Abstract
Many bacteria regulate gene expression through a cell-cell signaling process called quorum sensing (QS). In proteobacteria, QS is largely mediated by signaling molecules known as N-acylated L-homoserine lactones (AHLs) and their associated intracellular LuxR-type receptors. The design of non-native small molecules capable of inhibiting LuxR-type receptors, and thereby QS, in proteobacteria is an active area of research, and numerous lead compounds are AHL derivatives that mimic native AHL signals. Much of this past work has focused on the pathogen Pseudomonas aeruginosa, which controls an arsenal of virulence factors and biofilm formation through QS. The MexAB-OprM drug efflux pump has been shown to play a role in the secretion of the major AHL signal in P. aeruginosa, N-(3-oxododecanoyl) L-homoserine lactone. In the current study, we show that a variety of non-native AHLs and related derivatives capable of inhibiting LuxR-type receptors in P. aeruginosa display significantly higher potency in a P. aeruginosa Δ(mexAB-oprM) mutant, suggesting that MexAB-OprM also recognizes these compounds as substrates. We also demonstrate that the potency of 5,6-dimethyl-2-aminobenzimidazole, recently shown to be a QS and biofilm inhibitor in P. aeruginosa, is not affected by the presence or absence of the MexAB-OprM pump. These results have implications for the use of non-native AHLs and related derivatives as QS modulators in P. aeruginosa and other bacteria, and provide a potential design strategy for the development of new QS modulators that are resistant to active efflux.
Keywords: N-acyl l-homoserine lactone, anti-virulence, efflux pump, MexAB-OprM, Pseudomonas aeruginosa, quorum sensing
Introduction
Pseudomonas aeruginosa is an opportunistic pathogen responsible for life-threatening infections in immunocompromised patients, such as those suffering from AIDS, burn wounds, or cystic fibrosis.[1] These infections are often refractory to treatment with common antibiotics due to the emergence of multidrug-resistant (MDR) strains of P. aeruginosa, which has prompted an urgent need for the development of orthogonal treatment strategies. Targeting virulence phenotypes rather than general cell growth represents one potential strategy. Significant advances toward such “anti-virulence” therapies include the inhibition of biofilm formation,[2] toxin production,[3] bacterial adhesion factors,[4] and quorum sensing (QS).[5] Small molecules that target these pathways could find broad use as both research tools and new therapeutic agents. In particular, the design of QS inhibitors for P. aeruginosa and other bacterial pathogens has attracted significant attention over the past ~20 years.[6] The efficacy of such compounds as QS inhibitors in P. aeruginosa is the focus of the current study.
QS is widespread in bacteria and allows the coordination of gene expression with bacterial population density.[7] This intercellular communication pathway is mediated by small molecules or peptides (i.e., autoinducers) that vary in concentration as a function of cell number. At high cell densities, the signals reach a sufficient concentration to bind and activate QS receptors, which subsequently regulate transcription of primarily group-beneficial genes. Proteobacteria use N-acylated L-homoserine lactones (AHLs) as their main autoinducers. The AHL signals are generated by LuxI-type synthases and sensed by cytoplasmic LuxR-type receptors. P. aeruginosa has two LuxI/LuxR pairs, LasI/LasR and RhlI/RhlR, which produce and sense N-(3-oxododecanoyl) L-homoserine lactone (OdDHL) and N-butanoyl L-homoserine lactone (BHL), respectively. QscR, an additional “orphan” or “solo” LuxR-type receptor in P. aeruginosa, lacks an associated LuxI-type synthase; instead, it also recognizes the OdDHL signal produced by LasI. LasR is believed to play an important role in controlling virulence in P. aeruginosa and regulates the production of elastase B, exotoxin A, and the biosynthesis machinery for a number of metabolites related to host tissue breakdown.[8] Furthermore, clinical isolates of P. aeruginosa strains lacking a functional las system are less virulent in animal infection models, suggesting that successful LasR inhibition could significantly attenuate P. aeruginosa virulence.[9]
Our laboratory and others have synthesized and examined a range of non-native AHLs as LasR and QscR modulators in P. aeruginosa.[10] Of the >150 compounds in our in-house libraries of AHLs, we have identified a number of LasR inhibitors using an E. coli reporter strain to measure LasR-mediated transcriptional activation.[11] However, the potencies of these compounds in P. aeruginosa LasR reporter strains are generally muted in comparison.[12] Meijler and co-workers observed similar effects in their studies of both covalent and non-covalent inhibitors of LasR in related E. coli and P. aeruginosa reporter strains.[13] In general, the efficacy of small molecule drugs is often lower in P. aeruginosa relative to many other Gram-negative bacteria due to decreased membrane permeability, enhanced active efflux, or a combination of both factors,[14] which prompted us to consider the possibility that these features could also influence the potency of our synthetic LasR modulators.
In 1999, Iglewski and co-workers showed that OdDHL passively diffuses across the P. aeruginosa cell membrane (albeit at a ~10-fold slower rate than the shorter-chain autoinducer BHL) and that the presence of the efflux pump MexAB-OprM significantly reduces the intracellular concentration of OdDHL, suggesting that MexAB-OprM recognizes OdDHL as a substrate.[15] In concurrent work, Poole and co-workers demonstrated that a P. aeruginosa mutant capable of MexAB-OprM overexpression produced reduced levels of QS-regulated virulence factors, presumably due to low levels of intracellular OdDHL.[16] MexAB-OprM is a member of the resistance-nodulation-division (RND) family of efflux pumps, which are a main class of pumps in Gram-negative bacteria known to contribute to intrinsic and acquired resistance to exogenous compounds.[17] Given that RND pumps often possess broad substrate profiles, we reasoned that active efflux could play a role in reducing the potency of our AHL-derived LasR inhibitors in P. aeruginosa, and that, in particular, MexAB-OprM recognizes these non-native AHLs in a similar manner as it does OdDHL. Recent studies by Gotoh and co-workers—showing that MexAB-OprM can transport a small set of naturally occurring 3-oxo-acyl HLs (with 8–14 carbon acyl tails)—provide support for this hypothesis.[18][19] To our knowledge, systematic investigations into the effect of active efflux on AHL-derived and non-AHL QS inhibitors are yet to be reported.
Herein, we report our analysis of the potency of synthetic LasR antagonists in the presence and absence of the MexAB-OprM drug efflux pump. We generated a P. aeruginosa Δ(mexAB-oprM) mutant and used a GFP reporter of LasR activity to examine compound antagonism trends relative to a strain with an operative pump. We demonstrate that both OdDHL and non-native AHL analogues suffer significant decreases in potency when subjected to active efflux by MexAB-OprM, but a previously reported biofilm and LasR inhibitor in P. aeruginosa, 5,6-dimethyl-2-aminobenzimidazole (DMABI)[20] does not display this efflux-induced reduction in potency.
Results
P. aeruginosa MexAB-OprM reduces the potency of OdDHL
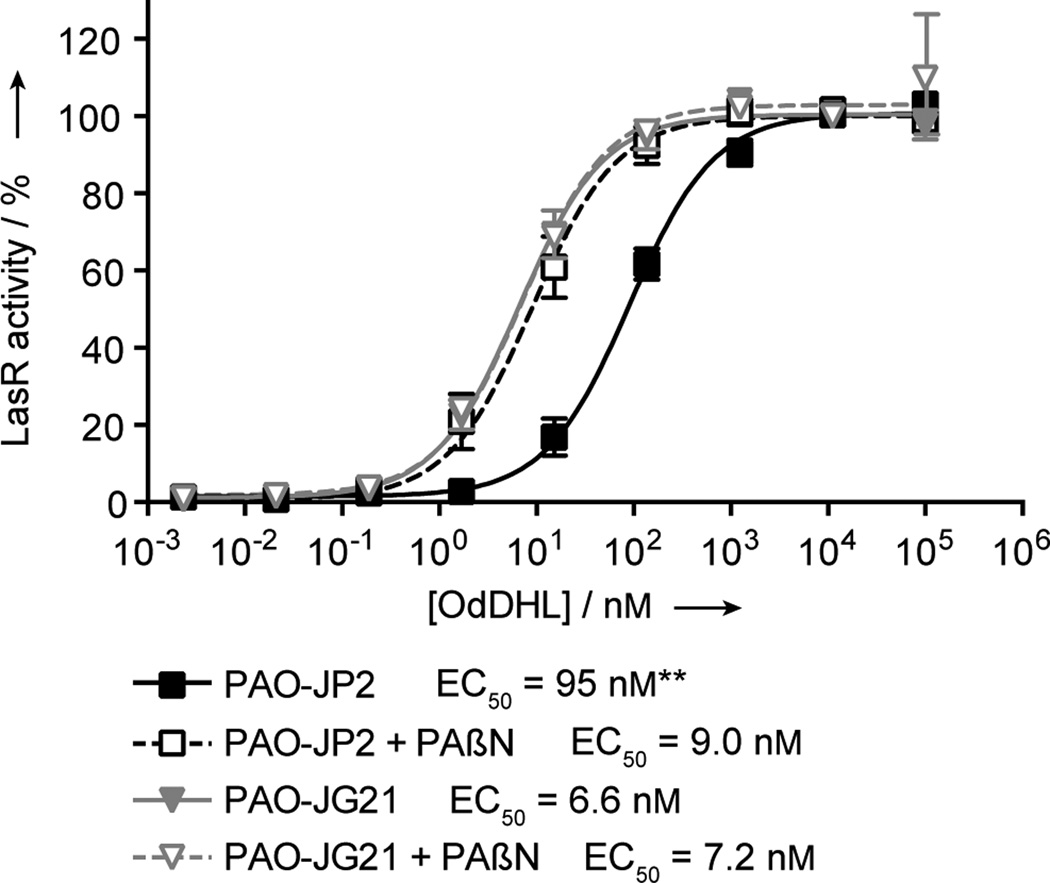
We began our study by comparing the potency of OdDHL in a P. aeruginosa mutant lacking both the AHL synthases LasI and RhlI (PAO-JP2; i.e., pump-active) and a mutant lacking both AHL synthases and the MexAB-OprM pump (PAO-JG21; i.e., pump-mutant). Both strains contained a functional LasR receptor and reported LasR activity via a plasI-LVAgfp reporter plasmid (see Experimental Section). Dose responses for LasR activation by OdDHL in the pump-active and pump-mutant P. aeruginosa strains were obtained. As we hypothesized, OdDHL was a more potent activator of LasR in the pump-mutant strain relative to the pump-active strain (Figure 1). The EC50 value shifted from 95 nM in the pump-active PAO-JP2 to 6.6 nM in the pump-mutant PAO-JG21. This > 10-fold increase in potency for OdDHL in the pump-mutant strain largely correlates with prior experiments by Iglewski and co-workers that showed at least a 3-fold increased cellular concentration of [3H]-OdDHL in a P. aeruginosa Δ(mexAB-oprM) mutant.[21]
Figure 1.
MexAB-OprM decreases the potency of OdDHL in P. aeruginosa. Dose responses are shown for OdDHL in pump-active PAO-JP2 and pump-mutant PAO-JG21 in the presence and absence of the non-specific efflux pump inhibitor PAβN. LasR activity was measured by expression of the lasI::gfp[LVA] transcriptional fusion. Both pump-active PAO-JP2 and pump-mutant PAO-JG21 displayed a similar dynamic range of response to OdDHL, but PAO-JG21 showed stronger LasR activity at lower OdDHL concentrations. The potency of OdDHL in pump-active PAO-JP2 in the presence of PAβN was comparable to the potency of OdDHL in pump-mutant PAO-JG21 in the absence of PAβN. No additive effect on the increased potency of OdDHL was observed in pump-mutant PAO-JG21 in the presence of PAβN. Error bars represent SEM of four trials for each strain. ** – p < 0.01 as measured by a t-test performed on the log(EC50) of each trial.
We next evaluated the possibility that other RND efflux pumps could be involved in active efflux of OdDHL in P. aeruginosa. The P. aeruginosa genome contains up to 12 RND efflux systems, seven of which have been directly shown to recognize a wide range of drugs.[14b, 22] If other RND pumps recognize OdDHL as a substrate, we reasoned that the addition of a non-specific inhibitor of RND-type pumps would result in a further decreased EC50 value for OdDHL. Dose responses for LasR activation by OdDHL in the pump-active PAO-JP2 were performed in the presence of the non-specific RND-type pump inhibitor, Phe-Arg-β-naphthylamide (PAβN; Figure 1).[23] The EC50 value for OdDHL in PAβN-treated pump-active PAO-JP2 was 9.0 nM, which was statistically indistinguishable from the EC50 value of 6.6 nM for OdDHL in the untreated pump-mutant PAO-JG21 (p > 0.60 as measured by a t-test). To ensure that the effects of PAβN inhibition and Δ(mexAB-oprM) mutation were not additive, an OdDHL dose response analysis was also performed in pump-mutant PAO-JG21 in the presence of PAβN. The EC50 value for OdDHL in this experiment was 7.2 nM, again indistinguishable from the values for the untreated pump-mutant and the PAβN-treated pump-active strain (Figure 1). These data support the hypothesis that OdDHL is not a substrate of other P. aeruginosa RND pumps.
P. aeruginosa Δ(mexAB-oprM) and E. coli LasR reporters respond similarly to non-native AHLs
As highlighted above, our prior biological evaluation of non-native AHL libraries revealed a number of potent LasR inhibitors using an E. coli reporter strain to measure LasR-mediated transcriptional activation.[11a–c] However, the activities of these compounds in P. aeruginosa LasR reporter strains were generally muted.[12] The increased potency of OdDHL in pump-mutant PAO-JG21 (shown above) suggested that efflux by MexAB-OprM might contribute to the discrepancies observed for AHL-induced modulation of LasR activity in P. aeruginosa as compared to that in E. coli. We thus performed side-by-side screens of our ~150-member non-native AHL library (shown in Figures S1–S4) for LasR antagonistic activity in the E. coli JLD271, pump-active PAO-JP2, and pump-mutant PAO-JG21 reporter strains in an attempt to resolve the source of these discrepancies. In these competitive antagonism assays, non-native compound was screened against OdDHL at its EC50 value in each of the reporter strains. To facilitate more rapid-throughput screening in E. coli, a new fluorescence-based reporter, pPROBE-KL, was designed for use in JLD271 (see Experimental Section). The plasmid pPROBE-KL harbors a lasI promoter that has been transcriptionally fused to gfp, allowing for the production of GFP upon activation of LasR.
The reporter gene assays yielded several interesting data trends (see Figures S5–S10 for full primary data). The LasR inhibitory activities of our non-native AHLs were modest in PAO-JP2, with only 11 AHLs showing ≥25% LasR inhibition (structures shown in Figure 3A; percent inhibition values listed in Table S1). In contrast, the AHLs were significantly more active in PAO-JG21, with 84 compounds showing ≥25% LasR inhibition, and 11 compounds showing ≥80% LasR inhibition (Table S2). The number of strong LasR inhibitors in E. coli was similar to that in PAO-JG21: 77 compounds showed ≥25% LasR inhibition, and 13 compounds showed ≥80% LasR inhibition (Table S3).[24]
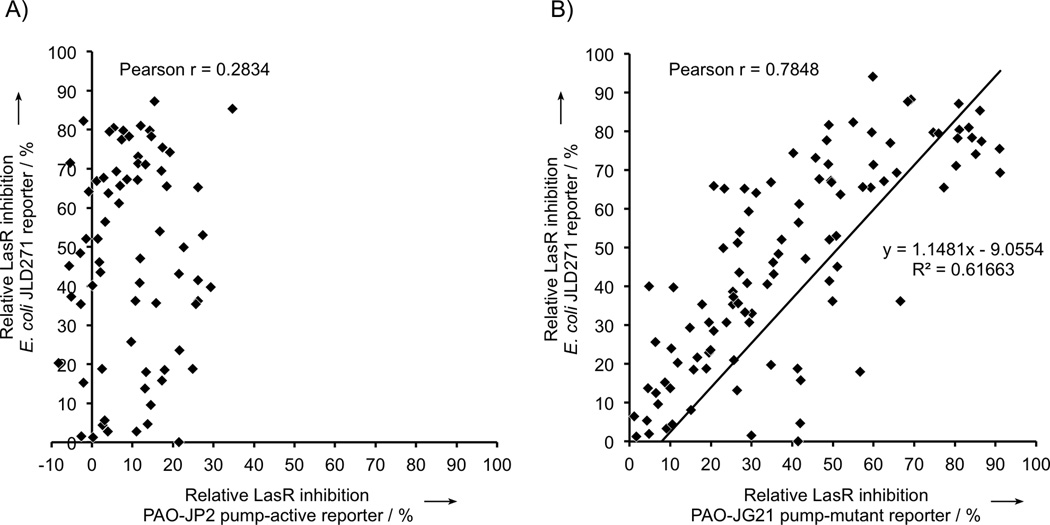
Analysis of these three sets of assay data revealed that the LasR antagonism trends observed for non-native ligands in E. coli and in the P. aeruginosa pump-mutant were strikingly conserved. Several of the strongest LasR inhibitors were shared amongst these two strains (e.g., AHLs E3, E5, E10, and E27; Tables S2–S3). To more quantitatively compare the three sets of assay data, we calculated Pearson product-moment correlation coefficients, which are commonly used to measure the linear dependence of two measured variables (Figure 2).[25] The LasR inhibitory activities of compounds in E. coli correlated much more strongly to pump-mutant PAO-JG21 (r = 0.7848) than to pump-active PAO-JP2 (r = 0.2834) (Figure 2B vs. Figure 2A). Additionally, a least-squares linear regression of the data in Figure 2B returned a trend line highly suggestive of one-to-one correspondence of LasR inhibitory activities. Notably, the strong correlation of antagonistic activities occurs despite the fact that membrane permeability and expression levels of LasR may not be similar in the E. coli and P. aeruginosa strains. These results suggest that—despite the differences between E. coli and P. aeruginosa—active efflux by MexAB-OprM is a primary cause of the discrepancies between the LasR inhibitory activities of our non-native AHLs in pump-active P. aeruginosa and the LasR inhibitory activities of the same compounds in the E. coli reporter.
Figure 2.
Comparative analyses of LasR inhibition data for non-native AHLs in P. aeruginosa and E. coli LasR reporter strains. Non-native AHLs show decreased LasR inhibition in A) pump-active P. aeruginosa PAO-JP2 as compared with pump-mutant P aeruginosa PAO-JG21, but inhibitory activities strongly correlate in B) E. coli JLD271 as compared with pump-mutant PAO-JG21. Linear correlations of data sets in A) and B) were compared by calculating the Pearson product-moment correlation coefficient (Pearson r). Each point represents a different non-native AHL. Data were obtained from three separate trials. Error bars are not included for clarity, but SEM of all primary screening data did not exceed ±10%.
MexAB-OprM reduces potency of non-native AHLs and related inhibitors
The results from the LasR antagonism screens above suggest that effective QS inhibitors in wild-type P. aeruginosa will require attributes that allow them to bypass the mechanism of MexAB-OprM efflux. As previously mentioned, evading efflux is an important issue in the development of effective antibiotics,[14a, 14d, 26] and in fact any small molecule agent acting via an intracellular target, in P. aeruginosa. While the primary data above indicated that non-native AHL activities were influenced by MexAB-OprM, we sought to determine if certain types of acyl chain structures rendered an AHL more or less susceptible to efflux by MexAB-OprM.
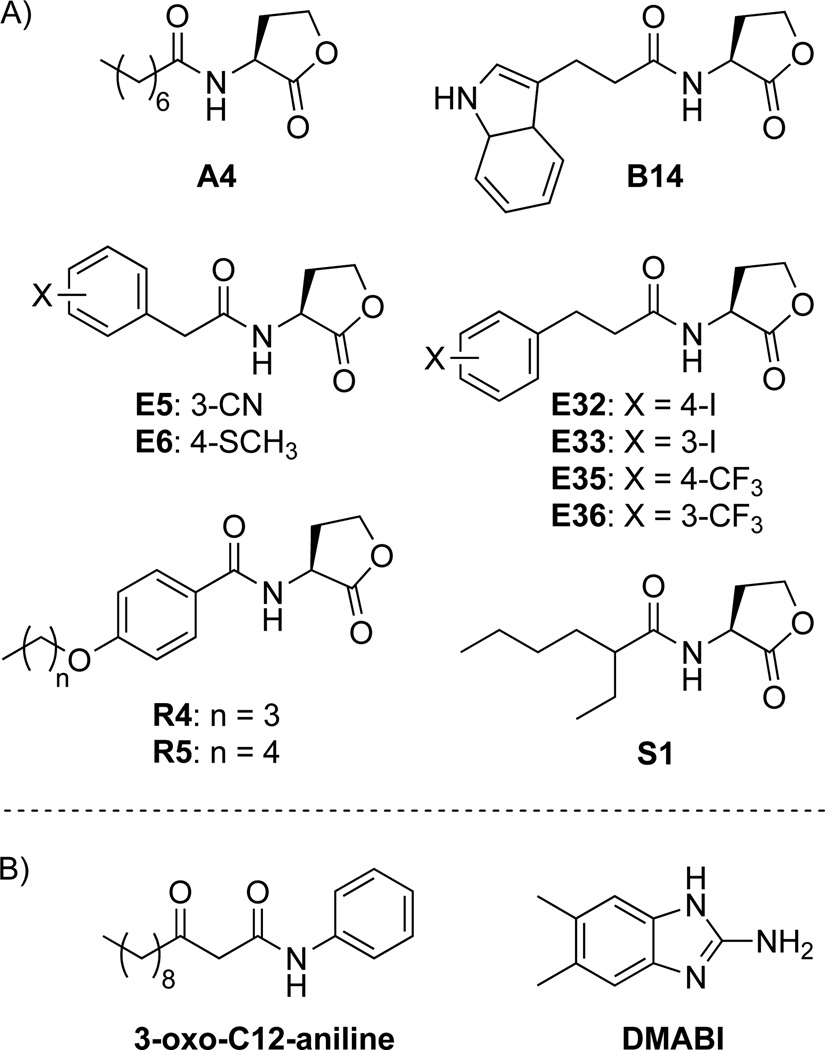
We reasoned that compounds acting as poor substrates of MexAB-OprM would likely show similar potencies in both pump-active and pump-mutant P. aeruginosa strains. To determine LasR inhibitory potencies in both strains, the compounds had to exhibit a full antagonism dose response curve in both the PAO-JP2 and PAO-JG21 reporter strains. Accordingly, we were limited to the compounds that inhibited LasR activity in the pump-active PAO-JP2 primary screen, which were the 11 AHLs capable of ≥ 25% LasR inhibition (shown in Scheme 1A). Dose–response analyses for LasR inhibition were performed in the presence of OdDHL at its EC50 in each strain. We also included in this dose-response analysis an OdDHL analogue, 3-oxo-C12-aniline, which lacks the hydrolysable lactone head group (Scheme 1B). Prior studies by our laboratory have shown that 3-oxo-C12-aniline is capable of inhibiting LasR activity by 30% in P. aeruginosa and by 55% in E. coli (using related reporter constructs).[27] Spring and co-workers have also reported that 3-oxo-C12-aniline is capable of strongly inhibiting pyocyanin production in wild-type P. aeruginosa, a well-known QS-controlled phenotype.[28] Because 3-oxo-C12-aniline inhibits P. aeruginosa pyocyanin production significantly better than non-native AHLs, while only showing modest activity in LasR reporter assays, we were interested to learn if 3-oxo-C12-aniline was able to evade efflux by MexAB-OprM.
Scheme 1.
Structures of compounds subjected to LasR antagonism dose-response analyses in this study. A) AHLs exhibiting ≥25% inhibition of LasR in the primary LasR antagonism screen in pump-active P. aeruginosa PAO-JP2. B) Non-AHL-based compounds chosen due to their activities in QS phenotypic screens in wild-type P. aeruginosa.
The IC50 values for the 11 AHLs and 3-oxo-C12-aniline in the pump-active PAO-JP2 and pump-mutant PAO-JG21 LasR reporter strains are listed in Table 1. Compounds E6, R4, R5 and S1 showed incomplete inhibition of LasR in pump-active PAO-JP2 at the solubility limit of the compound; thus, only a lower bound on the IC50 value could be determined. Overall, the potencies of the tested compounds were strongly affected by the presence of MexAB-OprM, with each of the IC50 values measured in pump-active PAO-JP2 being significantly different (p < 0.05) than the IC50 values measured in pump-mutant PAO-JG21. In fact, seven of the 12 compounds experienced a loss in potency of an even greater magnitude (up to 75-fold for AHL E33) than that observed for OdDHL (~10-fold; Figure 1). For certain AHLs, the fold-change in potency upon removal of MexAB-OprM was affected by subtle alterations in compound structure. For example, phenylacetanoyl HLs E5 and E6, differing in only their substituents at the 3- or 4- position on the aromatic ring, show clear differences in their potency shifts between the two strains (3.9 vs. ≥ 25 fold-changes, respectively). While the phenylpropionoyl HLs (E32, E33, E35, and E36) exhibited consistently high fold-changes in IC50 value upon deletion of MexAB-OprM (23–75), the position of the iodine substituent on the aromatic ring (i.e., in E32 and E33) appeared to cause a greater shift in potency relative to the trifluoromethyl substituent in E35 and E36. The more modest shifts in potency for E5 and 3-oxo-C12-aniline (3.9 and 6.5-fold changes, respectively) relative to other compounds in Table 1 suggest that they may represent scaffolds that partially evade efflux. This modest shift for 3-oxo-C12-aniline also suggests that factors apart from active efflux are contributing to its significantly heightened phenotypic inhibitory activity in P. aeruginosa relative to non-native AHLs. It may be the case that the absence of a hydrolysable lactone head group in 3-oxo-C12-aniline contributes to a longer half-life of the active inhibitor, increasing its observed efficacy in the long-term (> 12 h) pyocyanin assays.[28]
Table 1.
IC50 values for LasR inhibition by selected compounds in P. aeruginosa PAO-JP2 and PAO-JG21.a
| PAO-JP2 | PAO-JG21 | ||||
|---|---|---|---|---|---|
| Compound | IC50 (µM) | 95% CI (µM) | IC50 (µM) | 95% CI (µM) | Fold change of IC50 |
| A4 | 1.0b | 0.21–4.9 | 0.12b | 0.021–0.70 | 8.3 |
| B14 | 15b | 2.9–34 | 0.95b | 0.43–2.1 | 15 |
| E5 | 12 | 6.5–21 | 3.1 | 2.6–3.8 | 3.9 |
| E6 | ≥ 480 | - | 19 | 12–33 | ≥ 25 |
| E32 | 6.0 | 3.2–11 | 0.26b | 0.043–1.5 | 23 |
| E33 | 15b | 9.1–24 | 0.20b | 0.062–0.65 | 75 |
| E35 | 5.7b | 1.7–19 | 0.16b | 0.061–0.40 | 35 |
| E36 | 14b | 2.5–81 | 0.34b | 0.11–1.1 | 41 |
| R4 | ≥ 570 | - | 41 | 30–57 | ≥ 13 |
| R5 | ≥ 230 | - | 55 | 38–78 | ≥ 4.2 |
| S1 | ≥ 710 | - | 24 | 16–34 | ≥ 29 |
| 3-oxo-C12-aniline | 11 | 4.2–23 | 1.7 | 1.3–4.2 | 6.5 |
| DMABI | 2.3 | 1.3–4.1 | 1.4 | 0.81–2.5 | 1.6 |
Antagonism assays were performed in the presence of 100 nM OdDHL in PAO-JP2 and 10 nM OdDHL in PAO-JG21.
Antagonism dose response exhibited non-monotonic behavior, showing an increase in activity at high concentrations. We have previously identified this phenomenon,[11d] and studies are currently underway to identify the mechanistic basis for the trend.
DMABI is equally potent in pump-active and pump-mutant P. aeruginosa
The results above indicated that MexAB-OprM can recognize a wide variety of AHL derivatives with non-native acyl tails and 3-oxo-C12-aniline, with a non-native head group. In an attempt to uncover compound classes that resist efflux-induced reductions of potency and further explore the promiscuity of MexAB-OprM, we sought to test the potency of other QS modulators in P. aeruginosa with structures distinct from AHLs. Recently, our laboratory identified 2-aminobenzimidazoles (2-ABIs) as a potent class of biofilm inhibitors and dispersers in P. aeruginosa.[20] In particular, the compound 5,6-dimethyl-2-aminobenzimidazole (DMABI) is a potent inhibitor of biofilm growth in wild-type P. aeruginosa (IC50 at 24 h = 4 µM) and a modest (~40%) inhibitor of LasR activity in P. aeruginosa PAO-JP2 (harboring a plasI-LVAgfp reporter plasmid; Figure S-14). We tested the LasR inhibitory activity of DMABI in both the MexAB-OprM pump-active and pump-mutant LasR reporter strains of P. aeruginosa to determine the susceptibility of the 2-ABI scaffold to active efflux. While the maximum inhibition of LasR activity by DMABI was modest (40%), the IC50 values calculated from the dose–response inhibition curves in PAO-JP2 and PAO-JG21 were statistically indistinguishable (2.3 vs. 1.4, respectively; Table 1). This result indicates that the potency of DMABI is not affected by MexAB-OprM in P. aeruginosa. DMABI was also subjected to dose-response analyses in the presence of 25 µg/mL PAβN using both PAO-JP2 and PAO-JG21 LasR reporter strains. In each case, the presence of the non-specific pump inhibitor had no effect on DMABI potency (Figure S14), suggesting that DMABI is not recognized as a substrate by other RND-type pumps in P. aeruginosa.
Discussion
The results reported herein demonstrate that the RND efflux pump MexAB-OprM in P. aeruginosa has a significant effect on the potency of both native and non-native AHLs as LasR modulators. We first showed that OdDHL is approximately 10-fold more potent in a P. aeruginosa Δ(mexAB-oprM) mutant strain relative to a pump-active strain, which corroborates a previous report showing that deletion of MexAB-OprM increases the intracellular concentration of OdDHL.[15] Because MexAB-OprM has a clear influence on OdDHL potency, we suspected that similar perturbations could also occur with our non-natural AHLs and related analogs. Furthermore, we reasoned that the presence of the MexAB-OprM efflux pump in P. aeruginosa could account for the discrepancies that we have previously observed between the activities of our synthetic AHLs in P. aeruginosa versus those in E. coli LasR reporter strains.[12] We tested this hypothesis by performing side-by-side assays of our non-native AHL libraries in pump-active and pump-mutant P. aeruginosa LasR reporter strains and in an E. coli LasR reporter strain. We found that the LasR inhibitory activities of our compounds in E. coli much more strongly matched those in the pump-mutant P. aeruginosa compared to pump-active P. aeruginosa. These data suggest that the presence of the MexAB-OprM pump strongly affects the activities of non-native AHLs in P. aeruginosa. Although E. coli does express a homologous RND pump, AcrAB-TolC, this pump has a significantly different substrate profile and has not been shown to export any AHLs in E. coli.[14a, 29] Our experiments provide a clear illustration that LasR activity screens performed using E. coli reporters allow elucidation of SARs describing a compound’s ability to potentially bind to and modulate LasR, but they do not necessarily provide an accurate representation of the molecule’s intracellular availability in P. aeruginosa.
We next focused on identifying LasR inhibitors that might evade active efflux and therefore retain potency as LasR inhibitors in P. aeruginosa by performing dose-response analyses on 11 potent non-native AHL inhibitors in the pump-active and pump-mutant P. aeruginosa LasR reporter strains. We also examined the activity of an AHL analog that lacked the native lactone head group, 3-oxo-C12-aniline. The majority of these LasR inhibitors showed a significant loss of potency in the pump-active reporter (> 10-fold), despite possessing a wide variety of acyl tail functionalities (straight-chain alkyl, branched alkyl, aryl, and heterocyclic; see Scheme 1A), which suggests that MexAB-OprM is likely promiscuous across a broad range of AHL derivatives. Although none of the tested AHLs fully resisted potency effects due to active efflux, we noted that subtle changes in acyl chain structure for certain AHLs elicited varied responses to MexAB-OprM mutation (e.g., E5 vs. E6 and E32 vs. E33). RND-type pumps homologous to MexAB-OprM are present in a number of Gram-negative bacteria that use LuxR/LuxI-type QS circuits (e.g., Acinetobacter baumannii, Agrobacterium tumefaciens, Burkholderia spp., etc.).[14b] Though mutations of efflux pumps in some species have had varied effects on QS-controlled phenotypes,[30] our results have implications for future SAR studies of AHL-derived modulators of LuxR-type receptors performed in Gram-negative bacteria. It may be the case that variations in QS modulatory activity are a result of not only a compound’s differential ability to interact with a LuxR-type receptor, but also a compound’s differential susceptibility to active efflux.
Our observation that MexAB-OprM can recognize a range of AHLs as substrates prompted us to consider alternative chemical scaffolds that might inhibit QS while also bypassing MexAB-OprM efflux in P. aeruginosa. We recently identified a class of P. aeruginosa biofilm inhibitors known as 2-ABIs,[20] which were shown to inhibit LasR through an, as of yet, undetermined mechanism. Given their potent biofilm inhibitory activities in wild-type P. aeruginosa, we surmised that the compounds could be inherently resistant to active efflux. We tested the most potent lead compound, DMABI, and found that the presence of MexAB-OprM did not decrease the compound’s potency. We also measured the potency of DMABI in the presence of PAβN to determine if DMABI is recognized by any other RND pumps present in PAO-JG21. Co-incubation with PAβN showed no significant change in DMABI potency in either PAO-JP2 or PAO-JG21, confirming the QS-inhibitory efficacy of DMABI is not significantly impacted by any RND pump in P. aeruginosa.
We currently have three hypotheses to explain the mechanism by which DMABI evades a loss in potency due to efflux by P. aeruginosa: 1) MexAB-OprM and other RND-type pumps do not recognize DMABI as a substrate, 2) the diffusion rate of DMABI into the cell is so rapid that active efflux is negligible, or 3) DMABI acts upon an extracellular target. Regardless of the mechanism, the unique ability of DMABI to evade reduction of potency as a QS inhibitor will be further studied and leveraged to design more potent probes capable of QS inhibition in wild-type P. aeruginosa strains and even possibly for prevalent multidrug-resistant P. aeruginosa strains that overexpress RND-type pumps.[14b] Current efforts in our laboratory are focused on identifying the mechanism of biofilm and LasR inhibition by DMABI and designing new compounds that more strongly antagonize LasR, while retaining DMABI’s resistance to efflux by RND pumps.
Conclusions
The design of small molecules capable of blocking QS pathways, and thereby virulence, in the opportunistic pathogen P. aeruginosa is of significant interest. Numerous non-native AHLs and related analogs have been reported that are capable of modulating the activities of LuxR-type receptors in P. aeruginosa (LasR, RhlR, and QscR). In this study, we determined that a variety of non-native AHLs capable of inhibiting the LasR receptor in P. aeruginosa display significantly higher potency in a P. aeruginosa Δ(mexAB-oprM) mutant. These data suggest that the presence of the MexAB-OprM pump, and thus active efflux, strongly affects the activities of non-native AHLs in P. aeruginosa. We also demonstrated that while a variety of AHL analogues appear to be recognized as substrates by MexAB-OprM, a compound structurally distinct from the AHLs—DMABI—does not display this efflux-induced reduction in potency. To our knowledge, this study represents the first systematic investigation of the role of active efflux in the activity profiles of small molecule QS inhibitors in Gram-negative bacteria. Our results have implications for the use of QS modulators in P. aeruginosa and potentially other proteobacteria, and provide a prospective design strategy (based on 2-ABI derivatives) for the development of new QS modulators that may be resistant to active efflux.
Experimental Section
Bacterial strains and growth conditions
The bacterial strains and plasmids used in this study are listed in Table 2. Bacteria were grown at 37 °C in Luria-Bertani (LB) medium unless otherwise noted. P. aeruginosa strain PAO-JP2 harboring plasI-LVAgfp was grown in the presence of carbenicillin (300 µg/mL). As expected, the Δ(mexAB-oprM) strain PAO-JG21 was more sensitive to antibiotic selection and thus was grown in the presence of 100 µg/mL carbenicillin. E. coli strain JLD271 harboring plasmids pPROBE-KL and pJN105L was grown in the presence of kanamycin (50 µg/mL) and gentamicin (10 µg/mL). All overnight cultures were shaken at 200 rpm.
Table 2.
Bacterial strains and plasmids used in this study.
| Strain or plasmid | Relevant properties | Source or reference |
|---|---|---|
| Strains | ||
| P. aeruginosa | ||
| PAO-JP2 | PAO1 lasI::Tet rhlI::Tn501-2; HgR TcR | [33] |
| PAO-JG21 | PAO-JP2 Δ(mexA-mexB-oprM) | This study |
| E. coli | ||
| DH5α | E. coli strain for transformation | [34] |
| S17-1 | Mobilizer strain | [35] |
| JLD271 | K-12 ΔlacX74 sdiA271::Cam; ClR | [36] |
| Plasmids | ||
| pEX18Gm | GmR; oriT+
sacB+, gene replacement vector with MCS from pUC18; pEX100T backbone GmR |
[37] |
| pJG034 | pEX18Gm with markerless Δ(mexAB- oprM) |
This study |
| plasI-LVAgfp | lasI’-gfp[LVA] transcriptional fusion; CbR | [38] |
| pSC11 | Broad host range lasI-lacZ reporter; source of lasI DNA; ApR |
[39] |
| pPROBE-KT | Broad host range promotorless gfp transcriptional fusion vector; KmR |
[40] |
| pPROBE-KL | lasI’-gfp[LVA] transcriptional fusion; KmR | This study |
| pJN105L | Arabinose-inducible expression vector for lasR; pBBRMCS backbone; GmR |
[41] |
Chemicals
Non-native AHLs, 3-oxo-C12-aniline, and 5,6-dimethyl-aminobenzimidazole (DMABI) were synthesized as described previously.[11d, 20, 27, 31] Structures of the entire in-house AHL library evaluated in this study are shown in Figures S1–S4. N-(3-oxo)-dodecanoyl L-homoserine lactone (OdDHL) was purchased from Sigma-Aldrich. The pump inhibitor Phe-Arg-β-naphthylamide (PAβN) was purchased from Chem-Impex, International.
Construction of P. aeruginosa mexAB-oprM mutant (PAO-JG21)
To construct PAO-JG21, an 898-base pair (bp) upstream portion of mexA and a 521-bp downstream portion of oprM were amplified from P. aeruginosa PAO1 genomic DNA by PCR with the following primers: 5 ′–ACTAAGCTTCAATACATGGACGTCGGG–3′ (native HindIII site underlined) and 5′–ACTGAATTCGGAGATCGGCGACAGCACC–3′ (added EcoRI site underlined) for mexA; and 5′–ACTGAATTCGACCTGTCGACCACCGGCA–3′ (added EcoRI site underlined) and 5′–AGTTCTAGAGATGTCCGGGCGCCGT–3′ (added XbaI site underlined) for oprM. The EcoRI sites incorporated by the internal primers were digested and used to ligate the two portions together, yielding an in-frame deletion with an EcoRI scar. This construct was PCR amplified and ligated into the pEX18Gm vector using HindIII and XbaI sites to form pJG034. The pJG034 plasmid was introduced into E. coli S17-1::λpir by electroporation and transferred to P. aeruginosa PAO-JP2 by conjugation and citric acid + antibiotic selection on Vogel-Bonner minimal medium[32] supplemented with gentamicin (15 µg/ml). The resulting gentamicin-resistant merodiploid colonies were counterselected on LB agar plates supplemented with 5% sucrose, which yielded colonies that were gentamicin sensitive and sucrose resistant. The mexAB-oprM region of one mutant colony (PAO-JG21) was PCR amplified and sequenced to verify the markerless in-frame deletion.
Construction of pPROBE-KL
To construct plasmid pPROBE-KL, a 317-bp fragment spanning the upstream region of lasI (−282 to +35 relative to the lasI translational start codon) was amplified by PCR. The primers were 5′–GAATTAGGATCCGCAGGTTCTCGCCATTC–3′ (native BamHI site underlined) and 5′–GAATAGGAATTCTCGAACTCTTCGCGCCG–3′ (added EcoRI site underlined). The PCR-generated fragment was digested with EcoRI and BamHI and subsequently ligated with BamHI/EcoRI-digested pPROBE-KT to generate pPROBE-KL.
LasR reporter assay protocol
To evaluate the LasR inhibitory activities of our synthetic compounds in P. aeruginosa, PAO-JP2 or PAO-JG21 harboring the plasmid plasI-LVAgfp were grown overnight as described above. An appropriate amount of synthetic compound stock solution (or OdDHL stock solution, as a control) in DMSO was added to wells in 96-well microtiter plates, with final DMSO concentrations (after addition of cells) not exceeding 1%. The overnight culture was diluted 1:100 in fresh LB medium and was grown to an OD600 of 0.3. Subculture was treated with OdDHL (100 nM in PAO-JP2 or 10 nM in PAO-JG21) by adding the appropriate amount of an OdDHL stock solution in DMSO (10 mM or 1 mM). The subculture was then dispensed in 200 µL portions into each synthetic compound-treated well of the microtiter plate.
Plates were incubated at 37 °C for 6 h, and GFP production was monitored using a Biotek Synergy 2 plate reader (Excitation: 500 nm, Emission: 540 nm) and quantified with Gen5 1.05 software. The final OD600 of each well was measured to normalize GFP production to cell density. In LasR antagonism assays, the normalized fluorescence of each compound competing against OdDHL was reported relative to the normalized fluorescence of the OdDHL-only positive control. All synthetic compounds were tested in triplicate, and three separate trials were performed using unique cultures. Antagonism dose–response analyses were performed by testing compounds over a range of concentrations in this LasR reporter assay. IC50 values and 95% confidence intervals were calculated using GraphPad Prism 4 software. Dose response analyses of OdDHL were performed in a similar manner, except that the bulk subculture was not treated with OdDHL; instead, varying concentrations of OdDHL were added to the 96-well microtiter plates before addition of subculture.
To evaluate the LasR inhibitory activities of our synthetic compounds in E. coli, E. coli JLD271 harboring plasmids pPROBE-KL and pJN105L was grown overnight as described above. Antagonism assays were performed as those in P. aeruginosa with the following modifications: Overnight cultures were diluted 1:10 with fresh LB medium containing kanamycin (50 µg/mL) and gentamicin (10 µg/mL). Solid arabinose was added to the subculture to a final concentration of 0.5%, and a DMSO stock of OdDHL was added to the subculture to a final concentration of 2 nM. The subculture was immediately dispensed into compound-treated 96-well microtiter plates. The plates were shaken at 200 rpm for 4 h at 37 °C.
LasR agonism and antagonism assays in the presence of PAβN were performed by adding PAβN from a 50 mg/mL H2O stock to cultures immediately prior to plating to yield a final PAβN concentration of 25 µg/mL.
Supplementary Material
Acknowledgements
Financial support for this work was provided by the NIH (AI063326), Burroughs Wellcome Fund, and Johnson & Johnson. J.D.M. was supported in part by the NIH Biotechnology Training Program (T32 GM08349). J. P. G. was supported in part by the Department of Defense (DOD) Air Force Office of Scientific Research through a National Defense Science & Engineering Graduate (NDSEG) Fellowship (32 CFR 168a) and by the NIH Chemistry-Biology Interface Training Program (T32 GM008505). N.R.E was supported by a Ruth L. Kirschstein National Research Service Award (1F32 GM100728). We gratefully acknowledge Professors Brian Ahmer, Peter Greenberg, Barbara Iglewski, and Herbert Schweizer for donation of reporter strains and plasmids.
Footnotes
Supporting information for this article is available.
References
- 1.a) Lyczak JB, Cannon CL, Pier GB. Microb. Infect. 2000;2:1051–1060. doi: 10.1016/s1286-4579(00)01259-4. [DOI] [PubMed] [Google Scholar]; b) Richards MJ, Edwards JR, Culver DH, Gaynes RP. Crit. Care Med. 1999;27:887–892. doi: 10.1097/00003246-199905000-00020. [DOI] [PubMed] [Google Scholar]; c) Balasubramanian D, Schneper L, Kumari H, Mathee K. Nucleic Acids Res. 2013;41:1–20. doi: 10.1093/nar/gks1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kolodkin-Gal I, Romero D, Cao S, Clardy J, Kolter R, Losick R. Science. 2010;328:627–629. doi: 10.1126/science.1188628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hung DT, Shakhnovich EA, Pierson E, Mekalanos JJ. Science. 2005;310:670–674. doi: 10.1126/science.1116739. [DOI] [PubMed] [Google Scholar]
- 4.Firon N, Ashkenazi S, Mirelman D, Ofek I, Sharon N. Infect. Immun. 1987;55:472–476. doi: 10.1128/iai.55.2.472-476.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a) Hentzer M, Wu H, Andersen JB, Riedel K, Rasmussen TB, Bagge N, Kumar N, Schembri MA, Song Z, Kristoffersen P, Manefield M, Costerton JW, Molin S, Eberl L, Steinberg P, Kjelleberg S, Hoiby N, Givskov M. EMBO J. 2003;22:3803–3815. doi: 10.1093/emboj/cdg366. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Rasmussen TB, Givskov M. Int. J. Med. Microbiol. 2006;296:149–161. doi: 10.1016/j.ijmm.2006.02.005. [DOI] [PubMed] [Google Scholar]; c) Suga H, Smith KM. Curr. Opin. Chem. Biol. 2003;7:586–591. doi: 10.1016/j.cbpa.2003.08.001. [DOI] [PubMed] [Google Scholar]; d) Dong Y-H, Wang L-H, Xu J-L, Zhang H-B, Zhang X-F, Zhang L-H. Nature. 2001;411:813–817. doi: 10.1038/35081101. [DOI] [PubMed] [Google Scholar]
- 6.a) O'Connell KM, Hodgkinson JT, Sore HF, Welch M, Salmond GP, Spring DR. Angew. Chem. Int. Ed. 2013;52:10706–10733. doi: 10.1002/anie.201209979. [DOI] [PubMed] [Google Scholar]; b) Rutherford ST, Bassler BL. Cold Spring Harb. Perspect. Med. 2012;2:a012427. doi: 10.1101/cshperspect.a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a) Camilli A, Bassler BL. Science. 2006;311:1113–1116. doi: 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Waters CM, Bassler BL. Annu. Rev. Cell Dev. Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 8.a) Gambello MJ, Kaye S, Iglewski BH. Infect. Immun. 1993;61:1180–1184. doi: 10.1128/iai.61.4.1180-1184.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Fuqua WC, Winans SC, Greenberg EP. J. Bacteriol. 1994;176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.a) Lutter EI, Purighalla S, Duong J, Storey DG. Microbiology. 2012;158:2125–2132. doi: 10.1099/mic.0.054999-0. [DOI] [PubMed] [Google Scholar]; b) Zhu H, Bandara R, R. Conibear TC, Thuruthyil SJ, Rice SA, Kjelleberg S, Givskov M, P. Willcox MD. Invest. Ophthalmol. Vis. Sci. 2004;45:1897–1903. doi: 10.1167/iovs.03-0980. [DOI] [PubMed] [Google Scholar]; c) Wu H, Song Z, Givskov M, Doring G, Worlitzsch D, Mathee K, Rygaard J, Høiby N. Microbiology. 2001;147:1105–1113. doi: 10.1099/00221287-147-5-1105. [DOI] [PubMed] [Google Scholar]; d) Lesprit P, Faurisson F, Join-Lambert O, Roudot-Thoraval F, Foglino M, Vissuzaine C, Carbon C. Am. J. Respir. Crit. Care Med. 2003;167:1478–1482. doi: 10.1164/rccm.200207-736BC. [DOI] [PubMed] [Google Scholar]
- 10.a) Galloway W, Hodgkinson JT, Bowden SD, Welch M, Spring DR. Chem. Rev. 2011;111:28–67. doi: 10.1021/cr100109t. [DOI] [PubMed] [Google Scholar]; b) Geske GD, O'Neill JC, Blackwell HE. Chem. Soc. Rev. 2008;37:1432–1447. doi: 10.1039/b703021p. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Mattmann ME, Blackwell HE. J. Org. Chem. 2010;75:6737–6746. doi: 10.1021/jo101237e. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) O'Loughlin CT, Miller LC, Siryaporn A, Drescher K, Semmelhack MF, Bassler BL. Proc. Natl. Acad. Sci. U. S. A. 2013;110:17981–17986. doi: 10.1073/pnas.1316981110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.a) Geske GD, O'Neill JC, Miller DM, Wezeman RJ, Mattmann ME, Lin Q, Blackwell HE. ChemBioChem. 2008;9:389–400. doi: 10.1002/cbic.200700551. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Mattmann ME, Geske GD, Worzalla GA, Chandler JR, Sappington KJ, Greenberg EP, Blackwell HE. Bioorg. Med. Chem. Lett. 2008;18:3072–3075. doi: 10.1016/j.bmcl.2007.11.095. [DOI] [PubMed] [Google Scholar]; c) Geske GD, Mattmann ME, Blackwell HE. Bioorg. Med. Chem. Lett. 2008;18:5978–5981. doi: 10.1016/j.bmcl.2008.07.089. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Geske GD, O'Neill JC, Miller DM, Mattmann ME, Blackwell HE. J. Am. Chem. Soc. 2007;129:13613–13625. doi: 10.1021/ja074135h. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Geske GD, O'Neill JC, Blackwell HE. ACS Chem. Biol. 2007;2:315–319. doi: 10.1021/cb700036x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mattmann ME. Ph.D. thesis. Madison, WI: University of Wisconsin-Madison; 2010. [Google Scholar]
- 13.Amara N, Mashiach R, Amar D, Krief P, H. Spieser SA, Bottomley MJ, Aharoni A, Meijler MM. J. Am. Chem. Soc. 2009;131:10610–10619. doi: 10.1021/ja903292v. [DOI] [PubMed] [Google Scholar]
- 14.a) Kumar A, Schweizer HP. Adv. Drug Del. Rev. 2005;57:1486–1513. doi: 10.1016/j.addr.2005.04.004. [DOI] [PubMed] [Google Scholar]; b) Li X, Nikaido H. Drugs. 2009;69:1555–1623. doi: 10.2165/11317030-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Nikaido H, Pages JM. FEMS Microbiol. Rev. 2012;36:340–363. doi: 10.1111/j.1574-6976.2011.00290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Schweizer HP. Expert Opin. Drug Discovery. 2012;7:633–642. doi: 10.1517/17460441.2012.688949. [DOI] [PubMed] [Google Scholar]
- 15.Pearson JP, Van Delden C, Iglewski BH. J. Bacteriol. 1999;181:1203–1210. doi: 10.1128/jb.181.4.1203-1210.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evans K, Passador L, Srikumar R, Tsang E, Nezezon J, Poole K. J. Bacteriol. 1998;180:5443–5447. doi: 10.1128/jb.180.20.5443-5447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piddock LJV. Nat. Rev. Microbiol. 2006;4:629–636. doi: 10.1038/nrmicro1464. [DOI] [PubMed] [Google Scholar]
- 18.a) Minagawa S, Inami H, Kato T, Sawada S, Yasuki T, Miyairi S, Horikawa M, Okuda J, Gotoh N. BMC Microbiol. 2012;12:70. doi: 10.1186/1471-2180-12-70. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kondo A, Hirakata Y, Gotoh N, Fukushima K, Yanagihara K, Ohno H, Higashiyama Y, Miyazaki Y, Nishide K, Node M, Yamada Y, Kohno S, Kamihira S. Microbiol. Immunol. 2006;50:395–401. doi: 10.1111/j.1348-0421.2006.tb03806.x. [DOI] [PubMed] [Google Scholar]
- 19.Active efflux has also been implicated in other bacterial chemical signaling pathways, including Pseudomonas Quinolone Signaling. See: Lamarche MG, Déziel E. PLoS ONE. 2011;6:e24310. doi: 10.1371/journal.pone.0024310. Varga ZG, Armada A, Cerca P, Amaral L, Mior AA, Savka MA, Szegedi E, Kawase M, Motohashi N, Molnar J. in vivo. 2012;26:277–286.
- 20.Frei R, Breitbach AS, Blackwell HE. Angew. Chem. Int. Ed. 2012;51:5226–5229. doi: 10.1002/anie.201109258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Differences in the two assays and the mutant strains are likely origins for the different shifts of OdDHL potency/concentration observed in these two studies.
- 22.Schweizer HP. Gen. Mol. Res. 2003;2:48–62. [PubMed] [Google Scholar]
- 23.Lomovskaya O, Warren MS, Lee A, Galazzo J, Fronko R, Lee M, Blais J, Cho D, Chamberland S, Renau T, Leger R, Hecker S, Watkins W, Hoshino K, Ishida H, Lee VJ. Antimicrob. Agents Chemother. 2001;45:105–116. doi: 10.1128/AAC.45.1.105-116.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Some of the strongest LasR antagonists in pump-active PAO-JP2 displayed reduced LasR inhibitory activities in pump-mutant PAO-JG21 and E. coli JLD-271. After performing dose-response analyses on the compounds, we confirmed that those which displayed non-monotonic dose curves in pump-mutant PAO-JG21 (see Table 1) became potent agonists at 10 µM (see Figures S11-S13 for dose curves). These activity profiles can explain the reduced inhibitory actives observed for these compounds in the primary antagonism assay.
- 25.Rodgers JL, Nicewander WA. Am. Stat. 1988;42:59–66. [Google Scholar]
- 26.Walsh C. Nature. 2000;406:775–781. doi: 10.1038/35021219. [DOI] [PubMed] [Google Scholar]
- 27.McInnis CE, Blackwell HE. Biorg. Med. Chem. 2011;19:4812–4819. doi: 10.1016/j.bmc.2011.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morkunas B, Galloway WR, Wright M, Ibbeson BM, Hodgkinson JT, O'Connell KM, Bartolucci N, Della Valle M, Welch M, Spring DR. Org. Biomol. Chem. 2012;10:8452–8464. doi: 10.1039/c2ob26501j. [DOI] [PubMed] [Google Scholar]
- 29.Li X-Z, Nikaido H. Drugs. 2004;64:159–204. doi: 10.2165/00003495-200464020-00004. [DOI] [PubMed] [Google Scholar]
- 30.a) Mima T, Schweizer HP. Antimicrob. Agents Chemother. 2010;54:3113–3120. doi: 10.1128/AAC.01803-09. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Chan YY, Bian HS, Tan TM, Mattmann ME, Geske GD, Igarashi J, Hatano T, Suga H, Blackwell HE, Chua KL. J. Bacteriol. 2007;189:4320–4324. doi: 10.1128/JB.00003-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.a) Geske GD, Wezeman RJ, Siegel AP, Blackwell HE. J. Am. Chem. Soc. 2005;127:12762–12763. doi: 10.1021/ja0530321. [DOI] [PubMed] [Google Scholar]; b) Mattmann ME, Shipway PM, Heth NJ, Blackwell HE. ChemBioChem. 2011;12:942–949. doi: 10.1002/cbic.201000708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vogel HJ, Bonner DM. J. Biol. Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- 33.Pearson J, Pesci E, Iglewski B. J. Bacteriol. 1997;179:5756–5767. doi: 10.1128/jb.179.18.5756-5767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Durland RH, Eastman EM. Adv. Drug Del. Rev. 1998;30:33–48. doi: 10.1016/s0169-409x(97)00105-1. [DOI] [PubMed] [Google Scholar]
- 35.Simon R, O'Connell M, Labes M, Puhler A. Methods Enzymol. 1986;118:640–659. doi: 10.1016/0076-6879(86)18106-7. [DOI] [PubMed] [Google Scholar]
- 36.Lindsay A, M. Ahmer BM. J. Bacteriol. 2005;187:5054–5058. doi: 10.1128/JB.187.14.5054-5058.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. Gene. 1998;212:77–86. doi: 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 38.de Kievit TR, Gillis R, Marx S, Brown C, Iglewski BH. Appl. Environ. Microbiol. 2001;67:1865–1873. doi: 10.1128/AEM.67.4.1865-1873.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chugani SA, Whiteley M, Lee KM, D'Argenio D, Manoil C, Greenberg EP. Proc. Natl. Acad. Sci. U. S. A. 2001;98:2752–2757. doi: 10.1073/pnas.051624298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller WG, J. Leveau JH, Lindow SE. Mol. Plant-Microbe Interact. 2000;13:1243–1250. doi: 10.1094/MPMI.2000.13.11.1243. [DOI] [PubMed] [Google Scholar]
- 41.Lee JH, Lequette Y, Greenberg EP. Mol. Microbiol. 2006;59:602–609. doi: 10.1111/j.1365-2958.2005.04960.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.