Abstract
Background. Staphylococcus aureus is the most common cause of endovascular infections. The staphylococcal accessory regulator A locus (sarA) is a major virulence determinant that may potentially impact methicillin-resistant S. aureus (MRSA) persistence in such infections via its influence on biofilm formation.
Methods. Two healthcare-associated MRSA isolates from patients with persistent bacteremia and 2 prototypical community-acquired MRSA strains, as well as their respective isogenic sarA mutants, were studied for in vitro biofilm formation, fibronectin-binding capacity, autolysis, and protease and nuclease activities. These assays were done in the presence or absence of sub–minimum inhibitory concentrations (MICs) of vancomycin. In addition, these strain pairs were compared for intrinsic virulence and responses to vancomycin therapy in experimental infective endocarditis, a prototypical biofilm model.
Results. All sarA mutants displayed significantly reduced biofilm formation and binding to fibronectin but increased protease production in vitro, compared with their respective parental strains. Interestingly, exposure to sub-MICs of vancomycin significantly promoted biofilm formation and fibronectin-binding in parental strains but not in sarA mutants. In addition, all sarA mutants became exquisitely susceptible to vancomycin therapy, compared with their respective parental strains, in the infective endocarditis model.
Conclusions. These observations suggest that sarA activation is important in persistent MRSA endovascular infection, potentially in the setting of biofilm formation.
Keywords: sarA, biofilm formation, MRSA endocarditis
(See the editorial commentary by Herrmann on pages 1153–5.)
Staphylococcus aureus is the most common cause of endovascular infections, including infective endocarditis syndromes, which are associated with high morbidity and mortality [1]. The increase in prevalence of methicillin-resistant S. aureus (MRSA) strains in such infections and the relatively high rates of vancomycin clinical failures in these syndromes caused by MRSA strains whose vancomycin minimum inhibitory concentrations (MICs) fall within the susceptible range (ie, ≤2 μg/mL) have further complicated the management of these patients [2]. In addition, MRSA is a predominant cause of healthcare-associated (HA) infections [3]. However, life-threatening infections caused by community-acquired MRSA (CA-MRSA) in otherwise healthy people outside of healthcare settings have recently reached epidemic proportions globally [4].
The pathogenesis of S. aureus is complex and involves the coordinate expression of multiple gene products, including surface adhesins, exoproteins, and toxins. Of note, many staphylococcal virulence factors are controlled by a major regulatory locus called staphylococcal accessory regulator A (sarA) [5]. The sarA locus consists of 3 overlapping transcripts, driven by 3 distinct promoters: P1, P3, and P2. These DNA-binding proteins bind to AT-rich inverted repeat or palindromic sequences on respective target promoters to control the expression of multiple genes [5]. Importantly, laboratory-derived sarA mutants have exhibited diminished virulence in infection models, including infective endocarditis, osteomyelitis, and arthritis [6, 7].
Biofilm formation plays a critical role in the development of MRSA endovascular infections [8]. Treatment of these infections has become challenging because organisms within the biofilm are significantly more resistant to antimicrobial agents, as well as to clearance by the host defense system [9]. In addition, recent studies suggest that sublethal doses of selected antibiotics (eg, vancomycin) can induce biofilm formation in MRSA strains [10]. In this regard, SarA is an important positive regulator of biofilm development, in part because of its repressive activity on protease production [11–13]. To our knowledge, this study is the first to examine the impact of vancomycin on sarA-mediated biofilm formation in MRSA in vitro, as well as in vivo, in terms of intrinsic virulence and treatment outcomes in endovascular infections.
MATERIALS AND METHODS
MRSA Strains
Two HA-MRSA isolates, selected from a multinational clinical trial, were collected from patients demonstrating MRSA-positive blood cultures for ≥14 days despite receiving antibiotics to which the isolates were susceptible in vitro (Table 1) [16, 18]. In addition, 2 prototypical CA-MRSA strains, MW2 (USA400, reported to cause fatal infections in children [19]) and JE2 (LAC, USA300 derivative that was cured of its three plasmids, obtained from the Network on Antimicrobial Resistance in Staphylococcus aureus) were also chosen for this study. A site-specific mutant was obtained by transducing the sarA::Tn917LTV1 mutation from strain ALC637 (Newman, sarA:Tn917LTV1) [14] into strain 324-136, using phage 80α (erythromycinr). A sarA deletion in strains JE2 and 300-169 was achieved by transducing the sarA::kan mutation from ALC2543 (COL with a sarA::kan mutation) [15], using phage 85 (kanamycinr). The study MRSA strains were routinely grown in tryptic soy broth or on tryptic soy agar.
Table 1.
Staphylococcus aureus Strains Used in This Study
| Strains | Relevant Characteristic(s) | Reference |
|---|---|---|
| ALC637 | Newman sarA:Tn917LTV1, ermR | [14] |
| ALC2543 | COL sarA::kan | [15] |
| 300-169 | HA-MRSA (patient with persistent bacteremia) | [16] |
| 300-169 ΔsarA | 300-169 sarA::kan | This study |
| 324-136 | HA-MRSA (patient with persistent bacteremia) | [16] |
| 324-136 ΔsarA | 324-136 sarA::Tn917LTV1 | This study |
| MW2 | CA-MRSA, USA400 | [15] |
| ALC5415 | MW2 sarA::kan | [15] |
| JE2 | CA-MRSA, LAC, USA300 | [17] |
| JE2 ΔsarA | JE2 sarA::kan | This study |
Abbreviations: CA-MRSA, community-acquired methicillin-resistant S. aureus; HA-MRSA, healthcare-associated methicillin-resistant S. aureus.
Determination of Vancomycin MICs
MICs for vancomycin were determined by the standard broth microdilution method, as recommended by the Clinical and Laboratory Standards Institute [20].
In Vitro Time-Kill Curve of Vancomycin
The bactericidal activities of vancomycin (range, 1–5 × MICs) were compared between the parental strains and their respective sarA mutants by use of killing curve analyses [21].
Population Analyses Assay
Vancomycin population analyses were performed by standard protocols [22]. The range of vancomycin concentrations tested was 0.125–16 μg/mL to encompass sublethal-to-lethal drug levels, using an initial inoculum of approximately 109 colony-forming units (CFU)/mL. Population analysis curves were compared between each strain pair [8].
Biofilm Formation
Biofilm formation of the study MRSA strains was performed under static conditions [8, 23]. Briefly, MRSA cells from fresh culture plates were adjusted to a density of 0.5 McFarland standard and diluted 1:100 into brain heart infusion broth supplemented with 0.5% glucose; 200 μL of this suspension was transferred to 96-well tissue culture plates and incubated for 18 hours at 37°C. After incubation, the wells were washed, air dried, and stained with safranin (0.1% in distilled water). The adhering dye was dissolved in 30% acetic acid, and absorption was measured at OD490nm to quantify biofilm formation [8, 23].
Numerous investigations have shown that sub-MICs of some antibiotics can induce bacterial biofilm formation [8, 10, 24]. In this regard, we have recently reported that exposure to sub-MICs of vancomycin can promote MRSA biofilm formation, a process that may have clinical significance [8]. Therefore, the above static biofilm formation assay was performed in parallel with exposure to 0.5 times the MIC of vancomycin.
Biofilm Stability
Bacterial biofilms consist of carbohydrates, proteins, and extracellular DNA in variable proportions [9, 23]. Distinct biofilm chemical compositions are felt to contribute functionally and structurally to the organization of biofilm [9]. Therefore, differences in the stability of MRSA biofilm as a function of specific biochemical components were separately assessed in the presence of carbohydrate, protein, or DNA dispersal agents [23]. The supernatants of 18-hours-old biofilms generated by the study MRSA strains were replaced by fresh medium supplemented with either 10 mM sodium metaperiodate, 100 μg/mL proteinase K, or 140 U/mL RNase-free DNase I to assess relative carbohydrate, protein, and DNA content, respectively [23]. All incubations were performed for 2 hours at 37°C. Media without the above supplements served as respective controls. After treatment, the biofilms were quantified as described above.
Autolysis Assay
Standard Triton X-100–induced autolysis was performed as described elsewhere [15]. In addition, it has been reported that increased staphylococcal autolysis mediated by exposure to sub-MICs of vancomycin exposure can augment biofilm formation [10]. Thus, the MRSA strains were also examined for vancomycin-induced lysis. In brief, overnight cultured strains were adjusted to an OD580nm of 0.7, washed, exposed to 50 mM Tris-HCl (pH 7.2) containing either 0.05% Triton X-100 or 0.5 times the MIC of vancomycin, and incubated at 30°C with agitation (200 rpm). Staphylococcal lysis was measured during 24 hours of incubation by determining the changes in OD580nm.
Adherence to Fibronectin
For these assays, 6-well tissue culture plates were coated with purified human fibronectin (50 μg/mL) for 18 hours and then treated with 3% bovine serum albumin for 3 hours to prevent nonspecific adhesion [25]. Overnight-cultured MRSA cells grown in the presence or absence of 0.5 times the MIC of vancomycin were added to the plates (5×103CFU) and incubated for 1 hour [25]. Wells were then washed, and tryptic soy agar were added to each well, and overnight incubation at 37°C was performed. Adherence was expressed as the percentage (±SD) of the initial inoculum bound.
Protease and Nuclease Activity
MRSA extracellular proteases and nucleases are essential modulators of biofilms, being involved in the degradation of the biofilm matrix; they may also play a critical role in the biofilm-deficient phenotype in the sarA mutants [9, 26, 27]. Therefore, we tested overall extracellular protease and nuclease production of the study strains, using 1% caseinate agar (for proteolysis activity) and DNase agar (for nuclease activity) plates in the presence or absence of 0.5 times the MIC of vancomycin [27, 28].
Rabbit Model of MRSA Infective Endocarditis
A well-characterized rabbit model of catheter-induced aortic valve infective endocarditis with intravenous injection of approximately 105 CFU of each MRSA strain was used [16]. Rabbits were maintained in accordance with the American Association for Accreditation of Laboratory Animal Care criteria. The Institutional Animal Care and Use Committee of the Los Angeles Biomedical Research Institute at Harbor-UCLA Medical Center approved all animal study protocols. Twenty-four hours after infection, animals were randomized to receive no therapy (controls) or vancomycin (15 mg/kg or 7.5 mg/kg) intravenously twice daily for 3 days for HA- and CA-MRSA strain sets, respectively. Twenty-four hours after the last vancomycin dose, animals were euthanized, and their cardiac vegetations, kidneys, and spleen were removed and quantitatively cultured [16]. MRSA counts in target tissue are given as the mean log10 CFU/g of tissue (±SD).
Assessment of sarA Transcriptions In Vitro and Within Cardiac Vegetations In Vivo by Real-Time Quantitative PCR (qPCR)
For sarA transcriptions in vitro, RNA was isolated from cultures that were grown for 24 hours with or without exposure to 0.5 times the MIC of vancomycin for comparison with data generated in vivo within cardiac vegetation samples collected 24 hours after infection (see below) [16]. Briefly, 2 μg of DNase-treated RNA was transcribed into complementary DNA [16]. Real-time qPCR was performed using an ABI Prism 7000 instrument (Applied Biosystems) and the SYBR green PCR master kit (Applied Biosystems). The primers used to amplify sarA were qRT-sarA-F (5′-TCTTGTTAATGCACAACAACGTAA-3′) and qRT-sarA-R (5′-TGTTTGCTTCAGTGATTCGTTT-3′). gyrB was used to normalize for transcript quantification [16]. Relative quantification was calculated by the ΔΔCT method [29]. PCR experiments were performed using 2 biological replicates, each tested in triplicate.
For quantifying sarA transcripts within cardiac vegetations, catheterized animals were infected with each parental strain as described above. Twenty-four hours after infection, animals were randomized to receive no therapy or vancomycin as described above. Vancomycin was given for 1 day only (to ensure adequate numbers of MRSA cells within cardiac vegetations for RNA extractions). Twenty-four hours after receipt of the last vancomycin dose, animals were euthanized, and their cardiac vegetations were quick-frozen in liquid nitrogen. Total RNA was isolated using Tri reagent (Ambion) according to the manufacturer's instructions, including a 0.635 cm ceramic sphere in the cell disruption step [16]. Samples were then processed as described above. Real-time qPCR analysis for sarA expression was performed on at least 3 different animals for each group.
RESULTS
Vancomycin MICs
Vancomycin MICs were 0.5 μg/mL for the 2 HA-MRSA strains and 1.0 and 2.0 μg/mL for CA-MRSA strains MW2 and JE2, respectively. In addition, no differences were detected in vancomycin MICs between the parental strains and their respective sarA mutant strains.
In Vitro Time-Kill Curve of Vancomycin
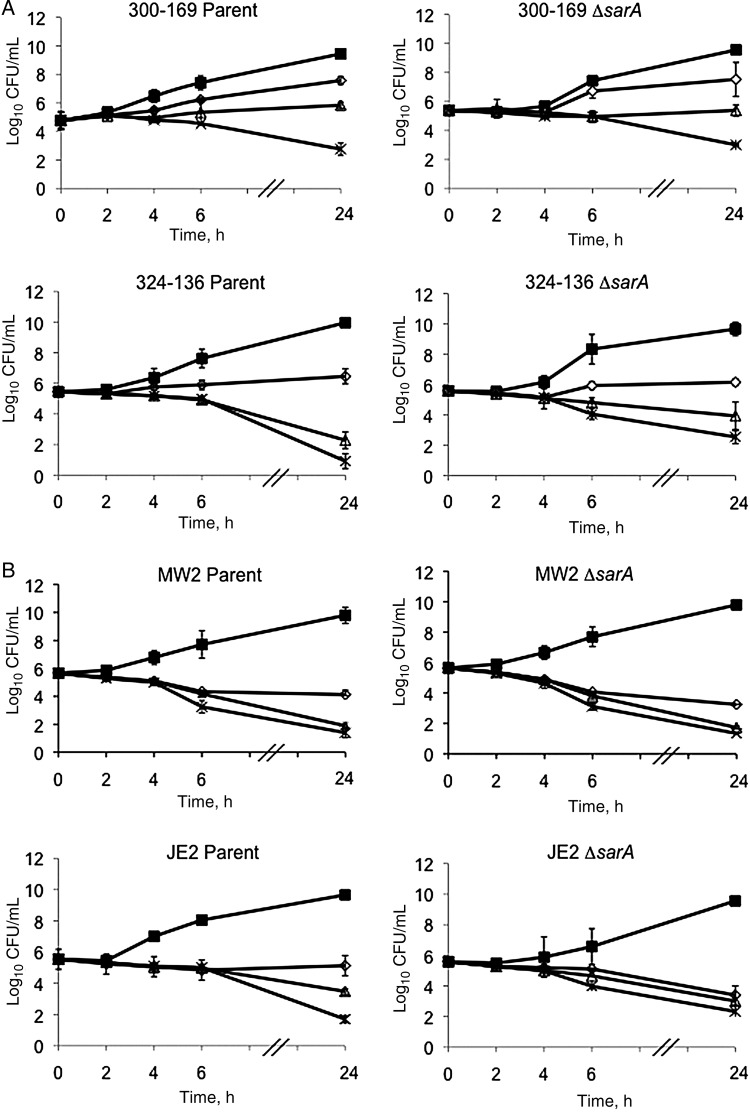
The results of the in vitro time-kill curve analysis of vancomycin are shown in Figure 1. No significant differences in vancomycin bactericidal effects were observed between parental and sarA mutant strains. In addition, no strains exhibited in vitro tolerance to exposure to 5 times the MIC of vancomycin (Figure 1).
Figure 1.
In vitro time-kill curves of vancomycin against healthcare-associated methicillin-resistant Staphylococcus aureus (HA-MRSA; 300–169 and 324–136; A) and community-acquired MRSA (CA-MRSA; MW2 and JE2; B) strain sets. ▪, control; ⋄, 1 times the minimum inhibitory concentration (MIC) of vancomycin; Δ, 2 times the MIC of vancomycin; ✱, 5 times the MIC of vancomycin.
Population Analyses
None of the study isolates revealed vancomycin-heteroresistant subpopulations (data not shown). In addition, there were no population curve shifts in comparing the parent and sarA mutant strains within each strain pair.
Biofilm Formation Under Static Conditions
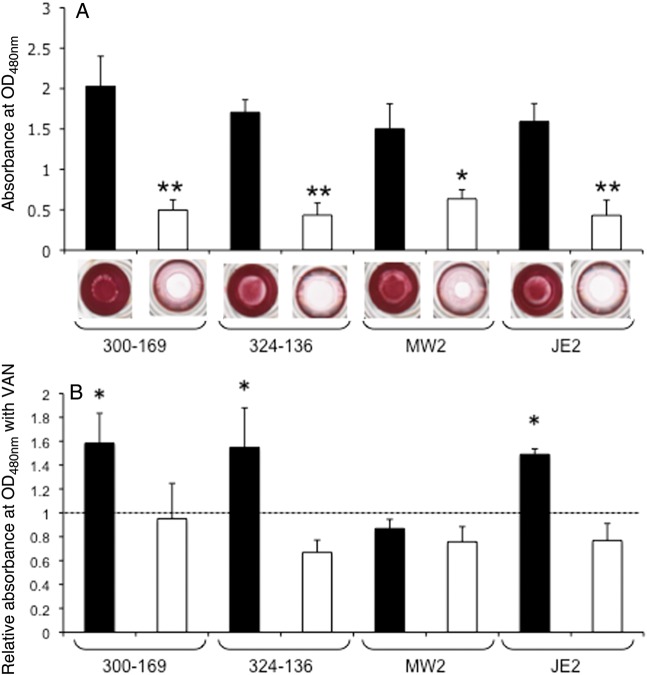
Consistent with previous reports [11, 12], all 4 sarA mutation were defective in biofilm formation as compared to their respective parental strains (Figure 2A). Interestingly, sublethal levels of vancomycin (ie, 0.5 times the MIC) significantly promoted biofilm formation in both HA-MRSA parental strains and in CA-MRSA parental strain JE2, compared with their respective parental controls without vancomycin exposure (Figure 2B). However, vancomycin exposure did not significantly change the biofilm formation profile in the MW2 parental strain (Figure 2B). Importantly, sublethal vancomycin exposure did not induce biofilm formation in any of the sarA mutants.
Figure 2.
Impact of sarA on biofilm formation. A, Biofilm formation of the wild types and their isogenic sarA mutants was quantified using a microtiter plate assay by solubilizing the safranin in 30% acetic acid, and absorption was measured at 490 nm. Results are shown as the mean of the A490nm ± SD from 3 independent experiments, each of which was done in triplicate. *P<.005 and **P<.0005 for the sarA mutants, compared with their respective parental strain. B, Effect of 0.5 times the MIC of vancomycin (VAN) on biofilm formation. Biofilm formation without VAN exposure was defined as being A490nm 1.0 and is represented by the dashed line. ▪, parental; □, sarA mutant. *P< .05, compared with control without VAN exposure.
Biofilm Stability
Treatment with sodium metaperiodate, proteinase K, or DNase I led to a similar significant reduction of biofilm formation in the 2 HA-MRSA parental strains, compared with their respective controls without exposure to any dispersal agent (Supplementary Figure 1). However, treatment with sodium metaperiodate did not cause a significant reduction of biofilm formation in 2 CA-MRSA parental strains, compared with their respective controls without sodium metaperiodate exposure, suggesting that biofilm formed by these CA-MRSA strains might have relatively lower carbohydrate content (compared with levels of protein and DNA; Supplementary Figure 1). For all sarA mutants, treatment with sodium metaperiodate had little effect on biofilm stability, whereas there was significant biofilm dispersion by treatment with proteinase K and DNase in 2 of the 4 sarA mutants, suggesting that these 2 sarA mutant strains have less carbohydrate content, compared with protein and DNA content (Supplementary Figure 1).
Cell Autolysis
No differences in autolysis profiles were observed between parental and sarA mutant strains in the presence of either Triton X-100 or 0.5 times the MIC of vancomycin (data not shown).
Adherence to Fibronectin
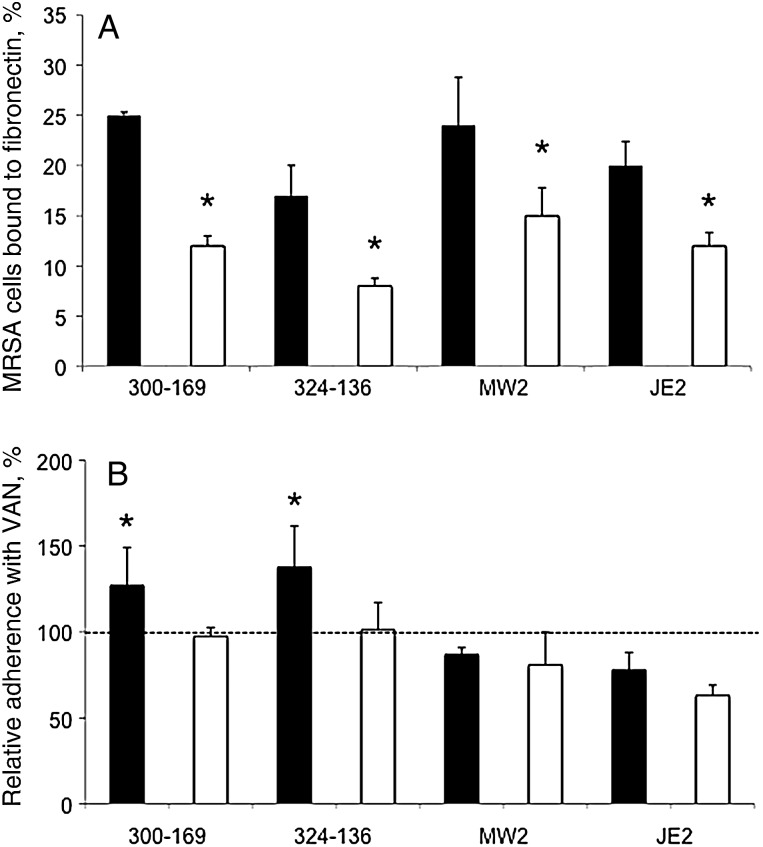
As anticipated from prior studies [25, 30], inactivation of sarA led to significantly reduced fibronectin binding in mutant strains, compared with their respective parental strain (P< .05; Figure 3A). Interestingly, in the presence of 0.5 times the MIC of vancomycin, 2 HA-MRSA parental strains showed significantly increased binding to fibronectin, compared with their respective controls, whereas the 2 CA-MRSA strains exhibited decreased fibronectin binding (although this did not reach statistical significance; Figure 3B). For all sarA mutants, fibronectin binding did not increase with vancomycin exposure (Figure 3B).
Figure 3.
Adherence of the study methicillin-resistant Staphylococcus aureus (MRSA) strain sets to immobilized human fibronectin in the absence (A) or presence (B) of 0.5 times the minimum inhibitory concentration of vancomycin (VAN). Controls without VAN exposure set up 100% adherence to fibronectin (B). ▪, parental strain; □, sarA mutant. *P< .05, compared with their respective parental strain (A) or with controls without VAN exposure (B).
Protease and Nuclease Activity
In accordance with previous studies showing that sarA negatively regulates protease and nuclease gene expression [12], the sarA mutants demonstrated increases in proteases and nuclease activity in all study strains except strain JE2 (in which nuclease activity did not change; Figure 4). Of note, although sub-MICs of vancomycin had a positive impact on biofilm formation as documented above, there were no differences in either protease or nuclease production induced by vancomycin (Figure 4).
Figure 4.
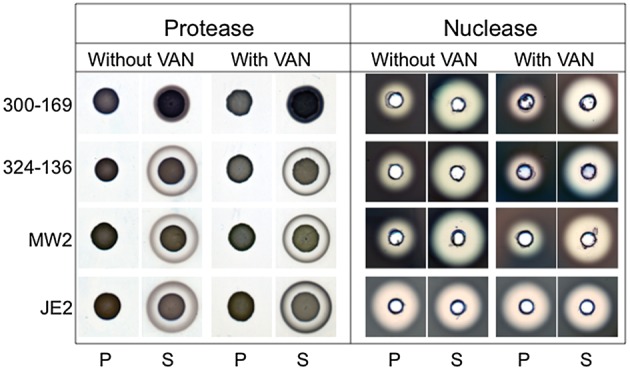
Production of extracellular protease and nuclease by the study strain sets in the absence or presence of 0.5 times the minimum inhibitory concentration of vancomycin (VAN). Results were assessed after overnight incubation of the study strains on caseinate and DNase agar plates for protease and nuclease, respectively. P, parental strain; S, sarA mutant strain.
Experimental Infective Endocarditis
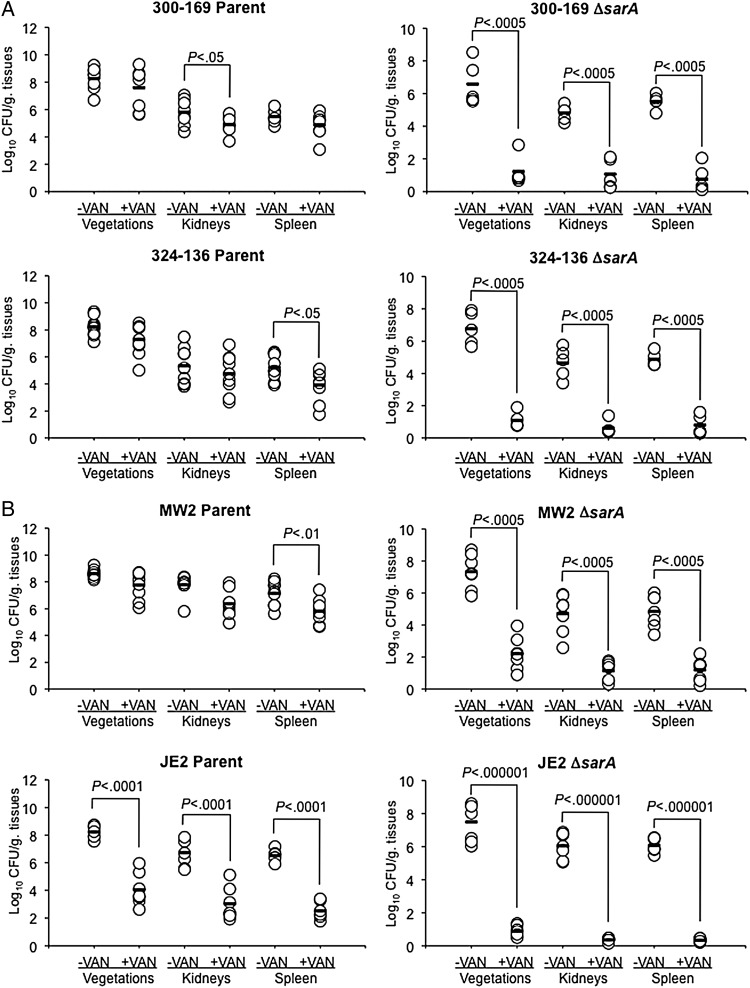
At the 105 CFU challenge inoculum, 3 of the 4 sarA mutants (excluding JE2) exhibited reduced virulence, compared with their parental strains, in terms of MRSA densities achieved within cardiac vegetations (but not kidneys or spleen; P< .05; Figure 5). Of note, vancomycin treatment resulted in dramatic and significant reductions of MRSA counts in all target tissues in animals infected with the sarA mutant strains (3.8–6.6 log10 CFU/g tissue reduction; P< .0005, compared with data for untreated sarA mutant controls; Figure 5). In addition, vancomycin treatment resulted in approximately 80% culture negativity for all target tissues among animals infected with sarA mutants in 300-169 and 324-136 strain backgrounds; also 18% and 100% of organ cultures were sterile in animals infected with sarA mutant in MW2 and JE2 backgrounds, respectively. In contrast, rabbits infected with 3 of the 4 parental strains (the 2 HA-MRSA strains and the MW2 strain, but not the JE2 strain) did not respond to vancomycin treatment, with residual target tissue MRSA densities in vancomycin-treated animals being similar to those in their respective untreated control groups (Figure 5).
Figure 5.
Methicillin-resistant Staphylococcus aureus (MRSA) densities in target tissues in the infective endocarditis model, with or without vancomycin (VAN) treatment (15 mg/kg and 7.5 mg/kg intravenously twice daily for 3 days for healthcare-associated MRSA [A] and community-acquired MRSA [B] strains, respectively). Each dot represents 1 rabbit. Horizontal black bars indicate mean values of observations.
sarA Transcriptions In Vitro and Within Cardiac Vegetations
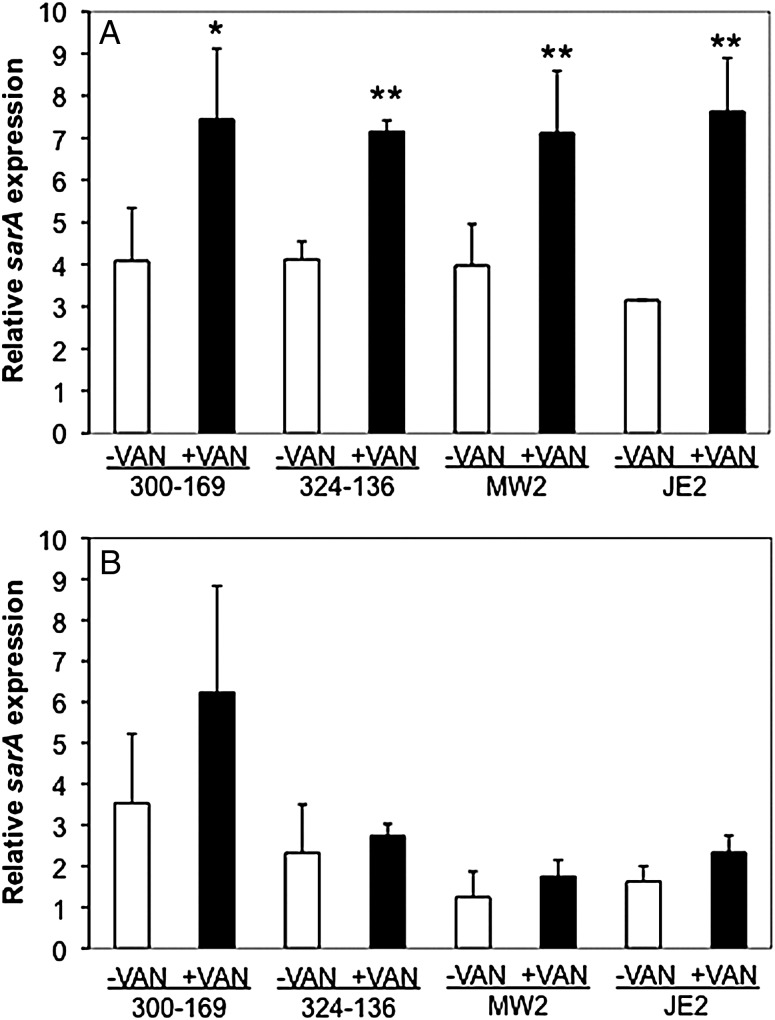
On exposure to 0.5 times the MIC of vancomycin in vitro, all parental strains exhibited significantly higher sarA transcript levels than those in their respective controls in the absence of vancomycin exposure (Figure 6A). For sarA expression within cardiac vegetations, higher sarA expression profiles were observed in the vancomycin treatment groups, compared with their respective controls without vancomycin therapy (Figure 6B). Interestingly, higher sarA expression was detected in HA-MRSA versus CA-MRSA parental strains without vancomycin treatment. However, these differences did not reach statistical significance (Figure 6B).
Figure 6.
Expression of sarA in all study methicillin-resistant Staphylococcus aureus (MRSA), parental strain with (+) or without (−) exposure in vitro to 0.5 times the minimum inhibitory concentration of vancomycin (VAN) exposure, after 24 hours (A) and within infected cardiac vegetations, with or without exposure to VAN, in the infective endocarditis model 24 hours after infection (B). Data were obtained by real-time quantitative polymerase chain reaction. Relative transcript levels of sarA represent the mean (+SD) of 2 biological replicates in vitro and of at least 3 animals per group in vivo (fold-changes vs gyrB). *P < .05 and **P < .005, compared with their respective controls without VAN exposure in vitro.
DISCUSSION
Biofilm formation is considered to be a major virulence factor in many S. aureus syndromes, including infective endocarditis [9, 26]. Biofilm formation not only facilitates bacterial colonization of host tissues, but also fosters resistance to bacterial clearance mediated by antimicrobial agents and by host immune responses [9, 26]. In addition, biofilms can serve as chronic foci of infection for metastatic spread of bacteria and release of toxins into the bloodstream, resulting in significant morbidity and mortality [9]. Therefore, biofilm formation has become a key target for the development of novel therapeutic tools against these life-threatening infections. In this study, we investigated the modulation of a central regulatory element that controls the production of many S. aureus virulence factors (including biofilm), sarA, as means of enhancing antimicrobial efficacy in a prototypical MRSA biofilm infection model (infective endocarditis).
In the current investigation, consistent with other reports, we observed that sarA mutants formed significantly less biofilm in vitro in all study strains, compared with their respective parental strains [11, 12]. It is well known that sarA has a global effect on many S. aureus virulence genes that seem to play a role in biofilm formation (eg, multiple extracellular proteases, nuclease, and fibronectin-binding proteins [FnBPs]) [31]. For instance, recent studies and our own current data show that the overall production of extracellular proteases is increased in sarA mutants, which may contribute to the reduced biofilm phenotype in such strains [27, 32, 33]. In addition, there is growing evidence that the release of extracellular DNA from bacterial cells undergoing autolysis during programmed cell death contributes to biofilm development [34, 35]. Therefore, an increased production of extracellular nuclease may contribute to the biofilm-deficient phenotype of sarA mutants [35, 36]. Moreover, nuclease (nuc) mutants form a very thick biofilm containing increased levels of matrix-associated extracellular DNA [34]. In this regard, in the current study, 3 of our 4 sarA mutants exhibited increased extracellular nuclease production, again correlating with reduced biofilm formation. Taken together, these results suggest that the regulatory role of sarA in repressing the production of extracellular enzymes (including proteases and nucleases) is one key factor leading to biofilm formation and stability [27].
S. aureus biofilm matrices consist of proteins, DNA, and polysaccharide (also called the polysaccharide intercellular adhesion [PIA] or poly-N-acetylglucosamine material) [9, 23]. Recently, it has become evident that the presence of PIA is not essential for biofilm development in many MRSA strains [30]. Interestingly, in these PIA-independent biofilms, FnBPs appear to substitute for PIA in driving biofilm formation [30]. In addition, this FnBP-mediated biofilm seems to be particularly frequent among highly virulent MRSA isolates, suggesting the importance of PIA-independent biofilm formation in such strains [30]. SarA is a positive regulator of FnBP production and subsequent biofilm formation in 2 complementary ways: (1) by enhancement of fnb gene expression and (2) by repression of extracellular proteases, which normally cleave and remodel surface adhesins such as FnBP [12, 13]. Thus, we tested the fibronectin-binding capacities of our study strains. Our results, in agreement with other studies [25, 37], found that all sarA mutants had significantly decreased capacity to bind to fibronectin. This reduced fibronectin-binding phenotype was correlated with increased protease production observed in sarA mutants, ostensibly contributing to reduced bacterial colonization at the early steps of endovascular infection.
Several groups have shown that sub-MICs of selected antibiotics, including vancomycin, can promote S. aureus biofilm formation in vitro [10, 38, 39]. In the current studies, we applied this concept to characterize the impact of sub-MICs of vancomycin on sarA-mediated biofilm formation in a model of persistent endovascular infection due to HA- and CA-MRSA strains. Vancomycin was chosen in this investigation because it remains a clinical mainstay as an anti-MRSA agent. We confirmed that sub-MICs of vancomycin significantly promoted biofilm formation in 3 of 4 MRSA parental strains (excluding MW2). In contrast, all sarA mutants demonstrated reduced biofilm formation in the presence or absence of sub-MICs of vancomycin in vitro. In addition, we demonstrated that sub-MICs of vancomycin significantly increased sarA gene expression in all study parental strains studied in vitro, compared with their respective controls without vancomycin exposure. This outcome was mirrored in vivo, because vancomycin treatment resulted a trend toward higher MRSA sarA expression within infective endocarditis cardiac vegetations, compared with findings for respective controls without vancomycin therapy. These phenotypic and genotypic differences in the impact of sub-MICs of vancomycin on biofilm formation and sarA expression paralleled in vivo outcomes. Thus, animals infected with parental MRSA strains were resistant to vancomycin treatment, whereas animals infected with sarA mutants were exquisitely susceptible to vancomycin therapy in the infective endocarditis model.
We recognize that additional mechanisms likely contribute to the differences in vancomycin-associated outcomes in vivo as noted above. For example, vancomycin has limited and slow penetration into biofilms [40, 41]. Therefore, one intriguing possibility is that sarA mutant strains form significantly thinner and/or less well-structured biofilms in vivo, compared with their respective parental strains. In turn, this latter phenotype may conceivably allow greater penetration of vancomycin into such defective biofilms. Furthermore, Hsu et al [10] reported that sublethal doses of vancomycin can induce more-robust biofilm formation through an enhanced autolysis- and extracellular DNA–dependent release in S. aureus. In contrast to their findings, however, the present studies were unable to detect any impact of sub-MICs of vancomycin on either autolysis or nuclease or protease production, suggesting these latter mechanisms were not in play in the current study strains.
Last, we noted that mutations in sarA had a definable, albeit modest effect on innate MRSA virulence, being observed only with in cardiac vegetations. This impact of sarA on infective endocarditis pathogenesis has been seen in prior studies in this model, using other S. aureus strains [6, 25, 42]. Of major importance, vancomycin therapy in the experimental infective endocarditis model was able to divulge the ability of sarA to blunt antimicrobial efficacies, presumably via a biofilm-dependent pathway. Interestingly, these in vivo results were observed despite similar vancomycin MICs and in vitro vancomycin killing kinetics between sarA mutants and their respective parental strains. These results suggest that sarA plays a key role in endovascular infections, especially in terms of antimicrobial therapy responsiveness, likely, at least in part, because of the decreased production of extracellular protease, resulting in augmented biofilm formation.
In summary, the present findings are, to our knowledge, the first to reveal the importance of sarA in 2 critical aspects of S. aureus pathogenesis in endovascular infections caused by clinical isolates: biofilm formation and target tissue persistence. Although the mechanism(s) of these phenomena are not entirely defined, these data support the notion that suppression of SarA has therapeutic potential in the important context of biofilm-associated infections due to HA-MRSA and CA-MRSA.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Financial support. This study was supported by the National Institutes of Health (grants R21AI097657 [to Y. Q. X.], RO1AI-39108 [to A. S. B.], and RO1AI91801 [to A. L. C.]) and the Department of Defense (grant W81XWH-12-2-0101 to M. R. Y.).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Fowler VG, Jr., Justice A, Moore C, et al. Risk factors for hematogenous complications of intravascular catheter-associated Staphylococcus aureus bacteremia. Clin Infect Dis. 2005;40:695–703. doi: 10.1086/427806. [DOI] [PubMed] [Google Scholar]
- 2.Rubinstein E. Staphylococcus aureus bacteraemia with known sources. Int J of Antimicrob Agents. 2008;32(Suppl 1):S18–20. doi: 10.1016/j.ijantimicag.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 3.Grundmann H, Aires-de-Sousa M, Boyce J, Tiemersma E. Emergence and resurgence of meticillin-resistant Staphylococcus aureus as a public-health threat. Lancet. 2006;368:874–85. doi: 10.1016/S0140-6736(06)68853-3. [DOI] [PubMed] [Google Scholar]
- 4.Chambers HF. Community-associated MRSA-resistance and virulence converge. N Engl J Med. 2005;352:1485–7. doi: 10.1056/NEJMe058023. [DOI] [PubMed] [Google Scholar]
- 5.Cheung AL, Nishina KA, Trotonda MP, Tamber S. The SarA protein family of Staphylococcus aureus. Int J Biochem Cell Bio. 2008;40:355–61. doi: 10.1016/j.biocel.2007.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiong YQ, Willard J, Yeaman MR, Cheung AL, Bayer AS. Regulation of Staphylococcus aureus alpha-toxin gene (hla) expression by agr, sarA, and sae in vitro and in experimental infective endocarditis. J Infect Dis. 2006;194:1267–75. doi: 10.1086/508210. [DOI] [PubMed] [Google Scholar]
- 7.Blevins JS, Elasri MO, Allmendinger SD, et al. Role of sarA in the pathogenesis of Staphylococcus aureus musculoskeletal infection. Infect Immun. 2003;71:516–23. doi: 10.1128/IAI.71.1.516-523.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abdelhady W, Bayer AS, Seidl K, et al. Reduced vancomycin susceptibility in an in vitro catheter-related biofilm model correlates with poor therapeutic outcomes in experimental endocarditis due to methicillin-resistant Staphylococcus aureus. Antimicrobial Agents Chemother. 2013;57:1447–54. doi: 10.1128/AAC.02073-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Otto M. Staphylococcal biofilms. Curr Top Microbiol Immun. 2008;322:207–28. doi: 10.1007/978-3-540-75418-3_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsu CY, Lin MH, Chen CC, et al. Vancomycin promotes the bacterial autolysis, release of extracellular DNA, and biofilm formation in vancomycin-non-susceptible Staphylococcus aureus. FEMS Immunol Med Microbiol. 2011;63:236–47. doi: 10.1111/j.1574-695X.2011.00846.x. [DOI] [PubMed] [Google Scholar]
- 11.Trotonda MP, Manna AC, Cheung AL, Lasa I, Penades JR. SarA positively controls bap-dependent biofilm formation in Staphylococcus aureus. J Bacteriol. 2005;187:5790–8. doi: 10.1128/JB.187.16.5790-5798.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mrak LN, Zielinska AK, Beenken KE, et al. saeRS and sarA Act Synergistically to Repress Protease Production and Promote Biofilm Formation in Staphylococcus aureus. PLoS One. 2012;7:e38453. doi: 10.1371/journal.pone.0038453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zielinska AK, Beenken KE, Mrak LN, et al. sarA-mediated repression of protease production plays a key role in the pathogenesis of Staphylococcus aureus USA300 isolates. Mol Microbiol. 2012;86:1183–96. doi: 10.1111/mmi.12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolz C, Pohlmann-Dietze P, Steinhuber A, et al. Agr-independent regulation of fibronectin-binding protein(s) by the regulatory locus sar in Staphylococcus aureus. Mol Microbiol. 2000;36:230–43. doi: 10.1046/j.1365-2958.2000.01853.x. [DOI] [PubMed] [Google Scholar]
- 15.Trotonda MP, Xiong YQ, Memmi G, Bayer AS, Cheung AL. Role of mgrA and sarA in methicillin-resistant Staphylococcus aureus autolysis and resistance to cell wall-active antibiotics. J Infect Dis. 2009;199:209–18. doi: 10.1086/595740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seidl K, Chen L, Bayer AS, Hady WA, Kreiswirth BN, Xiong YQ. Relationship of agr expression and function with virulence and vancomycin treatment outcomes in experimental endocarditis due to methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2011;55:5631–9. doi: 10.1128/AAC.05251-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spentzas T, Kudumula R, Acuna C, et al. Role of bacterial components in macrophage activation by the LAC and MW2 strains of community-associated, methicillin-resistant Staphylococcus aureus. Cell Immunol. 2011;269:46–53. doi: 10.1016/j.cellimm.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 18.Fowler VG, Jr., Boucher HW, Corey GR, et al. Daptomycin versus standard therapy for bacteremia and endocarditis caused by Staphylococcus aureus. N Engl J Med. 2006;355:653–65. doi: 10.1056/NEJMoa053783. [DOI] [PubMed] [Google Scholar]
- 19.CDC. From the Centers for Disease Contol and Prevention. Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus, Minnesota and North Dakota, 1997–1999. JAMA. 1999;282:1123–5. [PubMed] [Google Scholar]
- 20.NCCLS. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically: approved standard. 6th ed NCCLS document M7-A6; Wayne, PA. 2003. pp. 495–500. [Google Scholar]
- 21.Xiong YQ, Hady WA, Deslandes A, et al. Efficacy of NZ2114, a novel plectasin-derived cationic antimicrobial peptide antibiotic, in experimental endocarditis due to methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2011;55:5325–30. doi: 10.1128/AAC.00453-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones T, Yeaman MR, Sakoulas G, et al. Failures in clinical treatment of Staphylococcus aureus infection with daptomycin are associated with alterations in surface charge, membrane phospholipid asymmetry, and drug binding. Antimicrob Agents Chemother. 2008;52:269–78. doi: 10.1128/AAC.00719-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seidl K, Bayer AS, Fowler VG, Jr., et al. Combinatorial phenotypic signatures distinguish persistent from resolving methicillin-resistant Staphylococcus aureus bacteremia isolates. Antimicrob Agents Chemother. 2011;55:575–82. doi: 10.1128/AAC.01028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaplan JB. Antibiotic-induced biofilm formation. Int J Artif Organs. 2011;34:737–51. doi: 10.5301/ijao.5000027. [DOI] [PubMed] [Google Scholar]
- 25.Xiong YQ, Bayer AS, Yeaman MR, Van Wamel W, Manna AC, Cheung AL. Impacts of sarA and agr in Staphylococcus aureus strain Newman on fibronectin-binding protein A gene expression and fibronectin adherence capacity in vitro and in experimental infective endocarditis. Infect Immun. 2004;72:1832–6. doi: 10.1128/IAI.72.3.1832-1836.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boles BR, Thoendel M, Roth AJ, Horswill AR. Identification of genes involved in polysaccharide-independent Staphylococcus aureus biofilm formation. PLoS One. 2010;5:e10146. doi: 10.1371/journal.pone.0010146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beenken KE, Mrak LN, Griffin LM, et al. Epistatic relationships between sarA and agr in Staphylococcus aureus biofilm formation. PLoS One. 2010;5:e10790. doi: 10.1371/journal.pone.0010790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seidl K, Bayer AS, McKinnell JA, Ellison S, Filler SG, Xiong YQ. In vitro endothelial cell damage is positively correlated with enhanced virulence and poor vancomycin responsiveness in experimental endocarditis due to methicillin-resistant Staphylococcus aureus. Cell Microbiol. 2011;13:1530–41. doi: 10.1111/j.1462-5822.2011.01639.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Montgomery CP, Boyle-Vavra S, Daum RS. Importance of the global regulators agr and saeRS in the pathogenesis of CA-MRSA USA300 infection. PLoS One. 2010;5:e15177. doi: 10.1371/journal.pone.0015177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Neill E, Pozzi C, Houston P, et al. A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J Bacteriol. 2008;190:3835–50. doi: 10.1128/JB.00167-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dunman PM, Murphy E, Haney S, et al. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J Bacteriol. 2001;183:7341–53. doi: 10.1128/JB.183.24.7341-7353.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsang LH, Cassat JE, Shaw LN, Beenken KE, Smeltzer MS. Factors contributing to the biofilm-deficient phenotype of Staphylococcus aureus sarA mutants. PLoS One. 2008;3:e3361. doi: 10.1371/journal.pone.0003361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cassat J, Dunman PM, Murphy E, et al. Transcriptional profiling of a Staphylococcus aureus clinical isolate and its isogenic agr and sarA mutants reveals global differences in comparison to the laboratory strain RN6390. Microbiol. 2006;152:3075–90. doi: 10.1099/mic.0.29033-0. [DOI] [PubMed] [Google Scholar]
- 34.Mann EE, Rice KC, Boles BR, et al. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS One. 2009;4:e5822. doi: 10.1371/journal.pone.0005822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rice KC, Mann EE, Endres JL, et al. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. P Natl Acad Sci USA. 2007;104:8113–8. doi: 10.1073/pnas.0610226104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Izano EA, Amarante MA, Kher WB, Kaplan JB. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl Environ Microb. 2008;74:470–6. doi: 10.1128/AEM.02073-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kupferwasser LI, Yeaman MR, Nast CC, et al. Salicylic acid attenuates virulence in endovascular infections by targeting global regulatory pathways in Staphylococcus aureus. J Clin Invest. 2003;112:222–33. doi: 10.1172/JCI16876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mirani ZA, Jamil N. Effect of sub-lethal doses of vancomycin and oxacillin on biofilm formation by vancomycin intermediate resistant Staphylococcus aureus. J Basic Microb. 2011;51:191–5. doi: 10.1002/jobm.201000221. [DOI] [PubMed] [Google Scholar]
- 39.Kaplan JB, Izano EA, Gopal P, et al. Low levels of beta-lactam antibiotics induce extracellular DNA release and biofilm formation in Staphylococcus aureus. MBio. 2012;3:e00198–12. doi: 10.1128/mBio.00198-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh R, Ray P, Das A, Sharma M. Penetration of antibiotics through Staphylococcus aureus and Staphylococcus epidermidis biofilms. J Antimicrob Chemother. 2010;65:1955–8. doi: 10.1093/jac/dkq257. [DOI] [PubMed] [Google Scholar]
- 41.Jefferson KK, Goldmann DA, Pier GB. Use of confocal microscopy to analyze the rate of vancomycin penetration through Staphylococcus aureus biofilms. Antimicrob Agents Chemother. 2005;49:2467–73. doi: 10.1128/AAC.49.6.2467-2473.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Wamel W, Xiong YQ, Bayer AS, Yeaman MR, Nast CC, Cheung AL. Regulation of Staphylococcus aureus type 5 capsular polysaccharides by agr and sarA in vitro and in an experimental endocarditis model. Microb Pathogenesis. 2002;33:73–9. doi: 10.1006/mpat.2002.0513. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.