOI] [PubMed] [Google Scholar]
- Schmid F. X. Prolyl isomerase: enzymatic catalysis of slow protein-folding reactions. Annu Rev Biophys Biomol Struct. 1993;22:123–142. doi: 10.1146/annurev.bb.22.060193.001011. [DOI] [PubMed] [Google Scholar]
- Sosnick T. R., Mayne L., Hiller R., Englander S. W. The barriers in protein folding. Nat Struct Biol. 1994 Mar;1(3):149–156. doi: 10.1038/nsb0394-149. [DOI] [PubMed] [Google Scholar]
- Tanford C. Protein denaturation. C. Theoretical models for the mechanism of denaturation. Adv Protein Chem. 1970;24:1–95. [PubMed] [Google Scholar]
- Vershon A. K., Youderian P., Susskind M. M., Sauer R. T. The bacteriophage P22 arc and mnt repressors. Overproduction, purification, and properties. J Biol Chem. 1985 Oct 5;260(22):12124–12129. [PubMed] [Google Scholar]
- Waldburger C. D., Schildbach J. F., Sauer R. T. Are buried salt bridges important for protein stability and conformational specificity? Nat Struct Biol. 1995 Feb;2(2):122–128. doi: 10.1038/nsb0295-122. [DOI] [PubMed] [Google Scholar]