Abstract
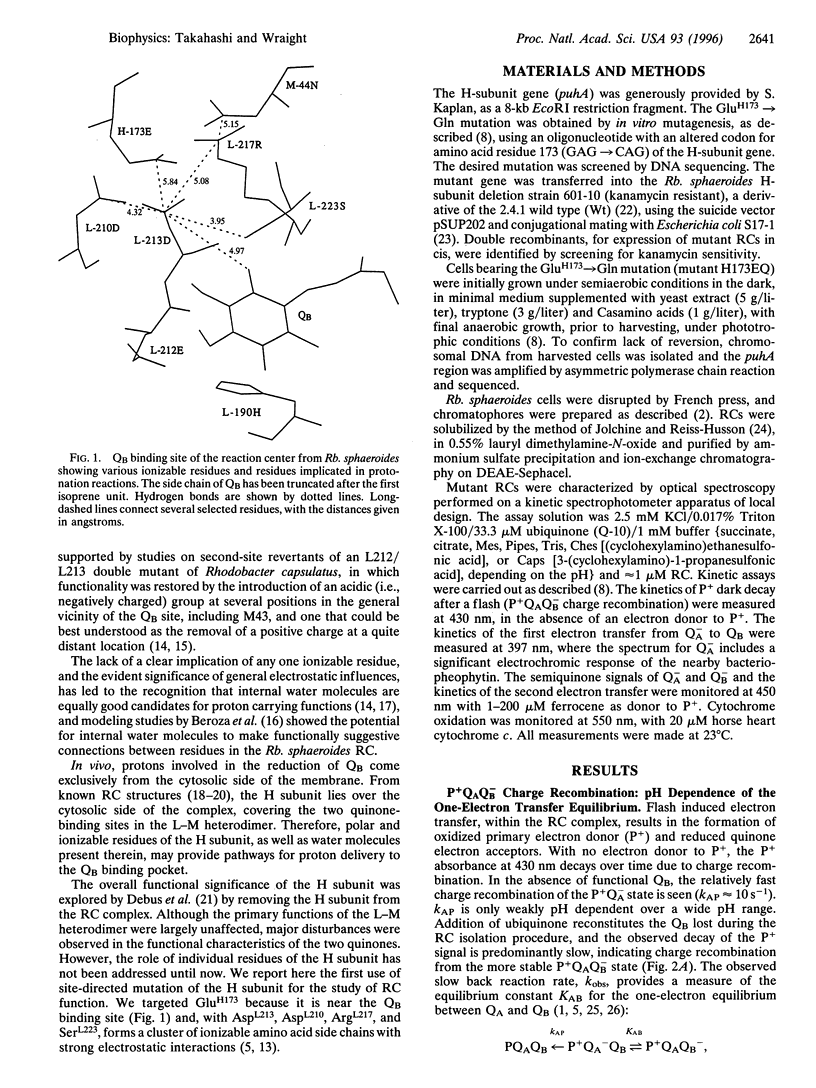
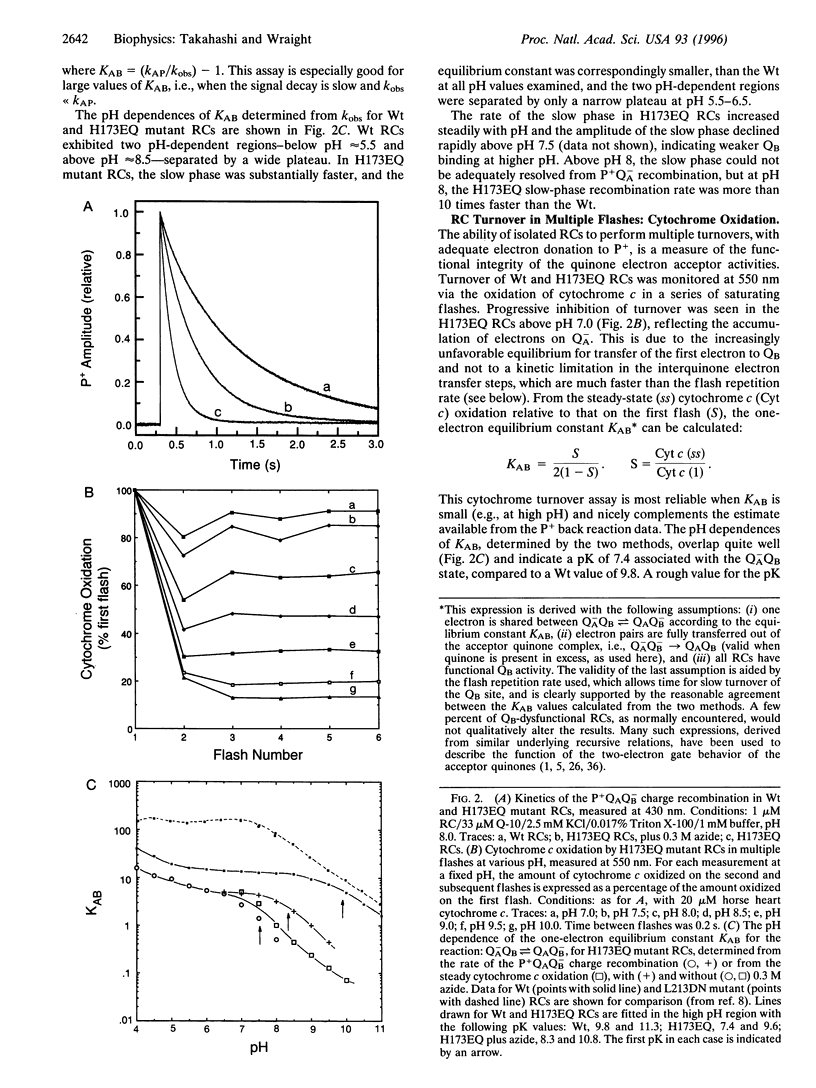
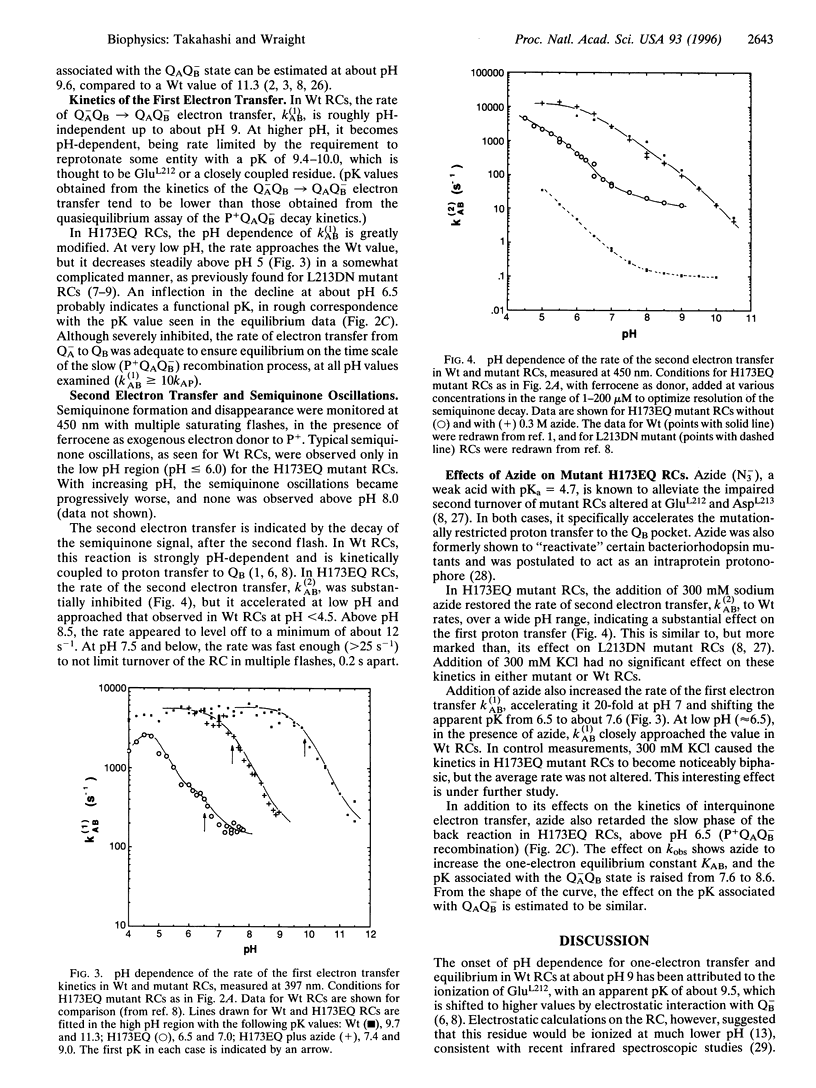
The x-ray crystallographic structure of the photosynthetic reaction center (RC) has proven critical in understanding biological electron transfer processes. By contrast, understanding of intraprotein proton transfer is easily lost in the immense richness of the details. In the RC of Rhodobacter (Rb.) sphaeroides, the secondary quinone (QB) is surrounded by amino acid residues of the L subunit and some buried water molecules, with M- and H-subunit residues also close by. The effects of site-directed mutagenesis upon RC turnover and quinone function have implicated several L-subunit residues in proton delivery to QB, although some species differences exist. In wild-type Rb. sphaeroides, Glu L212 and Asp L213 represent an inner shell of residues of particular importance in proton transfer to QB. Asp L213 is crucial for delivery of the first proton, coupled to transfer of the second electron, while Glu L212, possibly together with Asp L213, is necessary for delivery of the second proton, after the second electron transfer. We report here the first study, by site-directed mutagenesis, of the role of the H subunit in QB function. Glu H173, one of a cluster of strongly interacting residues near QB, including Asp L213, was altered to Gln. In isolated mutant RCs, the kinetics of the first electron transfer, leading to formation of the semiquinone, QB-, and the proton-linked second electron transfer, leading to the formation of fully reduced quinol, were both greatly retarded, as observed previously in the Asp L213 --> Asn mutant. However, the first electron transfer equilibrium, QA-QB <==> QAQB-, was decreased, which is opposite to the effect of the Asp L213 --> Asn mutation. These major disruptions of events coupled to proton delivery to QB were largely reversed by the addition of azide (N3-). The results support a major role for electrostatic interactions between charged groups in determining the protonation state of certain entities, thereby controlling the rate of the second electron transfer. It is suggested that the essential electrostatic effect may be to "potentiate" proton transfer activity by raising the pK of functional entities that actually transfer protons in a coupled fashion with the second electron transfer. Candidates include buried water (H3O+) and Ser L223 (serine-OH2+), which is very close to the O5 carbonyl of the quinone.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allen J. P., Feher G., Yeates T. O., Komiya H., Rees D. C. Structure of the reaction center from Rhodobacter sphaeroides R-26: protein-cofactor (quinones and Fe2+) interactions. Proc Natl Acad Sci U S A. 1988 Nov;85(22):8487–8491. doi: 10.1073/pnas.85.22.8487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroza P., Fredkin D. R., Okamura M. Y., Feher G. Electrostatic calculations of amino acid titration and electron transfer, Q-AQB-->QAQ-B, in the reaction center. Biophys J. 1995 Jun;68(6):2233–2250. doi: 10.1016/S0006-3495(95)80406-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bélanger G., Bérard J., Corriveau P., Gingras G. The structural genes coding for the L and M subunits of Rhodospirillum rubrum photoreaction center. J Biol Chem. 1988 Jun 5;263(16):7632–7638. [PubMed] [Google Scholar]
- Chang C. H., el-Kabbani O., Tiede D., Norris J., Schiffer M. Structure of the membrane-bound protein photosynthetic reaction center from Rhodobacter sphaeroides. Biochemistry. 1991 Jun 4;30(22):5352–5360. doi: 10.1021/bi00236a005. [DOI] [PubMed] [Google Scholar]
- Ermler U., Fritzsch G., Buchanan S. K., Michel H. Structure of the photosynthetic reaction centre from Rhodobacter sphaeroides at 2.65 A resolution: cofactors and protein-cofactor interactions. Structure. 1994 Oct 15;2(10):925–936. doi: 10.1016/s0969-2126(94)00094-8. [DOI] [PubMed] [Google Scholar]
- Hanson D. K., Baciou L., Tiede D. M., Nance S. L., Schiffer M., Sebban P. In bacterial reaction centers protons can diffuse to the secondary quinone by alternative pathways. Biochim Biophys Acta. 1992 Sep 25;1102(2):260–265. doi: 10.1016/0005-2728(92)90108-e. [DOI] [PubMed] [Google Scholar]
- Hanson D. K., Tiede D. M., Nance S. L., Chang C. H., Schiffer M. Site-specific and compensatory mutations imply unexpected pathways for proton delivery to the QB binding site of the photosynthetic reaction center. Proc Natl Acad Sci U S A. 1993 Oct 1;90(19):8929–8933. doi: 10.1073/pnas.90.19.8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hienerwadel R., Grzybek S., Fogel C., Kreutz W., Okamura M. Y., Paddock M. L., Breton J., Nabedryk E., Mäntele W. Protonation of Glu L212 following QB- formation in the photosynthetic reaction center of Rhodobacter sphaeroides: evidence from time-resolved infrared spectroscopy. Biochemistry. 1995 Mar 7;34(9):2832–2843. doi: 10.1021/bi00009a013. [DOI] [PubMed] [Google Scholar]
- Jolchine G., Reiss-Husson F. Comparative studies on two reaction center preparations from Rhodopseudomonas speheroides Y. FEBS Lett. 1974 Mar 15;40(1):5–8. doi: 10.1016/0014-5793(74)80881-1. [DOI] [PubMed] [Google Scholar]
- Kleinfeld D., Okamura M. Y., Feher G. Electron transfer in reaction centers of Rhodopseudomonas sphaeroides. I. Determination of the charge recombination pathway of D+QAQ(-)B and free energy and kinetic relations between Q(-)AQB and QAQ(-)B. Biochim Biophys Acta. 1984 Jul 27;766(1):126–140. doi: 10.1016/0005-2728(84)90224-x. [DOI] [PubMed] [Google Scholar]
- Maróti P., Hanson D. K., Baciou L., Schiffer M., Sebban P. Proton conduction within the reaction centers of Rhodobacter capsulatus: the electrostatic role of the protein. Proc Natl Acad Sci U S A. 1994 Jun 7;91(12):5617–5621. doi: 10.1073/pnas.91.12.5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel H., Epp O., Deisenhofer J. Pigment-protein interactions in the photosynthetic reaction centre from Rhodopseudomonas viridis. EMBO J. 1986 Oct;5(10):2445–2451. doi: 10.1002/j.1460-2075.1986.tb04520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel H., Weyer K. A., Gruenberg H., Dunger I., Oesterhelt D., Lottspeich F. The 'light' and 'medium' subunits of the photosynthetic reaction centre from Rhodopseudomonas viridis: isolation of the genes, nucleotide and amino acid sequence. EMBO J. 1986 Jun;5(6):1149–1158. doi: 10.1002/j.1460-2075.1986.tb04340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura M. Y., Feher G. Proton transfer in reaction centers from photosynthetic bacteria. Annu Rev Biochem. 1992;61:861–896. doi: 10.1146/annurev.bi.61.070192.004241. [DOI] [PubMed] [Google Scholar]
- Ovchinnikov YuA, Abdulaev N. G., Zolotarev A. S., Shmukler B. E., Zargarov A. A., Kutuzov M. A., Telezhinskaya I. N., Levina N. B. Photosynthetic reaction centre of Chloroflexus aurantiacus. I. Primary structure of L-subunit. FEBS Lett. 1988 Apr 11;231(1):237–242. doi: 10.1016/0014-5793(88)80739-7. [DOI] [PubMed] [Google Scholar]
- Paddock M. L., McPherson P. H., Feher G., Okamura M. Y. Pathway of proton transfer in bacterial reaction centers: replacement of serine-L223 by alanine inhibits electron and proton transfers associated with reduction of quinone to dihydroquinone. Proc Natl Acad Sci U S A. 1990 Sep;87(17):6803–6807. doi: 10.1073/pnas.87.17.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddock M. L., Rongey S. H., Feher G., Okamura M. Y. Pathway of proton transfer in bacterial reaction centers: replacement of glutamic acid 212 in the L subunit by glutamine inhibits quinone (secondary acceptor) turnover. Proc Natl Acad Sci U S A. 1989 Sep;86(17):6602–6606. doi: 10.1073/pnas.86.17.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddock M. L., Rongey S. H., McPherson P. H., Juth A., Feher G., Okamura M. Y. Pathway of proton transfer in bacterial reaction centers: role of aspartate-L213 in proton transfers associated with reduction of quinoneto dihydroquinone. Biochemistry. 1994 Jan 25;33(3):734–745. doi: 10.1021/bi00169a015. [DOI] [PubMed] [Google Scholar]
- Sockett R. E., Donohue T. J., Varga A. R., Kaplan S. Control of photosynthetic membrane assembly in Rhodobacter sphaeroides mediated by puhA and flanking sequences. J Bacteriol. 1989 Jan;171(1):436–446. doi: 10.1128/jb.171.1.436-446.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi E., Wraight C. A. Proton and electron transfer in the acceptor quinone complex of Rhodobacter sphaeroides reaction centers: characterization of site-directed mutants of the two ionizable residues, GluL212 and AspL213, in the QB binding site. Biochemistry. 1992 Jan 28;31(3):855–866. doi: 10.1021/bi00118a031. [DOI] [PubMed] [Google Scholar]
- Takahashi E., Wraight C. A. Small weak acids stimulate proton transfer events in site-directed mutants of the two ionizable residues, GluL212 and AspL213, in the QB-binding site of Rhodobacter sphaeroides reaction center. FEBS Lett. 1991 May 20;283(1):140–144. doi: 10.1016/0014-5793(91)80572-k. [DOI] [PubMed] [Google Scholar]
- Tittor J., Soell C., Oesterhelt D., Butt H. J., Bamberg E. A defective proton pump, point-mutated bacteriorhodopsin Asp96----Asn is fully reactivated by azide. EMBO J. 1989 Nov;8(11):3477–3482. doi: 10.1002/j.1460-2075.1989.tb08512.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wraight C. A. Electron acceptors of bacterial photosynthetic reaction centers. II. H+ binding coupled to secondary electron transfer in the quinone acceptor complex. Biochim Biophys Acta. 1979 Nov 8;548(2):309–327. doi: 10.1016/0005-2728(79)90138-5. [DOI] [PubMed] [Google Scholar]


