Abstract
Three-dimensional organotypic culture using reconstituted basement membrane matrix (rBM 3-D) is an invaluable tool to characterize morphogenesis of epithelial cells and to elucidate the tumor-modulating actions of extracellular matrix. microRNAs (miRNA) are a novel class of tumor modulating genes. A substantial amount of investigation of miRNAs in cancer is carried out using monolayer 2-D culture on plastic substratum, which lacks a consideration of the matrix-mediated regulation of miRNAs. In the current study we compared the expression of miRNAs in rBM 3-D and 2-D cultures of two lung adenocarcinoma cell lines. Our findings revealed a profound difference in miRNA profiles between 2-D and rBM 3-D cultures of lung adenocarcinoma cells. The rBM 3-D culture-specific miRNA profile was highlighted with higher expression of the tumor suppressive miRNAs (i.e., miR-200 family) and lower expression of the oncogenic miRNAs (i.e., miR-17~92 cluster and miR-21) than that of 2-D culture. Moreover, the expression pattern of miR-17, miR-21, and miR-200a in rBM 3-D culture correlated with the expression of their targets and acinar morphogenesis, a differentiation behavior of lung epithelial cells in rBM 3-D culture. Over-expression of miR-21 suppressed its target PTEN and disrupted acinar morphogenesis. In summary, we provide the first miRNA profile of lung adenocarcinoma cells in rBM 3-D culture with respect to acinar morphogenesis. These results indicate that rBM 3-D culture is essential to a comprehensive understanding of the miRNA biology in lung epithelial cells pertinent to lung adenocarcinoma.
Keywords: microRNA, three-dimensional organotypic culture, extracellular matrix, lung epithelial cells, morphogenesis
1. Introduction
The tumor microenvironment consists of extracellular matrix (ECM), growth factors, and inflammatory cytokines that are crucial determinants of tumorigenesis (McAllister and Weinberg, 2010). During tumorigenesis, cancer cells progressively loose differentiation response to basement membrane matrix, such as apical-basolateral polarity (Paszek et al., 2005; Lee et al., 2007). In vivo properties of epithelial cells, such as apical-basolateral polarity, have been faithfully modeled using the three-dimensional organotypic culture based on reconstituted basement membrane matrix Matrigel (rBM 3-D) (Paszek et al., 2005; Hebner et al., 2008). Therefore, rBM 3-D culture provides an invaluable platform to elucidate the molecular mechanisms that mediate the dysregulated cellular responses to basement membrane matrix during tumorigenesis, which cannot be accomplished using 2-D culture (2-D) (Vargo-Gogola and Rosen, 2007). The clinical significance of rBM 3-D culture is supported by a recent study that indicates the gene expression signature of rBM 3-D culture of breast cancer cells holds prognostic values for patients with breast cancer (Martin et al., 2008). Besides breast cancer, rBM 3-D culture bears promising utilization in elucidating molecular and cell biology of lung cancer cells. In rBM 3-D culture, primary human lung alveolar type II cells form alveolar acini in which a polarized monolayer of alveolar type II cells form a single central lumen and express cell type specific markers, such as surfactant proteins (Yu et al., 2007). Because lung adenocarcinoma originates from alveolar type II cells, it is plausible that disruption of alveolar acini in rBM 3-D reflects a pivotal dedifferentiating step during the course of lung tumorigenesis. In support of this notion, over-expression of the tumor suppressive peroxisome proliferator-activated receptor- gene can restore alveolar acini in rBM 3-D culture of H2122 cells, an aggressive and poorly differentiated human lung adenocarcinoma cell line, as well as inhibition of tumor growth of the grafted H2122 cells (Bren-Mattison et al., 2005).
A microRNA (miRNA) is a ~22 nucleotide long non-coding RNA that inhibits gene expression via binding to its complementary sequences within the 3′ untranslated region (3′ UTR) of its target mRNAs (Trang et al., 2008). Profiling miRNAs in human cancer specimens and cell lines reveals a growing number of oncogenic and tumor suppressive miRNAs. Among the tumor suppressive miRNAs, the miR-200 family is frequently silenced in cancer (Peter, 2009). The oncogenic miR-17~92 cluster and miR-21 are often up-regulated in cancer (Volinia et al., 2006). Despite tremendous growth of our understanding of miRNAs, the expression and function of miRNAs have not been investigated in rBM 3-D culture that provides unique advantage to elucidate the cell biology pertinent to tumorigenesis, such as epithelial cell polarity and cross talk between the tumor cells and their microenvironment (Vargo-Gogola and Rosen, 2007). Therefore, the current study is aimed to identify the rBM 3-D culture-specific miRNA expression with respect to acinar morphogenesis via comparison of expression profiles of miRNAs of lung adenocarcinoma cells in rBM 3-D and 2-D cultures.
2. Materials and Methods
2.1. Cell Culture
A549 cells, a human lung adenocarcinoma cell line, were purchased from ATCC and cultured in DMEM (Sigma, St. Louis MO) as previously described (Shan and Morris, 2005). A549LC cells, a more aggressive subpopulation of A549 cells, were established from A549 cells using a murine model of lung metastasis as we previously described (Bao et al., 2004). Briefly, A549 cells were injected via the jugular vein at 106 cells/mouse into adult female beige-SCID mice (Charles River, Wilmington MA). Four months after injection, lungs were inspected and one metastatic nodule was excised, disaggregated and established in culture. mK-ras-LE cells, a murine lung epithelial cell line, was established from a lung tumor bearing K-rasLA1 mouse and cultured in RPMI-1640 as previously described (Li et al., 2011; Johnson et al., 2001). A miR-21 over-expressing variant of mK-ras-LE (mK-ras-LE-miR-21) was generated using a retroviral vector expressing a 314 nucleotide fragment of pri-miR-21 from the pMSCV-Puro-GFP backbone vector as we previously described (Nguyen et al., 2012). A control variant of mK-ras-LE (mK-ras-LE-vec) was generated in parallel using the backbone vector.
2.2. rBM three-dimensional organotypic culture
Overlay rBM 3-D culture was carried out as described elsewhere (Li et al., 2011; Yu et al., 2007). Briefly, A549 and mK-ras-LE cells were seeded at 2 × 105 cells/well in a 6-well cell culture plate that was coated with Matrigel (BD Biosciences, Rockville MD). DMEM culture medium was supplemented with 4% of Matrigel and replaced every two days. According to the time course of morphogenesis of lung epithelial cells in rBM 3-D culture the morphology of cell colonies was monitored for 12 days and recorded using an inverse phase contrast microscope equipped with a digital camera (Li et al., 2011). The morphogenesis was also visualized by staining for filamentous actin using Alexa 594 conjugated phalloidin (Invitrogen, Carlsbad CA) followed by con-focal fluorescent microscopy analysis on a Bio-Rad Radiance 2100 system (Bio-Rad, Hercules CA) in 2-D and rBM 3-D cultures as we previously described (Li et al., 2011; Shan et al., 2008).
2.3. RNA Extraction and Analysis of mRNA and miRNA Expression
Total cell RNA was extracted using Trizol (Invitrogen, Carlsbad CA) from 2-D culture and from rBM 3-D culture on day 12 post-seeding as we previously described (Li et al., 2011). Quantitative RT-PCR (qRT-PCR) was carried out to determine the expression of miR-17, miR-92b, miR-21, miR-200a, primary miR-21 (pri-miR-21), and vacuole membrane protein 1 (VMP1) as described elsewhere (Nguyen et al., 2012). The miRNAs were quantified using TaqMan assays (Invitrogen, Carlsbad CA). The sequences of the primers for pri-miR-21 and VMP1 were listed in Supplementary Materials. The gene expression was normalized to the values of U6 for miRNAs and to the values of 36B4 for VMP1 and pri-miR-21. A fold change of each RNA was obtained by setting the values from the control group to one. All the measurements were repeated on the RNA samples collected from three independent experiments.
2.4. miRNA microarray analysis
The miRNA expression profiling service was provided by LC Sciences (Houston, TX) using μParaflo® technology and proprietary probe hybridization (Cy3 and Cy5 dendrimer dyes) that enables highly sensitive and specific direct detection of mouse miRNAs. The miRNA chip was designed based on Sanger Institute mouse miRbase version 14 that included 700 mouse miRNAs. The miRNA microarrays of each culture condition were conducted on the RNA samples collected from three independent experiments. As advised by the service provider, any miRNA with a fluorescent intensity value below 100 in all of the compared groups was excluded from the analysis. Multi-array log2 transformation, normalization, and t-test were performed to identify the differentially expressed miRNAs between any two selected groups using the geWorkbench genomic analysis software suite (http://www.geworkbench.org) (Floratos et al., 2010; Floratos et al., 2011). False Discovery Rate (FDR) was calculated using the Significance Analysis of Microarrays suite (Tusher et al., 2001). The miRNAs that exhibited a difference of P < 0.01 and FDA < 0.05 between any two compared groups were considered significantly differentially expressed. The differentially expressed miRNAs were plotted in a heatmap using the geWorkbench Suite.
2.5. Immunoblots
Total cell lysates were extracted from cell colonies isolated from 2-D and rBM 3-D culture using Laemmli buffer as we previously described (Li et al., 2011). The expression of the proteins of interest was determined using immunoblots. Cell Signaling Technology (Danvers, MA) provided the primary antibodies specific for β-actin, Zinc finger E-box-binding homeobox 2 (ZEB2), phosphatase and tensin homolog (PTEN). The IRDye800-conjugated secondary antibodies were used at a dilution of 1:15,000 and detected using an Odyssey Infrared Imaging System (Licor Biosciences, Lincoln, NE) (Shan et al., 2010). The results were representative of two independent experiments.
2.6. Statistical analysis
Statistical significance in the difference between any two selected groups was determined using the unpaired two-tailed Student T test (Prizm Version 5). A P value < 0.05 was considered significant.
3. Results
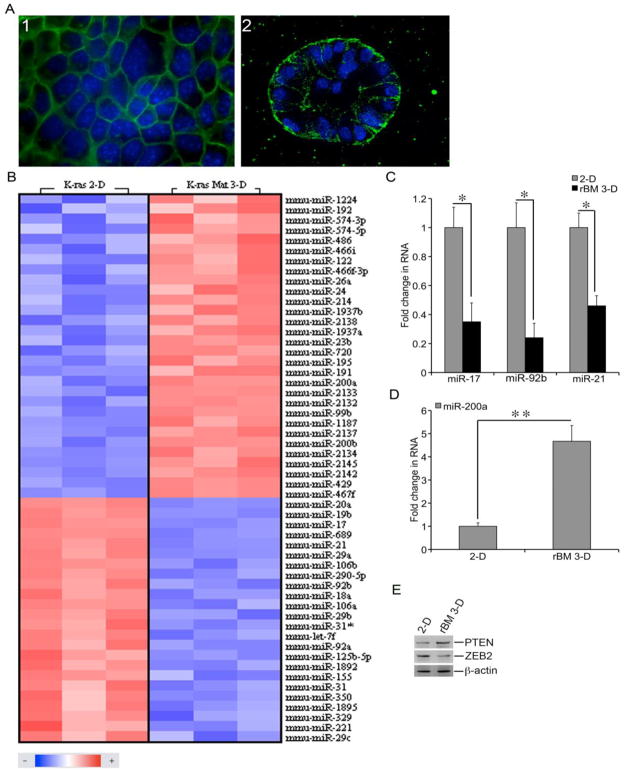
A recent study has uncovered a profound difference in global gene expression between 2-D and rBM 3-D cultures of breast cancer cells (Kenny et al., 2007). Because normal lung alveolar type II epithelial cells and well-differentiated lung adencarcinoma cells undergo acinar morphogenesis in rBM 3-D culture, we set to profile miRNA expression of the well-differentiated lung adenocarcinoma cells (Li et al., 2011; Bren-Mattison et al., 2005; Yu et al., 2007). We established rBM 3-D culture of m-K-ras-LE and A549 cells because they maintain the signature traits of alveolar type II cells (Li et al., 2011; Johnson et al., 2001; Wang et al., 2007). In 2-D culture, mK-ras-LE cells exhibited typical epithelial morphology as evidenced by cortical distribution of F-actin (Figure 1, A1). In rBM 3-D culture mK-ras-LE cells underwent acinar morphogenesis that featured a monolayer of polarized cells enclosing a single central lumen, a characteristic of normal alveolar type II cells (Figure 1, A2) (Johnson et al., 2001; Yu et al., 2007). To identify the rBM 3-D responsive miRNAs, we compared the miRNA profiles between 2-D and rBM 3-D cultures of mK-ras-LE cells using miRNA microarrays. Out of 154 mouse miRNAs considered for analysis, 54 miRNAs were significantly differentially expressed between 2-D and rBM 3-D cultures using the criteria as defined in the section of Materials and Methods (Figure 1C). Within this panel of miRNAs, 39 miRNAs exhibited a >2-fold difference with 22 miRNAs being higher and 17 miRNAs being lower in rBM 3-D than that in 2-D (Table 1). The elevated miRNAs featured several well-established tumor suppressive miRNAs, such as miR-192 and three members of the miR-200 family (miR-200a, miR-200b, and miR-429) (Kim et al., 2011; Wang et al., 2010; Wang et al. 2011; Peter, 2009). The repressed miRNAs featured several well-established oncogenic miRNAs, such as miR-21, miR-155, and seven members of the miR-17~92 cluster and miR-17 family (miR-17, miR-18a, miR-19b, miR-20a, miR-92b, miR-106a, and miR-106b) (Osada and Takahashi, 2011; Volinia et al., 2006; Mendell, 2008; Krichevsky and Gabriely, 2009). The differential expression of four miRNAs from Table 1 was further confirmed using qRT-PCR. The expression of the oncogenic miRNAs miR-17, miR-92b, and miR-21 was reduced in rBM 3-D culture to 35%, 24%, and 46% of their corresponding values in 2-D culture, respectively (Figure 1C). In contrast miR-200a exhibited 4.7-fold increase in rBM 3-D culture over that in 2-D culture (Figure 1D). Because PTEN is a common target of the miR-17~92 cluster and miR-21, we questioned whether the protein levels of PTEN were up-regulated in rBM 3-D culture as a consequence of a decrease in their suppressors miR-17~92 cluster and miR-21 (Meng et al., 2007; Xiao et al., 2008). As expected, the protein levels of PTEN were substantially higher in rBM 3-D than 2-D (Figure 1E). In contrast, the protein levels of ZEB2, a target of the miR-200 family, were lower in rBM 3-D than 2-D, which was consistent with the increased expression of the miR-200 family in rBM 3-D culture (Figure 1E) (Park et al., 2008). These findings demonstrate an inverse correlation between the rBM 3-D responsive miRNAs and their targets.
Figure 1.
Distinct miRNA profiles of mK-ras-LE cells in 2-D and rBM 3-D cultures. A) mK-ras-LE cells were grown in 2-D and rBM 3-D culture. Morphology of the cells was visualized using fluorescent staining for F-actin. The images of rBM 3-D culture were captured at the central planes of cell clusters using a confocal fluorescent microscope on day 12 post-seeding. The images were captured at 600x magnification. Representative images of each culture condition were displayed. B) Total cell RNA was extracted from mK-ras-LE cells in 2-D and rBM 3-D cultures. miRNA expression profiles were compared between the two culture conditions using miRNA microarrays. A heatmap of the significantly differentially expressed miRNAs was generated using geWorkbench suite (n = 3, P < 0.01, FDR < 0.05). C) The expression of miR-17, miR-92b, and miR-21 was measured using qRT-PCR in total cell RNA collected from 2-D and rBM 3-D cultures of mK-ras-LE cells. A fold change of each miRNA of interest was obtained by setting the values of each miRNA in 2-D culture to one and normalization to the values of U6. D) Similar to part C except that the expression of miR-200a was compared between 2-D and rBM 3-D cultures. E) The culture conditions were similar to part A. The cell lysates were collected from the cell colonies extracted from the culture. The protein levels of ZEB2, PTEN, and -actin were determined using immunoblots. The data were presented in averages and standard deviations obtained from three independent experiments. and indicated a P value < 0.05 and 0.01, respectively.
Table 1.
Disparate expression of miRNAs between rBM 3-D and 2-D cultures of mK-ras-LE cells. Total cell RNA was extracted from rBM 3-D and 2-D cultures. miRNA arrays were carried out and analyzed as described in Methods. A fold change of each miRNA was obtained by setting the values from 2-D culture to one. The results were average of three miRNA microarrays. The miRNAs bearing documented tumor-modulating properties were highlighted in bold.
| rBM 3-D/2-D
| |||
|---|---|---|---|
| Lower Expression | Higher Expression | ||
|
| |||
| Gene Name | Fold Change | Gene Name | Fold Change |
| mmu-miR-689 | 0.03 | mmu-miR-1937b | 2.06 |
| mmu-miR-19b | 0.09 | mmu-miR-99b | 2.17 |
| mmu-miR-106a | 0.09 | mmu-miR-2142 | 2.22 |
| mmu-miR-29b | 0.11 | mmu-miR-466f-3p | 2.25 |
| mmu-miR-20a | 0.19 | mmu-miR-200a | 2.33 |
| mmu-miR-18a | 0.23 | mmu-miR-2133 | 2.39 |
| mmu-miR-106b | 0.24 | mmu-miR-23b | 2.47 |
| mmu-miR-350 | 0.26 | mmu-miR-574-5p | 2.61 |
| mmu-miR-29c | 0.26 | mmu-miR-122 | 2.72 |
| mmu-miR-155 | 0.27 | mmu-miR-2137 | 2.75 |
| mmu-miR-17 | 0.30 | mmu-miR-2138 | 3.06 |
| mmu-miR-29a | 0.34 | mmu-miR-192 | 3.15 |
| mmu-miR-31* | 0.41 | mmu-miR-2134 | 3.23 |
| mmu-miR-21 | 0.42 | mmu-miR-195 | 3.35 |
| mmu-miR-92b | 0.42 | mmu-miR-191 | 3.40 |
| mmu-miR-290-5p | 0.46 | mmu-miR-2132 | 3.47 |
| mmu-let-7f | 0.47 | mmu-miR-467f | 3.71 |
| mmu-miR-466i | 4.01 | ||
| mmu-miR-200b | 4.20 | ||
| mmu-miR-1187 | 6.02 | ||
| mmu-miR-429 | 6.31 | ||
| mmu-miR-214 | 7.15 | ||
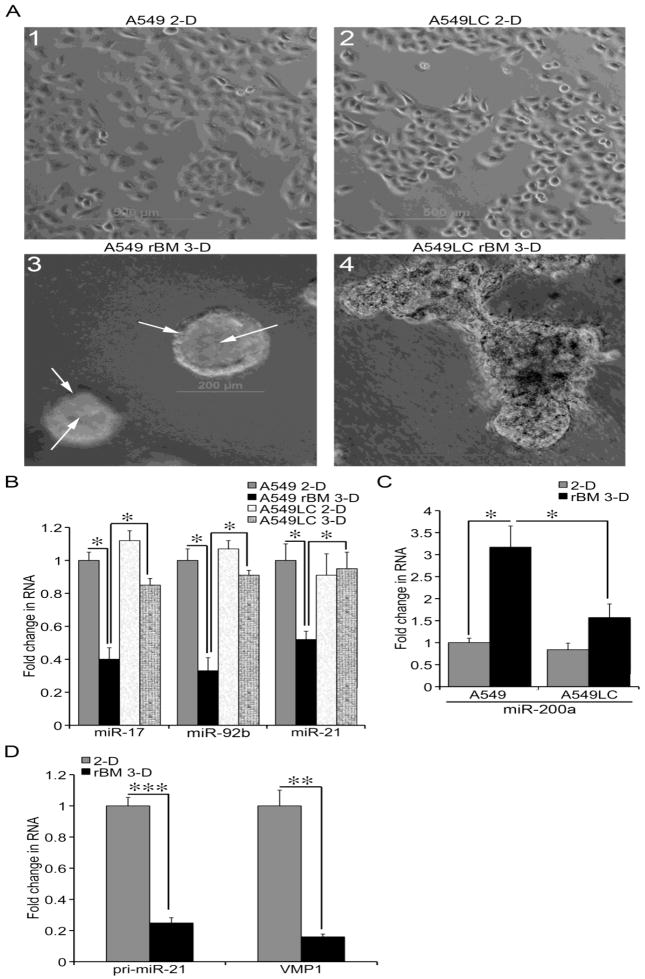
We then questioned whether human lung adenocarcinoma cells exhibited similar differential expression of miRNAs between 2-D and rBM 3-D cultures. We employed the well-differentiated A549 cells and its more aggressive derivative A549LC cells that were established as described in the section of Materials and Methods. When grafted subcutaneously into the flank of nude mice, A549 presented organized glandular structure that mimicked acini in rBM 3-D culture, whereas A549LC cells displayed disorganized architecture and lacked the glandular histology (unpublished observations). Moreover, the grafted A549LC cells doubled the growth of the implanted parental A549 cells in terms of tumor weight (unpublished observations). Despite the distinct histology and tumorigenic behavior in vivo, morphology of A549 and A549LC cells was nearly identical in 2-D culture (Figure 2, A1 & A2). However morphogenesis of A549 and A549LC cells was distinct in rBM 3-D culture (Figure 2, A3 & A4). Similar to normal alveolar type II epithelial cells, A549 cells formed acini, a polarized cell sphere with a single central lumen in rBM 3-D culture (Figure 2A3, as pointed by the arrows) (Li et al., 2011; Bren-Mattison et al., 2005; Yu et al., 2007). In contrast A549LC cells exhibited mass morphology that featured irregular cell clusters void of a central lumen, which resembled morphogenesis of H2122 cells, a poorly differentiated lung adenocarcinoma cell line in rBM 3-D culture (Figure 2A4) (Bren-Mattison et al., 2005). We then questioned whether the rBM 3-D culture-specific difference in morphology between A549 and A549LC cells correlated with differential expression of the tumor modulating miRNAs as revealed in mK-ras-LE cells (Table 1). To this end, we compared the expression of miR-17, miR-92b, miR-21, and miR-200a between A549 and A549LC cells in 2-D and rBM 3-D cultures. Congruent to their indistinguishable morphology in 2-D culture, the expression of these four miRNAs did not exhibit significant difference between A549 and A549LC cells (Figure 2, B & C). On the other hand, only A549, but not A549LC cells exhibited a substantial decrease in the oncogenic miR-17, miR-92b, and miR-21, and a substantial increase in the tumor suppressive miR-200a in rBM 3-D culture over that in 2-D culture (Figure 2, B & C). Biogenesis of miR-21 can be regulated at the levels of transcriptional production of pri-miR-21, processing from pri-miR-21 to pre-miR-21, and pre-miR-21 to mature 21 (Ribas and Lupold, 2010). To determine how rBM 3-D culture repressed the expression of miR-21, we compared the expression of pri-miR-21 between rBM 3-D and 2-D cultures. The primers for pri-miR-21 were designed to specifically amplify pri-miR-21, but not the overlapping 3′ UTR of its upstream gene, VMP1 (see Supplementary Materials). The RNA levels of pri-miR-21 in rBM 3-D cultured were reduced to 25% of that in 2-D culture (Figure 2D). Because the transcription start site of pri-miR-21 is located within the 3′ UTR of VMP1, we questioned whether the expression of VMP1 was co-regulated with pri-miR-21 in rBM 3-D culture (Ribas and Lupold, 2010). Therefore, we compared the expression of VMP1 between rBM 3-D and 2-D cultures. The primers of VMP1 were designed to specifically amplify VMP1, but not the overlapping pri-miR-21 (see Supplementary Materials). Similar to the response of pri-miR-21 to rBM, the mRNA levels of VMP1 were reduced to 16% of that in 2-D culture. These findings from mK-ras-LE, A549, and A549LC cells reveal a strong correlation between the miRNA expression profile and acinar morphogenesis in rBM 3-D culture.
Figure 2.
miRNA expression associated with aincar morphogenesis. A) A549 and A549LC cells were cultured in 2-D (A1 & A2) and rBM 3-D (A3 & A4) cultures. Morphology was recorded using an inverse phase contrast microscope. Polarized monolayer of the cells and the central lumen were pointed at by white arrows. B) The expression of miR-17, miR-92, and miR-21 was measured using qRT-PCR in total cell RNA collected from 2-D and rBM 3-D of A549 and A549LC cells. A fold change of each miRNA was obtained by setting the values of each miRNA from A549 cells in 2-D culture to one and normalization to the values of U6. C) Similar to part A except that the expression of miR-200a was compared across the groups. D) The culture conditions were similar to part A. The RNA levels of pri-miR-21 and VMP1 were measured in A549 cells using qRT-PCR. A fold change of each RNA was obtained by setting the values of RNA from A549 cells in 2-D culture to one and normalization to the values of 36B4. The data were presented in averages and standard deviations obtained from three independent experiments. , , and indicated a P value < 0.05, 0.01, and 0.001, respectively.
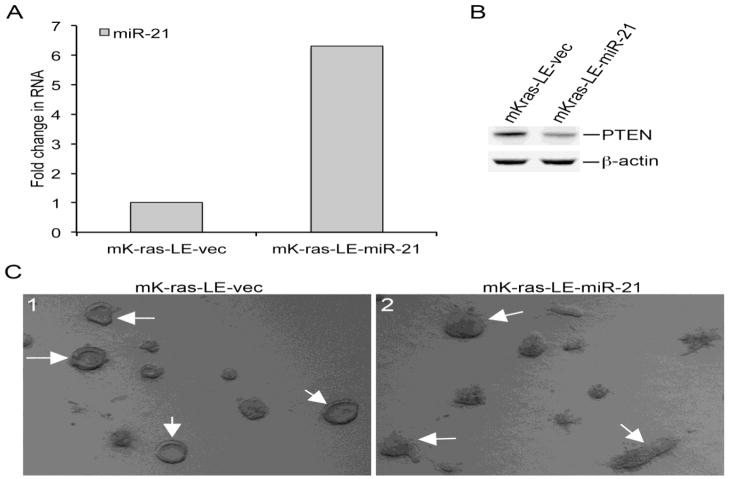
Among the rBM 3-D culture responsive miRNAs we chose to examine the role of miR-21 in acinar morphogenesis because miR-21 targets PTEN, an essential regulator of acinar morphogenesis (Fournier et al., 2009; Ma et al., 2011). Stable over-expression of miR-21 was established using retroviral transduction. The miR-21 overexpressing mK-ras-LE-miR-21 variant exhibited a more than 6-fold increase in the RNA levels of miR-21 over the control mK-ras-LE-vec variant (Figure 3A). Moreover, the protein levels of PTEN were substantially lower in mK-ras-LE-miR-21 than that in mK-ras-LE-vec in rBM 3-D culture (Figure 3B). A similar reduction of PTEN was also observed in 2-D culture of mK-ras-LE-miR-21 (data not shown). Consistent with a decrease in PTEN, mK-ras-LE-miR-21 cells failed to form acini in rBM 3-D and grew into irregular cell cluster void of a central lumen, whereas mK-ras-LE-vec underwent acinar morphogenesis (Figure 3C). These findings suggest that miR-21 regulates acinar morphogenesis via its target PTEN.
Figure 3.
Disruption of aincar morphogenesis by overexpression of miR-21. A) The RNA levels of miR-21 were measured in mK-ras-LE-miR-21 and mK-ras-LE-vec variants using qRT-PCR. A fold increase in miR-21 was determined by setting the values from mK-ras-LE-vec to one. B) Total cell lysates were extracted from mK-ras-LE-miR-21 and mK-ras-LE-vec variants in rBM 3-D culture. The protein levels of PTEN were determined using immunoblots. C) mK-ras-LE-miR-21 and mK-ras-LE-vec variants were monitored in rBM 3-D culture for 12 days. Morphogenesis of the mK-ras-LE variants in rBM 3-D culture was recorded using an inverse phase contrast microscope. Representative cell colonies were pointed at by white arrows.
4. Discussion
A recent gene expression profiling of mammary epithelial cells lines of diverse tumorigenic properties in 2-D and rBM 3-D cultures identifies the gene expression signatures of distinct morphogenesis (Kenny et al., 2007). Using a similar strategy, we establish a miRNA expression profile that correlates with acinar morphogensis of lung adenocarcinoma cells in rBM 3-D culture. The acinar morphogenesis associated miRNA profile provides unique and novel insight to guide future investigations of morphogenesis and tumorigenesis of lung epithelial cells as pioneered by several recent studies (Wu et al., 2011; Bren-Mattison et al., 2005; Yu et al., 2007).
In our comparison of the miRNA profiles between 2-D and rBM 3-D cultures of mK-ras-LE cells, a considerable portion of the differentially expressed miRNAs bear well-documented tumor-modulating activities (Figure 1 & Table 1). For instance, the tumor-promoting miR-17~92 cluster and miR-21 are substantially repressed in rBM 3-D culture of mK-ras-LE cells relative to that in 2-D culture (Figure 1 & Table 1) (Osada and Takahashi, 2011; Volinia et al., 2006; Mendell, 2008; Krichevsky and Gabriely, 2009). On the other hand, the tumor suppressive miR-200 family and miR-192 are activated in rBM 3-D culture (Figure 1 & Table 1) (Kim et al., 2011; Wang et al., 2010; Wang et al. 2011; Peter, 2009). Because rBM 3-D culture promotes differentiation of lung epithelial cells, a decrease in oncogenic miRNAs and an increase in tumor suppressive miRNAs strongly suggest that miRNAs regulate differentiation response of lung epithelial cells to basement membrane matrix (Wu et al., 2011; Bren-Mattison et al., 2005; Yu et al., 2007). Indeed, the difference in the expression of the miR-17~92 cluster, miR-21, and the miR-200 family between 2-D and rBM 3-D cultures matches the difference in the expression of their target genes between the two culture conditions (Figure 1E). Moreover over-expression of miR-21 alone is sufficient to suppress PTEN and disrupt acinar morphogenesis, which strongly suggests that down-regulation of miR-21 facilitates acinar morphogenesis via de-repression of the expression of PTEN (Figure 3). Correlation of an increase in members of miR-200 family with acinar morphogenesis also implicates a role of epithelial mesenchymal transition in acinar morphogenesis of lung epithelial cells because the miR-200 family is one of the most potent suppressors of epithelial-mesenchymal transition (Figure 1 & 2) (Peter, 2009). We further extrapolate that the expression profile of miR-21, miR-17, miR-92b, and miR-200a in rBM 3-D culture represents a miRNA signature of acinar morphogenesis because these four miRNA are differentially expressed 1) between 2-D and rBM 3-D cultures of mK-ras-LE cells; 2) between A549 cells of acinar morphogenesis and A549LC cells of mass morphogenesis (Figure 1 & 2). These findings warrant further investigation whether the identified miRNA candidates regulate acinar morphogenesis.
Besides differentiation promoting signals from rBM, rBM 3-D culture is also more compliant and less stiff than 2-D culture (Hebner et al., 2008; Butcher et al., 2009). A stiff and fibrotic tumor microenvironment promotes tumorigenesis and the aberrantly stiff extracellular matrix is a well-documented risk factor of breast cancer (Butcher et al., 2009). Recent advances in lung cancer research indicate a similar presence and function of a fibrotic tumor microenvironment. For instance, the expression of fibrogenic mediators TGF-β1 and fibronectin is up-regulated in human lung cancer and promote invasion and metastasis in experimental models of lung cancer (Toonkel et al., 2010; Han et al., 2006; Han and Roman, 2006; Soltermann et al., 2008). Thus, the differential expression of the tumor-modulating miRNAs between 2-D and rBM 3-D implicates that stiffness up-regulates the tumor-promoting miRNAs and down-regulates the tumor suppressive miRNAs to promote tumorigenesis. In support of this postulate, our recent report indicates that type I collagen, a stiffer matrix than rBM, can up-regulate the expression of miR-21 and the miR-17~92 cluster in rBM 3-D culture of mK-ras-LE and A549 cells (Li et al., 2011). Moreover, miR-21 is up-regulated in pulmonary fibrois induced by TGF-β1 and bleomycin (Liu et al., 2011). These findings raise the possibility that miRNAs transmit the tumor promoting actions of the stiff tumor microenvironment.
The expression of miR-21 can be regulated via diverse mechanisms. miR-21 can be transcriptionally activated by STAT3 and androgen receptors that bind to the cis elements in the miR-21 promoter (Ribas and Lupold, 2011). miR-21 can also be post-transcriptionally up-regulated in response to extracellular cues. The SMAD proteins promote processing of pri-miR-21 to pre-miR-21 in vascular smooth muscle cells exposed to TGF-β1 or BMP (Davis et al., 2008). Type I collagen is suggested to up-regulate miR-21 via promoting processing of pre-miR-21 to mature miR-21 in rBM 3-D culture (Li et al., 2011). In the current study, our results implicate that rBM 3-D culture transcriptionally represses the expression of miR-21 because the expression of pri-miR-21 is reduced to a greater extent than that of mature miR-21 in rBM 3-D culture over that in 2-D culture (Figure 2). Our results further suggest that miR-21 is co-regulated with its upstream protein-coding gene VMP1. VMP1 regulates a variety of cell behaviors including autophagy, cell adhesion, and membrane traffic (Calvo-Garrido et al., 2008). miR-21 can potentially regulate these cellular behaviors via targeting the protein-coding genes. For instance, miR-21 can indirectly regulate autophagy via targeting PTEN, a negative regulator of the Akt-mTOR axis that is crucial to autophagy (Janku et al.; Meng et al., 2007). Because autophagy has emerged as a pivotal regulator of acinar morphogenesis, it is plausible that co-regulated expression of VMP1, PTEN, and miR-21 participates in fine-tuning of authophagy in acinar morphogenesis (Debnath, 2008).
5. Conclusion
To the best of our knowledge, the current study provides the first miRNA expression profile that correlates with acinar morphogenesis of lung adenocarcinoma cells in rBM 3-D culture. Our findings have established a platform and a reference miRNA catologue to guide future investigations of miRNAs in acinar morphogenesis with respect to lung adenocarcinoma.
Supplementary Material
HIGH LIGHTS.
Distinct miRNA profiles between rBM 3-D and 2-D cultures of lung epithelial cells.
Enrichment of the tumor-modulating miRNAs in the rBM 3-D responsive miRNAs.
Correlation of tumor modulating miRNAs and acinar morphogenesis in rBM 3-D culture.
Acknowledgments
This work is supported NIH CA132603 awarded to G.M., B.S. and D.S. H.T.N. is supported in part by Developmental Funds of the Tulane Cancer Center. geWorkbench is an open source genomic analysis platform developed at Columbia University with funding from the NIH Roadmap Initiative (1U54CA121852-01A1) and the National Cancer Institute.
List of abbreviations
- ECM
extracellular matrix
- BM
basement membrane matrix
- rBM 3-D
reconstituted basement membrane matrix based three-dimensional organotypic culture
- miRNA
microRNA
- F-actin
filamentous actin
- qRT-PCR
quantitative RT-PCR
- pri-miR-21
primary miR-21
- VMP1
vacuole membrane protein 1
- 3′ UTR
3′ untranslated region
- ZEB2
Zinc finger E-box-binding homeobox 2
- PTEN
phosphatase and tensin homolog
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bao L, Jaligam V, Zhang XY, Kutner RH, Kantrow SP, Reiser J. Stable transgene expression in tumors and metastases after transduction with lentiviral vectors based on human immunodeficiency virus type 1. Hum Gene Ther. 2004;15:445–56. doi: 10.1089/10430340460745775. [DOI] [PubMed] [Google Scholar]
- Bren-Mattison Y, Van Putten V, Chan D, Winn R, Geraci MW, Nemenoff RA. Peroxisome proliferator-activated receptor-gamma (PPAR(gamma)) inhibits tumorigenesis by reversing the undifferentiated phenotype of metastatic non-small-cell lung cancer cells (NSCLC) Oncogene. 2005;24:1412–22. doi: 10.1038/sj.onc.1208333. [DOI] [PubMed] [Google Scholar]
- Butcher DT, Alliston T, Weaver VM. A tense situation: forcing tumour progression. Nat Rev Cancer. 2009;9:108–22. doi: 10.1038/nrc2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo-Garrido J, Carilla-Latorre S, Escalante R. Vacuole membrane protein 1, autophagy and much more. Autophagy. 2008;4:835–7. doi: 10.4161/auto.6574. [DOI] [PubMed] [Google Scholar]
- Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath J. Detachment-induced autophagy during anoikis and lumen formation in epithelial acini. Autophagy. 2008;4:351–3. doi: 10.4161/auto.5523. [DOI] [PubMed] [Google Scholar]
- Floratos A, Honig B, Pe’er D, Califano A. Using systems and structure biology tools to dissect cellular phenotypes. J Am Med Inform Assoc. 2010;19:171–5. doi: 10.1136/amiajnl-2011-000490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floratos A, Smith K, Ji Z, Watkinson J, Califano A. geWorkbench: an open source platform for integrative genomics. Bioinformatics. 2011;26:1779–80. doi: 10.1093/bioinformatics/btq282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier MV, Fata JE, Martin KJ, Yaswen P, Bissell MJ. Interaction of E-cadherin and PTEN regulates morphogenesis and growth arrest in human mammary epithelial cells. Cancer research. 2009;69:4545–52. doi: 10.1158/0008-5472.CAN-08-1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Khuri FR, Roman J. Fibronectin stimulates non-small cell lung carcinoma cell growth through activation of Akt/mammalian target of rapamycin/S6 kinase and inactivation of LKB1/AMP-activated protein kinase signal pathways. Cancer Res. 2006;66:315–23. doi: 10.1158/0008-5472.CAN-05-2367. [DOI] [PubMed] [Google Scholar]
- Han SW, Roman J. Fibronectin induces cell proliferation and inhibits apoptosis in human bronchial epithelial cells: pro-oncogenic effects mediated by PI3-kinase and NF-kappa B. Oncogene. 2006;25:4341–9. doi: 10.1038/sj.onc.1209460. [DOI] [PubMed] [Google Scholar]
- Hebner C, Weaver VM, Debnath J. Modeling morphogenesis and oncogenesis in three-dimensional breast epithelial cultures. Annu Rev Pathol. 2008;3:313–39. doi: 10.1146/annurev.pathmechdis.3.121806.151526. [DOI] [PubMed] [Google Scholar]
- Janku F, McConkey DJ, Hong DS, Kurzrock R. Autophagy as a target for anticancer therapy. Nat Rev Clin Oncol. 8:528–39. doi: 10.1038/nrclinonc.2011.71. [DOI] [PubMed] [Google Scholar]
- Johnson L, Mercer K, Greenbaum D, Bronson RT, Crowley D, Tuveson DA, Jacks T. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–6. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- Kenny PA, Lee GY, Myers CA, Neve RM, Semeiks JR, Spellman PT, Lorenz K, Lee EH, Barcellos-Hoff MH, Petersen OW, Gray JW, Bissell MJ. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol Oncol. 2007;1:84–96. doi: 10.1016/j.molonc.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Veronese A, Pichiorri F, Lee TJ, Jeon YJ, Volinia S, Pineau P, Marchio A, Palatini J, Suh SS, Alder H, Liu CG, Dejean A, Croce CM. p53 regulates epithelial-mesenchymal transition through microRNAs targeting ZEB1 and ZEB2. J Exp Med. 2011;208:875–83. doi: 10.1084/jem.20110235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky AM, Gabriely G. miR-21: a small multi-faceted RNA. J Cell Mol Med. 2009;13:39–53. doi: 10.1111/j.1582-4934.2008.00556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GY, Kenny PA, Lee EH, Bissell MJ. Three-dimensional culture models of normal and malignant breast epithelial cells. Nat Methods. 2007;4:359–65. doi: 10.1038/nmeth1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Nguyen HT, Zhuang Y, Lin Y, Flemington EK, Guo W, Guenther J, Burow ME, Morris GF, Sullivan D, Shan B. Post-transcriptional up-regulation of miR-21 by type I collagen. Mol Carcinog. 2011;50:563–70. doi: 10.1002/mc.20742. [DOI] [PubMed] [Google Scholar]
- Liu G, Friggeri A, Yang Y, Milosevic J, Ding Q, Thannickal VJ, Kaminski N, Abraham E. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J Exp Med. 207:1589–97. doi: 10.1084/jem.20100035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Kumar M, Choudhury SN, Becker Buscaglia LE, Barker JR, Kanakamedala K, Liu MF, Li Y. Loss of the miR-21 allele elevates the expression of its target genes and reduces tumorigenesis. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:10144–9. doi: 10.1073/pnas.1103735108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin KJ, Patrick DR, Bissell MJ, Fournier MV. Prognostic breast cancer signature identified from 3D culture model accurately predicts clinical outcome across independent datasets. PLoS One. 2008;3:e2994. doi: 10.1371/journal.pone.0002994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister SS, Weinberg RA. Tumor-host interactions: a far-reaching relationship. J Clin Oncol. 2010;28:4022–8. doi: 10.1200/JCO.2010.28.4257. [DOI] [PubMed] [Google Scholar]
- Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–22. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–58. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HT, Li C, Lin Z, Zhuang Y, Flemington EK, Burow ME, Lin Y, Shan B. The microRNA expression associated with morphogenesis of breast cancer cells in three-dimensional organotypic culture. Oncology reports. 2012;28:117–26. doi: 10.3892/or.2012.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osada H, Takahashi T. let-7 and miR-17-92: small-sized major players in lung cancer development. Cancer Sci. 2011;102:9–17. doi: 10.1111/j.1349-7006.2010.01707.x. [DOI] [PubMed] [Google Scholar]
- Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, Hammer DA, Weaver VM. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–54. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Peter ME. Let-7 and miR-200 microRNAs: guardians against pluripotency and cancer progression. Cell Cycle. 2009;8:843–52. doi: 10.4161/cc.8.6.7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribas J, Lupold SE. The transcriptional regulation of miR-21, its multiple transcripts, and their implication in prostate cancer. Cell Cycle. 2010;9:923–9. doi: 10.4161/cc.9.5.10930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan B, Hagood JS, Zhuo Y, Nguyen HT, MacEwen M, Morris GF, Lasky JA. Thy-1 attenuates TNF-alpha-activated gene expression in mouse embryonic fibroblasts via Src family kinase. PLoS One. 2010;5:e11662. doi: 10.1371/journal.pone.0011662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan B, Morris GF. Binding sequence-dependent regulation of the human proliferating cell nuclear antigen promoter by p53. Exp Cell Res. 2005;305:10–22. doi: 10.1016/j.yexcr.2004.09.033. [DOI] [PubMed] [Google Scholar]
- Shan B, Yao TP, Nguyen HT, Zhuo Y, Levy DR, Klingsberg RC, Tao H, Palmer ML, Holder KN, Lasky JA. Requirement of HDAC6 for transforming growth factor-beta1-induced epithelial-mesenchymal transition. J Biol Chem. 2008;283:21065–73. doi: 10.1074/jbc.M802786200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltermann A, Tischler V, Arbogast S, Braun J, Probst-Hensch N, Weder W, Moch H, Kristiansen G. Prognostic significance of epithelial-mesenchymal and mesenchymal-epithelial transition protein expression in non-small cell lung cancer. Clin Cancer Res. 2008;14:7430–7. doi: 10.1158/1078-0432.CCR-08-0935. [DOI] [PubMed] [Google Scholar]
- Toonkel RL, Borczuk AC, Powell CA. Tgf-beta signaling pathway in lung adenocarcinoma invasion. J Thorac Oncol. 5:153–7. doi: 10.1097/JTO.0b013e3181c8cc0c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trang P, Weidhaas JB, Slack FJ. MicroRNAs as potential cancer therapeutics. Oncogene. 2008;27(Suppl 2):S52–7. doi: 10.1038/onc.2009.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargo-Gogola T, Rosen JM. Modelling breast cancer: one size does not fit all. Nat Rev Cancer. 2007;7:659–72. doi: 10.1038/nrc2193. [DOI] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Herman-Edelstein M, Koh P, Burns W, Jandeleit-Dahm K, Watson A, Saleem M, Goodall GJ, Twigg SM, Cooper ME, Kantharidis P. E-cadherin expression is regulated by miR-192/215 by a mechanism that is independent of the profibrotic effects of transforming growth factor-beta. Diabetes. 2010;59:1794–802. doi: 10.2337/db09-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Haviland DL, Burns AR, Zsigmond E, Wetsel RA. A pure population of lung alveolar epithelial type II cells derived from human embryonic stem cells. Proc Natl Acad Sci U S A. 2007;104:4449–54. doi: 10.1073/pnas.0700052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Peng B, Wang D, Ma X, Jiang D, Zhao J, Yu L. Human tumor microRNA signatures derived from large-scale oligonucleotide microarray datasets. Int J Cancer. 2011;129:1624–34. doi: 10.1002/ijc.25818. [DOI] [PubMed] [Google Scholar]
- Wu X, Peters-Hall JR, Bose S, Pena MT, Rose MC. Human bronchial epithelial cells differentiate to 3D glandular acini on basement membrane matrix. Am J Respir Cell Mol Biol. 2011;44:914–21. doi: 10.1165/rcmb.2009-0329OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL, Rajewsky K. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nature immunology. 2008;9:405–14. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Fang X, Ewald A, Wong K, Hunt CA, Werb Z, Matthay MA, Mostov K. Formation of cysts by alveolar type II cells in three-dimensional culture reveals a novel mechanism for epithelial morphogenesis. Mol Biol Cell. 2007;18:1693–700. doi: 10.1091/mbc.E06-11-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.