Abstract
The molecular basis of nephron progenitor cell renewal and differentiation into nascent epithelial nephrons is an area of intense investigation. Defects in these early stages of nephrogenesis lead to renal hypoplasia, and eventually hypertension and chronic kidney disease. Terminal nephron differentiation, the process by which renal epithelial precursors cells exit the cell cycle and acquire physiological functions is equally important. Failure of terminal epithelial cell differentiation results in renal dysplasia and cystogenesis. Thus, a better understanding of the transcriptional frameworks which regulate early and late renal cell differentiation is of great clinical significance. In this review, we will discuss evidence implicating the Mdm2-p53 pathway in cell fate determination during development. The emerging central theme from loss- and gain-of-function studies is that tight regulation of p53 levels and transcriptional activity is absolutely required for nephrogenesis. We will also discuss how post-translational modifications of p53 (e.g., acetylation and phosphorylation) alter the spatiotemporal and functional properties of p53 and thus cell fate during kidney development. Mutations and polymorphisms in the Mdm2-p53 pathway are present in more than 50% of cancers in humans. This raises the question of whether sequence variants in the Mdm2-p53 pathway increase the susceptibility to renal dysgenesis, hypertension or chronic kidney disease. With the advent of whole exome sequencing and other high throughput technologies, this hypothesis is testable in cohorts of children with renal dysgenesis.
Keywords: Nephrogenesis, terminal differentiation, mdm2, p53, p73
Introduction
p53, p63, and p73 are members of a family of transcription factors that plays a key role in cell fate determination [1]. Although tumor suppression is the original and traditional function assigned to the p53 gene family, it is now clear that the three proteins have overlapping as well as distinct biological roles [1]. The tumor suppressor protein, p53, prevents aberrant growth by acting as a transcription factor for cell cycle (e.g., p21Waf-1/Cip-1), apoptosis (e.g., Noxa, Puma, and Bax) and DNA repair (e.g. GADD45) genes. p53 activates transcription of its target genes through binding to a consensus DNA element which consists of two copies of the inverted repeat sequence [RRRC(A/T)(T/A)GYYY] separated by 0–13 nucleotides [2]. Wild-type p53 also represses the transcription of some genes. The repression can be mediated via specific DNA binding, selective targeting of histone deacetylases to the regulatory regions or by an effect on the basal transcription machinery [3]. Chromatin immunoprecipitation coupled to Next Gen DNA sequencing in primary and cancer cell lines has shown that non-traditional targets, e.g., metabolic, autophagy and oxidative genes, are among the novel p53-regulated pathways in the human genome, hence the pleomorphic effects of p53 on cancer, development, ageing, and metabolism [4–6].
Human p53 consists of 393 amino acids and is composed of three regions: an N-terminal activation domain that interacts with co-activators and other proteins (e.g., its inhibitor, MDM-2), a core DNA-binding domain and C-terminal activation/oligomerization domains. Mutations in the p53-regulated pathway are found in more than 50% of all human cancers [7].
Regulation of p53 activity
The principal mechanisms that govern p53 activity are exerted at the posttranslational level. These include regulation of p53 protein stability, subcellular localization, and posttranslational modifications that allow activation of the DNA binding activity of p53. It is now established that tight regulation of p53 levels and activity is absolutely required for organ development and terminal epithelial differentiation.
Murine Double Minute (MDM2)
One of the central components in the regulation of p53 is the p53-interacting protein MDM2 [8–11] (Fig. 2). MDM2 is a direct transcriptional target for p53 and so establishes a feedback loop in which p53 drives expression of its own negative regulator. This loop serves to dampen p53 activity in normal unstressed cells and contributes to limiting or reversing the p53 response following the removal of the initiating stress signals. MDM2 inhibits p53 functions via several different mechanisms: MDM2 interacts with and conceals the N-terminal transcriptional activation domain of p53. This binding of MDM2 to p53 also inhibits p53 ability to contact transcriptional co-activators such as CBP/p300. In addition, MDM2 plays a major role in the regulation of p53 protein stability [10, 12–17]. MDM2 is a RING-finger protein that functions as an E3 ubiquitin ligase directing p53 to proteosomal degradation. Also, Mdm-2 shuttles p53 from the nucleus to the cytoplasm where it is then degraded [18].
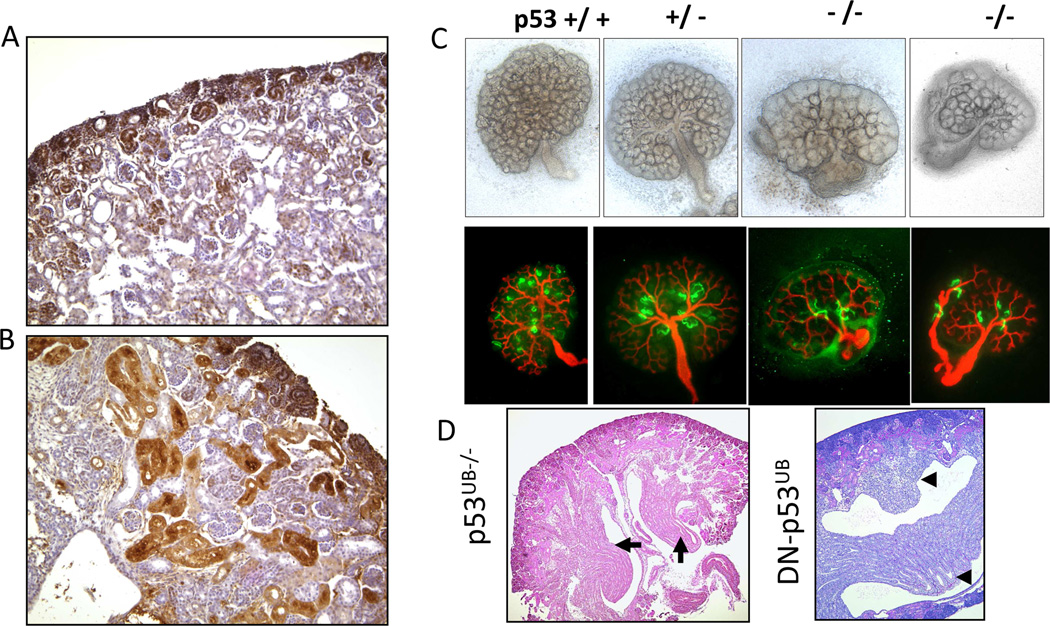
Fig. 2.
p53+/+ (A) p53−/− (B) kidney sections at postnatal day 1 exhibit ectopic PCNA staining within the differentiation zone. (C) E11.5 p53−/− metanephroi cultured in vitro for 72 hrs show expanded peripheral domain which contains ureteric bud (UB) tips but few proximal tubules consistent with hypoplasia. (D) Duplex collecting system is another prominent phenotype see in mice with targeted disruption of p53 in the UB or transgenic mice overexpressing a mutant p53 lacking the C-terminus in the UB. Black arrows/arrowheads point to the duplex papillae. Adapted from J Am Soc Nephrol. 2009;20:2328; Am J Physiol Renal Physiol. 2002;283:F727 (used with permission).
Post-translational modifications of p53
Phosphorylation
In response to DNA damage, p53 is phosphorylated at multiple sites at the N- and C-terminal by a number of kinases, and most of these phosphorylation sites are conserved between human and mouse p53 (e.g., S15, S20, S392) [19]. It has been proposed that a distinctive combination of phosphorylated residues could be required for further modifications, leading to maximal activation [20]. It is also possible that a unique phosphorylation pattern induced by specific stimuli could determine the cellular response. Until recently, the specific residues phosphorylated during organ development were unknown. However, we now know that p53 is subject to phosphorylation in a developmental and cell-specific manner [21].
Acetylation
The p300/CREB-binding protein (CBP) and p300/CBP-associated factor (PCAF) acetylate the carboxyl terminus of p53 at lysines 373 and 382 (p300/CBP) and at Lys320 (PCAF) [19]. The recruitment of these co-activators stabilizes p53 and augments sequence-specific DNA binding following DNA damage. Deacetylation of C-terminal lysine in p53 by histone deacetylases (HDACs) promotes ubiquitylation of these lysines by Mdm2, indicating that Mdm-2 and HDACs cooperate in the negative regulation of p53 stability and transcriptional activity [22]. Differential acetylation “cassettes” in C-terminus of p53 have distinct effects on cell fate [23]. For example, acetylation of lysine 320 (K320) prevents phosphorylation of crucial serines in the N-terminus of p53, only allows activation of p21 and promotes survival after DNA damage. In contrast, acetylation of K373 enhances p53-mediated activation of pro-apoptosis genes. In addition, differential acetylation of these lysine residues appears to modulate the ability of p53 to interact with co-activators or co-repressors [23]. It should be noted that unacetylated p53 (which accumulates following MDM2 inactivation) is fully capable of inducing apoptosis but is defective in cell cycle arrest, further supporting a selective role for acetylation in the regulation of p53 functions [24]. In conclusion, p53 phosphorylation and acetylation may constitute a “post-translational code” which determines which proteins are recruited and thus the cell fate.
p53 as a developmental factor
In addition to its traditional role as a tumor suppressor, p53 has well established roles in development and differentiation, which can be summarized as follows [25]:
Developmental expression patterns
In developing mouse embryos, a good correlation exists between p53 levels and the fraction of embryonic cells undergoing proliferation. Later in development, p53 expression is restricted to a few organs undergoing differentiation such as kidney, intestines, thymus, and spleen [26–28].
p53-induced cell differentiation
Introduction of wild-type p53 into undifferentiated cells can result in progression from an undifferentiated to a more differentiated state. This effect has been demonstrated in a variety of cell lineages such as myogenic cells, osteogenic cells, keratinocytes, neurons, thyroid cells, and renal epithelial cells. In contrast, the normal differentiation pathways can be blocked by introduction of dominant negative forms of p53 [29, 30].
Activation of p53 during cellular differentiation
The p53 protein is stabilized and its DNA binding and transcriptional activities are induced during skeletal muscle differentiation in vitro [31]. In the skeletal muscle cell lineage, p53 cooperates with MyoD in the transcriptional induction of the muscle creatine kinase gene, whereas expression of a dominant negative p53 protein in muscle cells reduces the expression of endogenous creatine kinase [31, 32]. In the renal epithelial cell lineage, p53 forms an enhanceosome-like complex with CREB and Kruppel-like factor-4 and recruits CREB-Binding Protein (CBP/p300) in the transcriptional activation of the bradykinin B2 receptor (Bdkrb2), a terminal nephron differentiation gene [33].
p53 depletion or inactivation affects embryonic development in Xenopus and mice
Considering the central role of p53 as a regulator of cellular proliferation and homeostasis, as well as cell death and premature senescence, the viability of p53−/− mice and the absence of overt developmental defects came as a surprise [34]. Although embryonic development is grossly unaffected in p53−/− mice, there are reports of neural tube closure defects in approximately 16% of female p53−/− mice and a reduced reproductive capacity in both genders [35]. This is in striking contrast to the phenotype following p53 depletion in Xenopus embryos which leads to gastrulation failure and defects in mesoderm formation due to impaired TGF-β/Nodal/activin gene responses [36]. A possible explanation for the different p53 knockout phenotype in Xenopus and mice is that p53 is only one member of a family of related genes. In addition to p53, early mouse embryos express the p53 family members p63 and p73 that might compensate for the loss of p53, while in Xenopus p53 is solely responsible for early embryogenesis as p73 is not found in the lower vertebrates and Xenopus p63 is only expressed at later stages during organogenesis [36]. There is also evidence that p53 deficiency on CBA genetic background is embryonic lethal, which can be rescued by inclusion of genes from 129/SvJ background [37].
During the organogenesis and differentiation stages in mice, p53 acquires unique functions in some organs, such as the kidney, liver, eye and testes. Hepatocytes from adult livers of p53−/− mice divide over 2.5 times the rate of normal mice and show altered morphology in vivo and persistent α-fetoprotein gene expression [38]. The latter gene is normally repressed by p53 [39]. Also, p53−/− mice have a high frequency of persistent hyperplastic primary vitreous and cataracts and a testicular giant cell degenerative syndrome [40, 41]. Collectively, inhibition of p53 expression/activity interferes with numerous developmental programs, and the outcome may depend on the presence or absence of compensation by other family members and on the stage of development.
Unopposed p53 activity is detrimental to normal growth and development
There is substantial evidence that excess or unopposed p53 activity is detrimental to embryonic development. For example, overexpression of p53 in Xenopus embryos is associated with delayed or arrested cleavage of injected cells with subsequent distortions of cell movements, induction processes and tissue differentiation [42]. Transgenic mice overexpressing p53 in testes exhibit profound apoptosis of developing spermatids in a dose-dependent fashion [43]. Importantly, transgenic overexpression of p53 or deletion of Mdm2 from the ureteric bud [21, 44] in mice interferes with collecting duct growth and branching leading to renal hypoplasia and impaired differentiation [45]. Genetic evidence that tight control of p53 by Mdm-2 is crucial for normal embryogenesis is indicated by the fact that p53 deletion rescues the embryonic lethality in Mdm-2 and Mdm-4 deficient mice [46, 47] and XRCC4-deficient mice [48]. Targeted deletion of Mdm-2 from bone, using genetic crosses of collagen-3-Cre and Mdm2-floxed mice, revealed that unopposed p53 activity interferes with chondrocyte differentiation and results in skeletal dysgenesis and excessive apoptosis; the abnormal phenotype is rescued by germline deletion of p53 [49, 50]. The negative regulation of p53 by mdm-2 is so potent that 20% reduction in mdm-2 levels in mice is sufficient to cause increased radiosensitivity, growth impairment, and increased apoptosis in both lymphocytes and epithelial cells [50]. In humans, a single nucleotide polymorphism (SNP309) in the MDM2 promoter increases the affinity of the transcriptional activator Sp1, resulting in higher levels of MDM2 gene expression and subsequent attenuation of the p53 pathway. Also, SNP309 is associated with accelerated tumor formation in both hereditary and sporadic cancers [51].
Role of p53 and p73 in terminal nephron differentiation
During early nephrogenesis, p53 is expressed in nephron progenitors, nascent nephrons and UB branches [4, 52] (Fig. 1). During the latter stages of nephrogenesis, p53 is enriched in differentiating epithelial cells expressing renal function genes (RFGs) (Fig. 1), such as receptors for vasoactive hormones (bradykinin B2 receptor [Bdkrb2], angiotensin type 1a [agtr1a]), the sodium pump (Na,K-ATPase α1), and epithelial sodium (ENac) and water channels (AQP2). Moreover, p53 binds to and transactivates the promoters of RFGs [30]. In contrast, expression of a dominant negative mutant p53 inhibits endogenous RFG expression. Binding of endogenous p53 to the promoters of RFGs coincides with the differentiation process and is attenuated once renal epithelial cells are fully differentiated [30]. In the case of the Bdkrb2 gene, p53 recruits two other transcription factors, CREB and KLF-4, to bind adjoining sequences, as well as the co-activator CBP/p300. This transcriptional complex functions as an enhanceosome [33]. Angiotensin II stimulates CREB phosphorylation on Serine 133 and its interaction with p53 on the Bdkrb2 promoter. The co-factors associated with p53 that are required for transactivation of other RFGs are unknown. The physiological relevance of these finding was later demonstrated in p53−/− mice, which exhibit defects in terminal nephron differentiation, including ectopic tubular proliferation and disorganized spatial expression of RFGs [30, 53] (Fig. 2A,B).
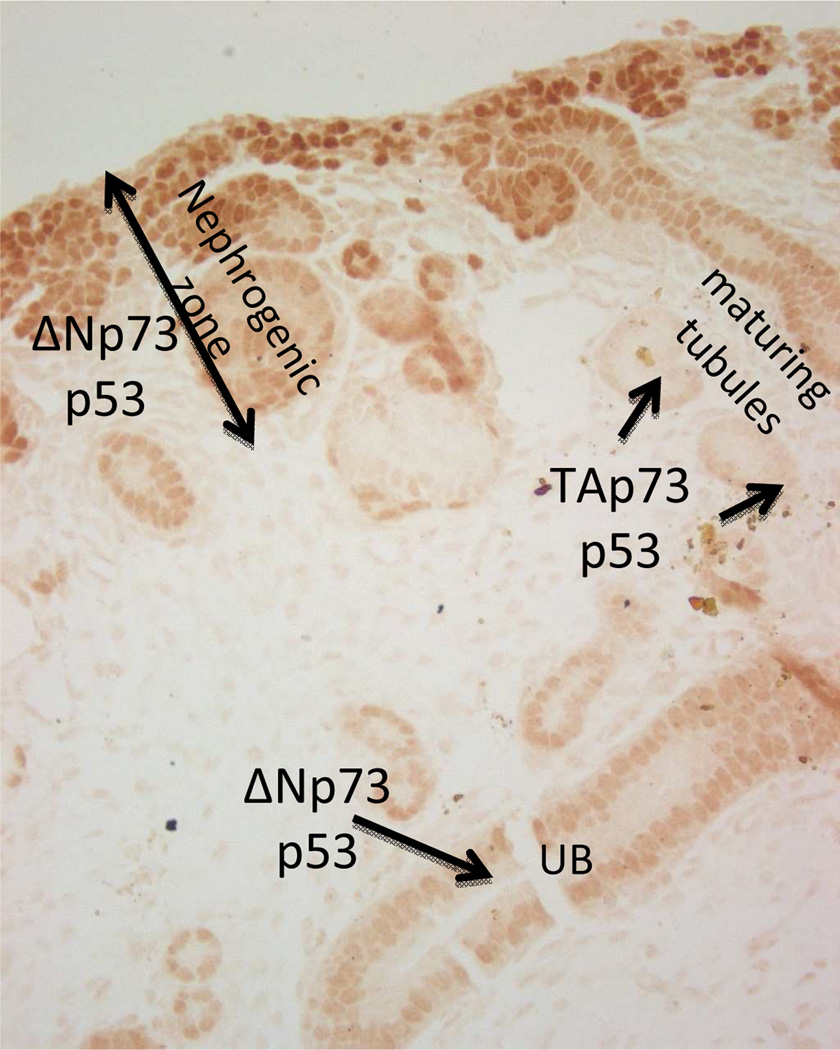
Fig. 1.
Localization of p53 and p73 isoforms in the developing kidney. Section from E17.5 mouse kidney stained for Pax2 (brown nuclear staining). ×20
In a recent study, we reported that p53 undergoes phosphorylation on N- and C-terminal serines as well as C-terminus acetylation during kidney development. These post-translational modifications are associated with differential effects on p53 transcriptional activity and nephron-segment specific expression [21]. For example, Compared to wild-type p53, mutant p53(S/A15) displays significantly reduced transcriptional effects on the AQP2, BdkrB2, and Agtr1a promoter reporter constructs even after correction for p53 protein levels. p53(T/A18) and p53(S/A20) mutant constructs display lower transcriptional activity of AQP2 but not BdkrB2 promoter constructs; however, both mutants exhibit lower repression of Agtr1a.
The p53 family member, p73, also plays a role in epithelial and CNS development [54]. Alternate promoter usage results in p73 isoforms that either contain the amino terminus transactivation domain (P1 5’ promoter; transactivation (TA) domain isoforms) or lack this domain (P2 intronic promoter; amino terminus-truncated (ΔN) isoforms). The ΔN isoforms act in a dominant negative fashion to inhibit TAp73- and p53-mediated transactivation. Importantly, although the TAp73 isoforms induce cell cycle arrest or apoptosis, the ΔN isoforms are pro-survival and anti-apoptotic. We have examined the role of p73 in nephron differentiation [55]. TAp73 and ΔNp73 exhibit reciprocal spatiotemporal expression and functions during nephrogenesis. TAp73 is predominantly expressed in the collecting ducts in an overlapping manner with AQP-2 and in differentiating proximal tubules. Moreover, the endogenous AQP-2 promoter is occupied by TAp73 in a developmentally regulated manner. In transient transfection assays, TAp73 activates AQP-2 and Bdkrb2 promoters independent of p53. These findings indicate that TAp73 can compensate functionally for the absence of p53. On the other hand, p73 is not sufficient to fully compensate for p53 during kidney development since p53-null mice have defects in terminal nephron differentiation. In contrast to p53 and TAp73, ΔNp73 isoforms are induced early in development and are preferentially expressed in Pax-2 positive nephron progenitors and immature collecting ducts [55]. Functionally, ΔNp73 antagonizes p53-mediated transcription of B2R and AQP-2. The spatiotemporal switch from ΔNp73 to TAp73 during nephrogenesis may participate in the specification of terminal differentiation gene expression.
p53 and Mdm2 in the ureteric bud lineage
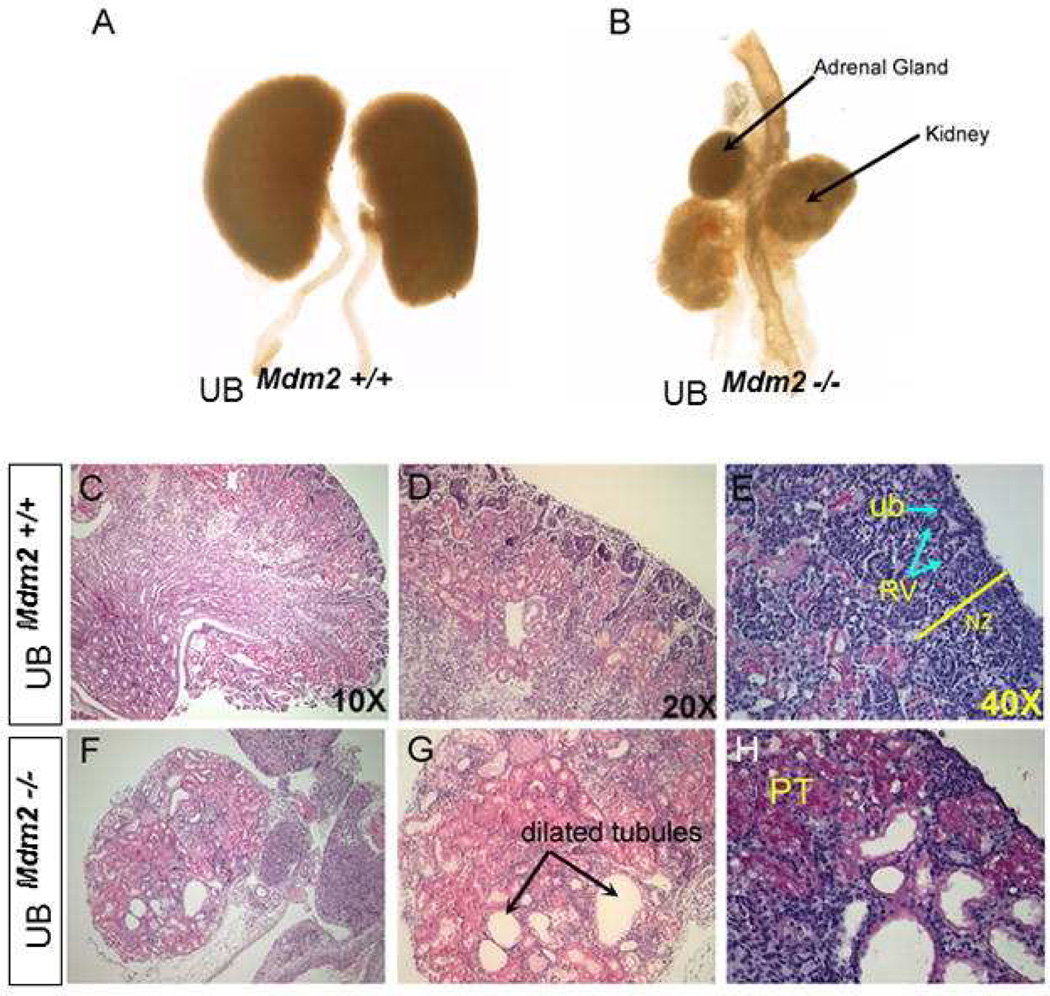
We examined the effects of p53 inactivation or overexpression on early stages of kidney development [4, 44, 52]. p53−/− embryos bred on C57Bl6 background exhibit a spectrum of congenital abnormalities of the kidney and urinary tract, most prominently double ureters/collecting systems and hypoplastic metanephroi (Fig. 2C). Interestingly, transgenic expression of dominant negative p53 or conditional inactivation of p53 in the UB recapitulates the duplex phenotype (Fig. 2D). p53 inactivation causes enhanced sensitivity to glial cell line-derived neurotrophic factor (GDNF), which explains ectopic budding. Conversely, deletion of Mdm2 from the UB lineage results in accumulation of p53, cell cycle arrest, apoptosis and suppression of Wnt9b expression. Hence, Mdm2-null kidneys display severe hypodysplasia and low nephron number (Fig. 3). Taken together, these results support the conclusion that tight regulation of p53 activity is required for normal kidney development.
Fig. 3. Ureteric bud-specific deletion of Mdm2 results in bilaterally small, hypodysplastic kidneys.
Gross morphology of P1 kidneys isolated from wild type (A) and conditional homozygous null mutant (B) littermates. Note; the contralateral adrenal gland in Panel B was lost during dissection. (C–H) Histological staining of kidney sections at P1 reveals severe structural anomalies in UBmdm2−/− mice. The mutant kidneys have poor tissue organization and numerous dilated tubules resembling cysts occupy the bulk of the kidney. The cortical and medullary zones are not identifiable. C, D, F, and G represent Hematoxylin and Eosin staining while E and H represent Periodic Acid Schiff staining. Ub: ureteric bud; RV: renal vesicle; PT: proximal tubule, NZ: nephrogenic zone. Adapted from Dev Biol. 2011; 353:354 (used with permission).
In summary, Mdm2 is required to restrain p53 activity during nephrogenesis. p53, in turn, plays multiple roles in kidney development, which include terminal differentiation of the collecting duct, transcriptional regulation of renal function genes, ureteric bud outgrowth and branching, and renewal and differentiation of nephron progenitors.
Acknowledgments
The authors would like to thank the Tulane Hypertension and Renal Center of Excellence Molecular and Imaging Core. This work was supported by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grant R01DK62550 and RO1DK56264 to SED and by National Institutes of Health National Center for Research Resources (NIH-NCRR) Center of Biomedical Research Excellence grant support P20RR017659 to ZS. K. Aboudehen current address is UT Southwestern, Department of Medicine, Dallas, TX.
References
- 1.Dotsch V, Bernassola F, Coutandin D, Candi E, Melino G. p63 and p73, the ancestors of p53. Cold Spring Harb Perspect Biol. 2010;2:a004887. doi: 10.1101/cshperspect.a004887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 3.Mack DH, Vartikar J, Pipas JM, Laimins LA. Specific repression of TATA-mediated but not initiator-mediated transcription by wild-type p53. Nature. 1993;363:281–283. doi: 10.1038/363281a0. [DOI] [PubMed] [Google Scholar]
- 4.Saifudeen Z, Liu J, Dipp S, Yao X, Li Y, McLaughlin N, Aboudehen K, El-Dahr SS. A p53-Pax2 pathway in kidney development: implications for nephrogenesis. PLoS One. 2012;7:e44869. doi: 10.1371/journal.pone.0044869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tuteja G, White P, Schug J, Kaestner KH. Extracting transcription factor targets from ChIP-Seq data. Nucleic Acids Res. 2009;37:e113. doi: 10.1093/nar/gkp536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, Liu J, Zhao XD, Chew JL, Lee YL, Kuznetsov VA, Sung WK, Miller LD, Lim B, Liu ET, Yu Q, Ng HH, Ruan Y. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 7.Albrechtsen N, Dornreiter I, Grosse F, Kim E, Wiesmuller L, Deppert W. Maintenance of genomic integrity by p53: complementary roles for activated and non-activated p53. Oncogene. 1999;18:7706–7717. doi: 10.1038/sj.onc.1202952. [DOI] [PubMed] [Google Scholar]
- 8.Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24:2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- 9.Freedman DA, Wu L, Levine AJ. Functions of the MDM2 oncoprotein. Cell Mol Life Sci. 1999;55:96–107. doi: 10.1007/s000180050273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 11.Lozano G, Montes de Oca Luna R. MDM2 function. Biochim Biophys Acta. 1998;1377:M55–M59. doi: 10.1016/s0304-419x(97)00037-1. [DOI] [PubMed] [Google Scholar]
- 12.Fuchs SY, Fried VA, Ronai Z. Stress-activated kinases regulate protein stability. Oncogene. 1998;17:1483–1490. doi: 10.1038/sj.onc.1202184. [DOI] [PubMed] [Google Scholar]
- 13.Ashcroft M, Vousden KH. Regulation of p53 stability. Oncogene. 1999;18:7637–7643. doi: 10.1038/sj.onc.1203012. [DOI] [PubMed] [Google Scholar]
- 14.Ashcroft M, Kubbutat MH, Vousden KH. Regulation of p53 function and stability by phosphorylation. Mol Cell Biol. 1999;19:1751–1758. doi: 10.1128/mcb.19.3.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kubbutat MH, Ludwig RL, Ashcroft M, Vousden KH. Regulation of Mdm2-directed degradation by the C terminus of p53. Mol Cell Biol. 1998;18:5690–5698. doi: 10.1128/mcb.18.10.5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lohrum MA, Vousden KH. Regulation and activation of p53 and its family members. Cell Death Differ. 1999;6:1162–1168. doi: 10.1038/sj.cdd.4400625. [DOI] [PubMed] [Google Scholar]
- 17.Lain S. Protecting p53 from degradation. Biochem Soc Trans. 2003;31:482–485. doi: 10.1042/bst0310482. [DOI] [PubMed] [Google Scholar]
- 18.Roth J, Dobbelstein M, Freedman DA, Shenk T, Levine AJ. Nucleo-cytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus rev protein. Embo J. 1998;17:554–564. doi: 10.1093/emboj/17.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bode AM, Dong Z. Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer. 2004;4:793–805. doi: 10.1038/nrc1455. [DOI] [PubMed] [Google Scholar]
- 20.Saito S, Yamaguchi H, Higashimoto Y, Chao C, Xu Y, Fornace AJ, Jr, Appella E, Anderson CW. Phosphorylation site interdependence of human p53 post-translational modifications in response to stress. J Biol Chem. 2003;278:37536–37544. doi: 10.1074/jbc.M305135200. [DOI] [PubMed] [Google Scholar]
- 21.Aboudehen K, Hilliard S, Saifudeen Z, El-Dahr SS. Mechanisms of p53 activation and physiological relevance in the developing kidney. Am J Physiol. 2012;302:F928–F940. doi: 10.1152/ajprenal.00642.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li M, Luo J, Brooks CL, Gu W. Acetylation of p53 inhibits its ubiquitination by Mdm2. J Biol Chem. 2002;277:50607–50611. doi: 10.1074/jbc.C200578200. [DOI] [PubMed] [Google Scholar]
- 23.Knights CD, Catania J, Giovanni SD, Muratoglu S, Perez R, Swartzbeck A, Quong AA, Zhang X, Beerman T, Pestell RG, Avantaggiati ML. Distinct p53 acetylation cassettes differentially influence gene-expression patterns and cell fate. J Cell Biol. 2006;173:533–544. doi: 10.1083/jcb.200512059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. Acetylation is indispensable for p53 activation. Cell. 2008;133:612–626. doi: 10.1016/j.cell.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi J, Donehower LA. p53 in embryonic development: maintaining a fine balance. Cell Mol Life Sci. 1999;55:38–47. doi: 10.1007/s000180050268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rogel A, Popliker M, Webb CG, Oren M. p53 cellular tumor antigen: analysis of mRNA levels in normal adult tissues, embryos, and tumors. Mol Cell Biol. 1985;5:2851–2855. doi: 10.1128/mcb.5.10.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Louis JM, McFarland VW, May P, Mora PT. The phosphoprotein p53 is down-regulated post-transcriptionally during embryogenesis in vertebrates. Biochim Biophys Acta. 1988;950:395–402. doi: 10.1016/0167-4781(88)90136-4. [DOI] [PubMed] [Google Scholar]
- 28.Schmid P, Lorenz A, Hameister H, Montenarh M. Expression of p53 during mouse embryogenesis. Development. 1991;113:857–865. doi: 10.1242/dev.113.3.857. [DOI] [PubMed] [Google Scholar]
- 29.Almog N, Rotter V. Involvement of p53 in cell differentiation and development. Biochim Biophys Acta. 1997;1333:F1–F27. doi: 10.1016/s0304-419x(97)00012-7. [DOI] [PubMed] [Google Scholar]
- 30.Saifudeen Z, Dipp S, El-Dahr SS. A role for p53 in terminal epithelial cell differentiation. J Clin Invest. 2002;109:1021–1030. doi: 10.1172/JCI13972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tamir Y, Bengal E. p53 protein is activated during muscle differentiation and participates with MyoD in the transcription of muscle creatine kinase gene. Oncogene. 1998;17:347–356. doi: 10.1038/sj.onc.1201929. [DOI] [PubMed] [Google Scholar]
- 32.Porrello A, Cerone MA, Coen S, Gurtner A, Fontemaggi G, Cimino L, Piaggio G, Sacchi A, Soddu S. p53 regulates myogenesis by triggering the differentiation activity of pRb. J Cell Biol. 2000;151:1295–1304. doi: 10.1083/jcb.151.6.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saifudeen Z, Dipp S, Fan H, El-Dahr SS. Combinatorial control of the bradykinin B2 receptor promoter by p53, CREB, KLF-4, and CBP: implications for terminal nephron differentiation. Am J Physiol Renal Physiol. 2005;288:F899–F909. doi: 10.1152/ajprenal.00370.2004. [DOI] [PubMed] [Google Scholar]
- 34.Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Jr, Butel JS, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 35.Sah VP, Attardi LD, Mulligan GJ, Williams BO, Bronson RT, Jacks T. A subset of p53-deficient embryos exhibit exencephaly. Nat Genet. 1995;10:175–180. doi: 10.1038/ng0695-175. [DOI] [PubMed] [Google Scholar]
- 36.Cordenonsi M, Dupont S, Maretto S, Insinga A, Imbriano C, Piccolo S. Links between tumor suppressors: p53 is required for TGF-beta gene responses by cooperating with Smads. Cell. 2003;113:301–314. doi: 10.1016/s0092-8674(03)00308-8. [DOI] [PubMed] [Google Scholar]
- 37.Dey DC, Bronson RP, Dahl J, Carroll JP, Benjamin TL. Accelerated development of polyoma tumors and embryonic lethality: different effects of p53 loss on related mouse backgrounds. Cell Growth Differ. 2000;11:231–237. [PubMed] [Google Scholar]
- 38.Dumble ML, Knight B, Quail EA, Yeoh GC. Hepatoblast-like cells populate the adult p53 knockout mouse liver: evidence for a hyperproliferative maturation-arrested stem cell compartment. Cell Growth Differ. 2001;12:223–231. [PubMed] [Google Scholar]
- 39.Lee KC, Crowe AJ, Barton MC. p53-mediated repression of alpha-fetoprotein gene expression by specific DNA binding. Mol Cell Biol. 1999;19:1279–1288. doi: 10.1128/mcb.19.2.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reichel MB, Ali RR, D'Esposito F, Clarke AR, Luthert PJ, Bhattacharya SS, Hunt DM. High frequency of persistent hyperplastic primary vitreous and cataracts in p53-deficient mice. Cell Death Differ. 1998;5:156–162. doi: 10.1038/sj.cdd.4400326. [DOI] [PubMed] [Google Scholar]
- 41.Rotter V, Schwartz D, Almon E, Goldfinger N, Kapon A, Meshorer A, Donehower LA, Levine AJ. Mice with reduced levels of p53 protein exhibit the testicular giant-cell degenerative syndrome. Proc Natl Acad Sci U S A. 1993;90:9075–9079. doi: 10.1073/pnas.90.19.9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoever M, Clement JH, Wedlich D, Montenarh M, Knochel W. Overexpression of wild-type p53 interferes with normal development in Xenopus laevis embryos. Oncogene. 1994;9:109–120. [PubMed] [Google Scholar]
- 43.Allemand I, Anglo A, Jeantet AY, Cerutti I, May E. Testicular wild-type p53 expression in transgenic mice induces spermiogenesis alterations ranging from differentiation defects to apoptosis. Oncogene. 1999;18:6521–6530. doi: 10.1038/sj.onc.1203052. [DOI] [PubMed] [Google Scholar]
- 44.Hilliard S, Aboudehen K, Yao X, El-Dahr SS. Tight regulation of p53 activity by Mdm2 is required for ureteric bud growth and branching. Dev Biol. 2011;353:354–366. doi: 10.1016/j.ydbio.2011.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Godley LA, Kopp JB, Eckhaus M, Paglino JJ, Owens J, Varmus HE. Wild-type p53 transgenic mice exhibit altered differentiation of the ureteric bud and possess small kidneys. Genes Dev. 1996;10:836–850. doi: 10.1101/gad.10.7.836. [DOI] [PubMed] [Google Scholar]
- 46.Jones SN, Roe AE, Donehower LA, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378:206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 47.Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 48.Gao Y, Ferguson DO, Xie W, Manis JP, Sekiguchi J, Frank KM, Chaudhuri J, Horner J, DePinho RA, Alt FW. Interplay of p53 and DNA-repair protein XRCC4 in tumorigenesis, genomic stability and development. Nature. 2000;404:897–900. doi: 10.1038/35009138. [DOI] [PubMed] [Google Scholar]
- 49.Lengner CJ, Steinman HA, Gagnon J, Smith TW, Henderson JE, Kream BE, Stein GS, Lian JB, Jones SN. Osteoblast differentiation and skeletal development are regulated by Mdm2-p53 signaling. J Cell Biol. 2006;172:909–921. doi: 10.1083/jcb.200508130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mendrysa SM, McElwee MK, Michalowski J, O'Leary KA, Young KM, Perry ME. mdm2 Is critical for inhibition of p53 during lymphopoiesis and the response to ionizing irradiation. Mol Cell Biol. 2003;23:462–472. doi: 10.1128/MCB.23.2.462-473.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bond GL, Hu W, Bond EE, Robins H, Lutzker SG, Arva NC, Bargonetti J, Bartel F, Taubert H, Wuerl P, Onel K, Yip L, Hwang SJ, Strong LC, Lozano G, Levine AJ. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell. 2004;119:591–602. doi: 10.1016/j.cell.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 52.Saifudeen Z, Dipp S, Stefkova J, Yao X, Lookabaugh S, El-Dahr SS. p53 regulates metanephric development. J Am Soc Nephrol. 2009;20:2328–2337. doi: 10.1681/ASN.2008121224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saifudeen Z, Marks J, Du H, El-Dahr SS. Spatial repression of PCNA by p53 during kidney development. Am J Physiol Renal Physiol. 2002;283:F727–F733. doi: 10.1152/ajprenal.00114.2002. [DOI] [PubMed] [Google Scholar]
- 54.Yang A, Walker N, Bronson R, Kaghad M, Oosterwegel M, Bonnin J, Vagner C, Bonnet H, Dikkes P, Sharpe A, McKeon F, Caput D. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature. 2000;404:99–103. doi: 10.1038/35003607. [DOI] [PubMed] [Google Scholar]
- 55.Saifudeen Z, Diavolitsis V, Stefkova J, Dipp S, Fan H, El-Dahr SS. Spatiotemporal switch from DeltaNp73 to TAp73 isoforms during nephrogenesis: impact on differentiation gene expression. J Biol Chem. 2005;280:23094–23102. doi: 10.1074/jbc.M414575200. [DOI] [PubMed] [Google Scholar]