Abstract
Evidence implicating dysregulation of the IRE1/XBP-1s arm of the unfolded protein response (UPR) in cancer pathogenesis (e.g., multiple myeloma) has prompted the development of IRE1 RNase inhibitors. Here, effects of cyclin-dependent kinase inhibitor, SCH727965 (dinaciclib), on the IRE1 arm of the UPR were examined in human leukemia and myeloma cells. Exposure of cells to extremely low (e.g., nM) concentrations of SCH727965, a potent inhibitor of CDKs 1/2/5/9, diminished XBP-1s and Grp78 induction by the ER stress-inducers thapsigargin (Tg) and tunicamycin (Tm), while sharply inducing cell death. SCH727965, in contrast to IRE1 RNase inhibitors, inhibited the UPR in association with attenuation of XBP-1s nuclear localization and accumulation rather than transcription, translation, or XBP-1 splicing. Notably, in human leukemia cells, CDK1 and CDK5 shRNA knock-down diminished Grp78 and XBP-1s up-regulation while increasing Tg lethality, arguing for a functional role for CDK1/5 in activation of the cytoprotective IRE1/XBP-1s arm of the UPR. In contrast, CDK9 or CDK2 inhibitors or shRNA knockdown failed to down-regulate XBP-1s or Grp78. Furthermore, IRE1, XBP-1, or Grp78 knockdown significantly increased Tg lethality, as observed with CDK1/5 inhibition/knockdown. Finally, SCH727965 diminished myeloma cell growth in vivo in association with XBP-1s down-regulation. Together, these findings demonstrate that SCH727965 acts at extremely low concentrations to attenuate XBP-1s nuclear accumulation and Grp78 up-regulation in response to ER stress inducers. They also highlight a link between specific components of the cell cycle regulatory apparatus (e.g., CDK1/5) and the cytoprotective IRE1/XBP-1s/Grp78 arm of the UPR that may be exploited therapeutically in UPR-driven malignancies.
Keywords: SCH727965 (dinaciclib), X-box–binding protein-1 (XBP-1), Grp78/BiP, unfolded protein response
INTRODUCTION
The endoplasmic reticulum (ER) represents a major site for the synthesis and folding of secreted and membrane-bound cellular proteins. Various environmental stimuli, including oxidative injury, perturbations in Ca+2 homeostasis, or increased energy requirements interfere with protein folding, resulting in ER stress, a condition which, if prolonged, can be lethal to the cell. To circumvent ER stress-mediated cell death, cells engage a complex, evolutionarily conserved process known as the unfolded protein response (UPR) (1). The function of the UPR is to relieve stress induced by the accumulation of unfolded proteins and to restore normal ER homeostasis. While the UPR generally represents an adaptive, pro-survival mechanism, it is a dynamic process, and over time, like ER stress, can promote cell death (1-3). The UPR consists of three arms which act through distinct yet interconnected mechanisms to diminish the mis/unfolded protein burden. Specifically, ER stress leads to disassociation of Grp78 from three ER transmembrane receptors: PERK (PKR-like ER kinase), ATF-6 (activating transcription factor-6), and IRE-1 (inositol-requiring enzyme-1), resulting in activation of these UPR pathways (4). Among these three signaling proteins, IRE1 uniquely contains both a kinase and RNase (endonuclease) domain. Following its dimerization, IRE1 endonuclease activity splices the mRNA of the transcription factor XBP-1, removing a 26-bp segment from the full-length XBP-1 messenger RNA, creating a translational frame shift leading to the expression of active XBP-1s (5) which, upon nuclear translocation, encodes additional chaperone proteins as well as proteins involved in ER accelerated degradation (ERAD), further diminishing the protein load (6, 7). XBP-1s is a major mediator of UPR responses (7, 8), is highly expressed in several diseases, and has been implicated in myelomagenesis (9). Grp78, also up-regulated in many cancer cell types, confers resistance to diverse chemotherapeutic agents and has been implicated in leukemia progression (10, 11). Consequently, XBP-1 and Grp78 are considered promising UPR targets for therapeutic intervention in cancer therapy. These considerations have prompted extensive efforts to develop XBP-1 inhibitors, including agents that act as IRE1 RNase inhibitors and block XBP-1 splicing e.g STF-083010, MK0186893, and MKC-3946 (12-14). Indeed, such agents have been shown to induce MM cell death and to potentiate the activity of anti-myeloma agents that trigger ER stress (12-14). However, the high concentrations (e.g., 10-100 μM) of these agents generally required to inhibit XBP-1s expression could limit their potential.
The observation that de-regulation of cell-cycle control due to aberrant CDK activity is a common feature of most cancers prompted the development of CDK inhibitors such as the pan-CDK inhibitor flavopiridol, the first of its class to enter the clinic (15). More recently, newer-generation CDK inhibitors have been developed, including dinaciclib (SCH727965) (16), an inhibitor of CDKs 1, 2, 5, and 9, which display greater potency than flavopiridol. Although SCH727965 has shown promising pre-clinical activity against a variety of tumor cell types (16), and is currently undergoing phase I/II clinical trials in several malignancies, the mechanisms responsible for its anti-tumor activity remain to be fully elucidated. Furthermore, connections between the ER stress response and the cell cycle regulatory apparatus are not well understood. Here we report that extremely low concentrations (e.g., 2-10 nM) of the CDK inhibitor SCH727965, or either CDK1 or CDK5 knock-down, disrupt the IRE1 arm of the UPR in human myeloma and leukemia cells through a novel mechanism i.e., inhibition of XBP-1s nuclear localization and accumulation, rather than XBP1 splicing as observed with IRE1 endonuclease inhibitors (12-14), accompanied by down-regulation of Grp78. Significantly, these events increase the lethality of established ER stress inducers (e.g., thapsigargin, tunicamycin). Such observations argue that specific CDKs (e.g., CDK1 and CDK5) play key roles in the UPR, thus identifying a link between components of the cell cycle regulatory apparatus and the ER stress response. These findings also suggest that in certain neoplastic cells, CDK inhibitors like SCH727965 may disable the IRE1 arm of the UPR and in so doing, potentiate the activity of agents which elicit this cytoprotective process.
MATERIALS AND METHODS
Cell lines
BaF3/Bcr-Abl was obtained as previously described (17). Jurkat, U937, K562 (myeloid leukemia cells), RPMI-8226, U266, H929, J558 (multiple myeloma cells) were purchased from ATCC (Manassas, VA) or DSMZ (Braunschweig, Germany). J558 cells were cultured in ATCC-formulated Dulbecco's Modified Eagle's Medium (ATCC # 30-2002) supplemented with 10% horse serum. All other cell lines were cultured in RPMI1640 medium containing 10% FBS. Older frozen stocks were authenticated by DNA profiling (STR analysis) using Promega PowerPlexx16HS assay with 15 autosomal loci plus X/Y. All lines were frozen within two months of receipt, and fresh aliquot were thawed before lines reached six months in culture.
Patient samples
Peripheral blood was obtained with informed consent from all patients; these studies have been approved by the VCU IRB. Mononuclear cells were isolated by Ficoll-Hypaque (Sigma–Aldrich, St. Louis, MO) density gradient separation as described previously (18).
Reagents
SCH727965 ((S)-(−)-2-(1-{3-ethyl-7-[(1-oxy-pyridin-3-ylmethyl)amino] pyrazolo[1,5-a]pyrimidin-5-yl}piperidin-2-yl)ethanol); structure shown in Supplementary Fig. S1A) was provided by Merck (Whitehouse Station, N.J) in association with the Cancer Treatment and Evaluation Program (CTEP), NCI. STF-083010, thapsigargin and tunicamycin were purchased from Sigma-Aldrich (St. Louis, MO). MG-132, RO-3306 and Polybrene were purchased from Santa Cruz Biotechnology (Dallas, TX) and EMD Millipore (Billerica, MA) respectively. PHA-848125, PD-0332991, AZD5438, JNJ7706621, and PHA793887 were purchased from Selleck Chemicals (Houston, TX). All compounds were dissolved in dimethyl sulfoxide (DMSO) for in vitro study.
Plasmids
IRE alpha-pcDNA3.EGFP (Addgene #13009, Fumihiko Urano) (19) was a gift from Addgene (Cambridge, MA). P3xFLAG-CML-10 was purchased from Sigma-Aldrich (St. Louis, MO). Knockdown CDK1-pLKO.1, CDK2-pLKO.1, CDK5-pLKO.1, IRE1-pLKO.1 and CDK9-pLKO.1 were purchased from Thermo Scientific (Waltham, MA). Luciferase/pLKO.1 or scramble shRNA/pLKO.1 was used as control. shXBP-1(s)-pSR was constructed by inserting the target sequence for human XBP1 (5’GGAACAGCAAGTGGTAGATTT 3’) into pSUPER.retro.puro (Oligoengine, Seattle, WA) according to the manufacture's protocol. Similarly shGRP78-pSR was constructed by inserting the target sequence (5’GCTCGACTCGAATTCCAAAGA 3’) and (5’GGTCAACTTGATTGAGATTTG 3’) into pSUPER.retro.puro.
Transfection
Plasmids IRE1 alpha/pcDNA3, shGRP78-pSR, shXBP1-pSR were transfected by Amaxa nucleofector according to the manufacturer's protocol (Lonza, Walkersville, MD). Knockdown of CDK1, CDK2, CDK5, CDK9 and IRE1/pLKO.1 were followed the Addgene protocol. Briefly, cocktails of pLKO.1 shRNA (3μg), psPAX2 (1.5 μg), pMD2.G (0.5 μg), OPTI-MEM 40 μl (Invitrogen (Life Technologies, Grand Island, NY) # 31985) and FuGENE®6 12 μl (Roche Applied Science, Indianapolis, IN, # 1181443001) were mixed at appropriate concentrations and dropped evenly via pipette onto 6 ml of HEK-293T cells in Petrie dishes. The harvested media (containing viral production) was collected at 24 and 48 h, and then mixed with Lenti-X concentrator (Clontech, Mountain View, CA, # 631231), centrifuged, dissolved in a small amount of RPMI, and stored at −80°C. Target cells were added to the lentiviral particle solution with polybrene (1-10 μg/ml). After 48hr, the cells were collected for experiments.
Nuclear and cytoplasmic extraction
Nuclear fractions were prepared by using the nuclear extraction kit (Active Motif, Carlsbad, CA). Briefly, after drug treatment, cells were pelleted and lysed by vigorous vortex in hypotonic buffer for 15 min. The samples were then centrifuged at 14,000 × g for 1 min; the supernatant was considered cytoplasmic. Insoluble pellets were further lysed in complete lysis buffer for 30 min, and nuclear extracts (supernatant) were collected after a 10-min centrifugation at 14,000 × g. Both cytoplasmic and nuclear fractions were quantified and subjected to Western blot analysis.
Polymerase Chain Reaction
Total RNA was extracted using Trizol® (Life Technologies, Grand Island, NY) # 15596-018) and followed a standard protocol. PCR was amplified using SuperScript III one-step RT-PCR system with Platinum Taq polymerase (Life Technologies, Grand Island, NY # 12574-018). Primer sets for human XBP-1, Grp78 were XBP-1/PCR/F 5’ CCTGGTTGCTGAAGAGGAGC; XBP-1/PCR/R 5’ CCATGGGGAGATGTTCTGGAG 3’; GRP78/PCR/F 5’ TAGCGTATGGTGCGTCTGTC 3’; GRP78/PCR/R 5’ TTTGTCAGGGGTCTTTCACC 3’. Primer set for mouse XBP-1 was XBP1-S 5’ GAACCAGGAGTTAAGAACACG 3’; XBP-1-R 5’ AGGCAACAGTGTCAGAGTCC 3’. The program for the thermal cycler was 45°C 30 min, 94°C 2min, 40 cycles of 94°C 15 sec, 58°C 30sec, 68°C 1 min and then 68°C for 5 min.
Assessment of apoptosis
The extent of apoptosis was evaluated by either annexin V-fluorescein isothiocyanate staining (BD Biosciences (Pharmingen), San Jose, CA) or 7-aminoactinomycin D (Sigma–Aldrich, St. Louis, MO) by flow cytometry as described previously (18).
Confocal Microscopy
Briefly, cells were fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100. XBP-1s was detected by anti-XBP1 (Santa Cruz Biotechnology, Dallas, TX) and amplified by Alexa Fluor 488–conjugated secondary Abs (Molecular Probes, Eugene, OR (Life Technologies). Slides were mounted in mounting medium containing DAPI (Southern Biotechnology Associates, Birmingham, AL). Slides were analyzed using a Zeiss LSM 700 confocal microscope and software (Carl Zeiss, Thornwood, NY (Zen 2011)).
Immunoblot (Western blot)
Western blot analysis was carried out as previously described (18). The following were used as primary antibodies: IRE1, p-eIF2α (Ser51), p-JNK, p70-S6K (Ser421/424), p70-S6K (T389), eIF4E (Ser209), 4E-BP1 (Thr70), 4E-BP1 (Ser65), caspase-3, cleaved PARP (Cell Signaling Technology, Danvers, MA); XBP-1(s), Grp78, CDK1, CDK9 (Santa Cruz Biotechnology, Dallas, TX); Mcl-1 (BD Biosciences (Pharmingen), San Jose, CA); Actin, FLAG (M2), α-tubulin (Sigma–Aldrich, St. Louis, MO). Blots were stripped and reprobed with actin or tubulin antibodies to ensure equal loading and transfer of proteins.
Animal studies
Animal studies were conducted under an approved protocol by the Virginia Commonwealth University Institutional Animal Care and Use Committee. Athymic NCr-nu/nu mice (NCI) were subcutaneously inoculated in the right rear flank with 3 × 106 J558 cells. SCH727965 was dissolved in 20% Hydroxypropyl B-cyclodextrin. Once tumors became apparent, mice were treated 4 days per week with SCH727965 45mg/kg administered via i.p injection. Control animals were injected with equal volumes of vehicle. Mice were monitored for tumor growth every other day by caliper measurement. Tumor volumes were calculated using the formula (length × width2)/2. When tumor length or width reached 20 mm, mice were euthanized in accordance with institutional guidelines.
Statistical analysis
The significance of differences between experimental conditions was determined by using the Student's t-test.
RESULTS
SCH727965 blocks induction of XBP-1s, Grp78 and downstream targets
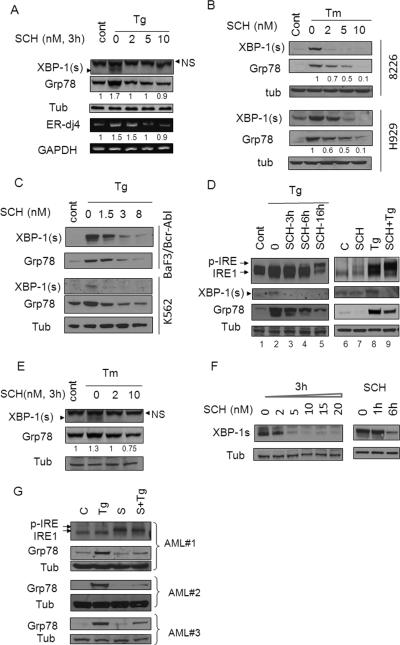
U937 cells exposed (3h) to the ER stress-inducer Tg displayed a clear induction of XBP-1s accompanied by increased expression of two of its downstream targets e.g., ERjd4 mRNA and Grp78 protein (8) (Figure 1A). Notably, each of these events (i.e., XBP-1s, Grp78, and ERdj4 up-regulation) was blocked by co-administration of SCH727965 (2-10 nM). Similarly, SCH727965 markedly attenuated XBP-1s and reduced Grp78 expression in Tm or Tg-treated 8226 and H929 multiple myeloma cells (Figure 1B and supplementary Fig. S1B) as well as in K562 and BaF3/Bcr-Abl leukemia cells (Figure 1C). Time course studies indicated that in U937 cells, SCH727965 (2 nM) prevented up-regulation of XBP-1s and Grp78 at 3h, and further reductions were observed over the ensuing 16h (Figure 1D, left panel). Similarly, co-administration of SCH727965 (2 nM) and Tg for 16h blocked Tg-induced XBP-1s and Grp78 induction in K562 cells (Figure 1D, right panel). SCH727965-mediated reductions in XBP-1s and Grp78 expression were also observed in U937 cells exposed to another ER stress inducer, tunicamycin (Figure 1E). In addition, exposure of J558 multiple myeloma cells, which display high basal XBP-1s expression (20), to SCH727965 induced dose and time-dependent declines in XBP-1s protein expression (Figure 1F). Finally, exposure to SCH727965 (1.6 nM) blocked Tg-mediated Grp78 induction in three primary AML patient samples (Figure 1G). Together, these findings indicate that very low (e.g., nM) concentrations of SCH727965 effectively block induction of multiple IRE1 targets in human myeloma and leukemia cells exposed to ER stress inducers.
Figure 1. SCH727965 blocks induction of XBP-1s and downstream targets.
(A) U937 cells were exposed to Tg (0.5-2.5 μM) with or without SCH727965 at the indicated concentrations for 3h, after which cells were lysed and Western blot (WB) or RT-PCR analysis performed as described in Material and Methods. For these and subsequent WB studies, each lane was loaded with 20 μg of protein; blots were then stripped and reprobed with tubulin antibodies to ensure equivalent loading and transfer. Results are representative of 3 independently performed experiments. (B) 8226, H929 cells were exposed to Tm (2.5μg/ml) alone or in combination with SCH727965 (2-10 nM) for 4h, after which cells were lysed and analysed. (C) K562, BaF3/Bcr-abl cells were exposed to Tg alone or in combination with SCH727965 (1.5-8 nM) for 6h, after which cells were lysed and analysed. (D) U937 cells were exposed to Tg (50nM) alone or in combination with SCH727965 (2 nM) for varying intervals (left panel). K562 cells were exposed to Tg (500 nM) alone or in combination with SCH727965 2 nM for 16h after which cell extracts were analyzed by Western-blot (right panel). (E) U937 cells were exposed to Tunicamycin (Tm) with or without SCH727965 at the indicated concentrations for 3h, after which cells were lysed and Western blot analysis performed. (F) J558 cells, exhibiting high basal expression of XBP-1s, were exposed to SCH727965 at the indicated concentrations for 3h, or SCH727965 3 nM for varying intervals, after which cells were analyzed by Western-blot. (G) Three primary patient samples (AML) were exposed to SCH727965 1.6 nM alone or in combination with 50 nM Tg for 16h, after which the cells were analyzed by Western blot. Values below the blots represent densitometric ratios relative to controls. NS = non-specific.
SCH727965 does not inhibit IRE1 activation or XBP-1s splicing
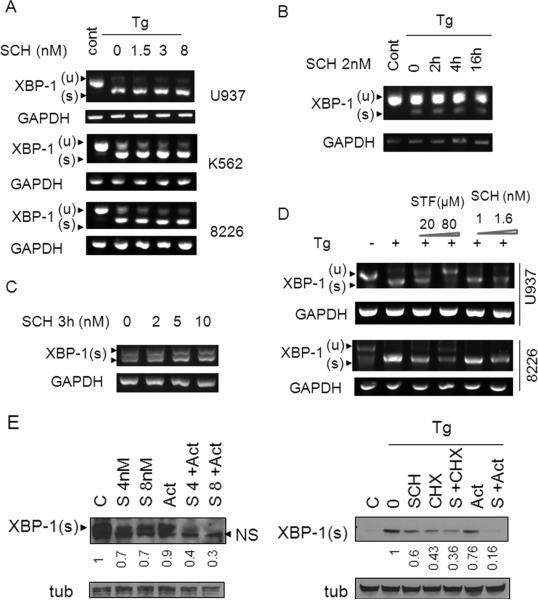
Previous studies have identified several compounds which inhibit IRE1 activity and as a consequence, XBP-1s splicing (12-14). Consequently, several approaches were employed to compare the effects of SCH727965 to those of known IRE1 inhibitors with respect to inhibition of XBP-1s generation. First, U937 cells were exposed to Tg in the presence or absence of SCH727965 (1.5-8 nM), after which XBP-1s transcription was monitored by RT-PCR. As shown in Figure 2A, exposure to SCH727965 failed to block XBP-1s mRNA induction in multiple Tg-treated leukemia and myeloma cell lines e.g., U937, K562, 8226 and BaF3/T315I. Furthermore, the absence of XBP-1s mRNA inhibition persisted over the entire 16h SCH727965 exposure interval (Figure 2B). Consistent with these findings, SCH727965 also failed to diminish XBP-1s mRNA expression in J558 cells, which constitutively express high levels of this protein (Figure 2C). Notably, the failure of SCH727965 to block XBP-1s mRNA induction by Tg contrasted sharply with the actions of bona fide endonuclease inhibitors such as STF-083010 (21), which markedly inhibited XBP-1s mRNA formation in several cell lines (Figure 2D). Secondly, as shown in Figure 1D (lines 8), Tg induced IRE1 activity, reflected by increased splicing of XBP-1s and IRE1 phosphorylation/dimerization in K562 cells. Interestingly, co-administration of SCH727965 with Tg resulted in further increases in IRE1 activation, manifested by IRE1 phosphorylation/ dimerization and up-regulation of its downstream target p-JNK (22), although XBP-1s expression was completely abrogated (Figure 1D, lines 8 and data not shown). These results argue that SCH727965 does not inhibit XBP-1s by blocking IRE1 activation. Finally, SCH727965 sharply down-regulated XBP-1s expression in cells ectopically-expressing IRE1 to an equivalent extent as observed in empty-vector control cells (Supplementary Fig. 1B), implying that SCH727965 inhibits XBP-1s formation through an IRE1-independent process. Collectively, these findings support the notion that SCH727965 opposes the induction of XBP-1s by ER stress-inducers through a fundamentally different mechanism from that of IRE1 endonuclease inhibitors.
Figure 2. SCH 727965 does not inhibit XBP-1s transcription.
(A) U937, K562, or 8226 cells were exposed to Tg and SCH as in Figure 1A above, after which mRNA was extracted and analyzed by RT-PCR. (B) U937 cells were exposed to Tg (50 nM) and SCH727965 (2 nM) for the indicated intervals, after which mRNA was extracted and subsequently analyzed by RT-PCR. (C) J558 cells were treated with 2-10 nM SCH727965 for 3h, after which RNA was extracted, followed by analysis by RT-PCR. (D) 8226 and K562 cells were stimulated with Tg alone or in combination with STF (20-80μM) or SCH727965 (1-1.6 nM) for 16h after which mRNA was extracted and analyzed by RT-PCR. (E) J558 cells were treated with SCH727965 (4 or 8 nM) with or without actinomycin (2.5μg/ml) for 1h (upper panel). Alternatively, 8226 cells were stimulated with Tg (1 μM) for 1h and subsequently treated with 8 nM SCH727965 with or without CHX (2μg/ml) or actinomycin (2.5μg/ml) for 1h (lower panel). Following treatment, cells were lysed and analyzed by Western blot. Values below the blots represent densitometric ratios relative to controls.
To gain further insights into the mechanism(s) by which SCH727965 diminishes XBP-1s up-regulation, J558 cells were exposed to SCH727965 (4 or 8 nM) in the presence or absence of the transcription inhibitor actinomycin (2.5 μg/ml), after which XBP-1s expression was examined. Both SCH727965 and actinomycin, administered individually, moderately (SCH727965) or modestly (actinomycin) reduced XBP-1s expression. However, combined treatment produced considerably more pronounced reductions in XBP-1s levels (Figure 2E, left panel). Similar results were obtained in 8226 multiple myeloma cells exposed to Tg in conjunction with SCH727965 and actinomycin (Figure 2E, right panel). Taken together, these observations argue against the possibility that SCH727965 blocks XBP-1s induction through a transcriptional mechanism.
Down-regulation of XBP-1s by SCH727965 involves a post-translational mechanism rather than inhibition of the general translation regulatory apparatus
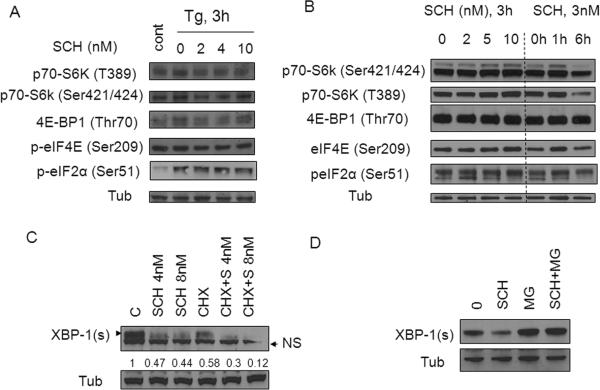
Effects of SCH727965 were then examined with respect to events associated with translational regulation. Exposure of Tg-treated U937 cells to SCH727965 (2-10 nM) failed to modify phosphorylation of p70-S6K, eIF4E, 4EPB1, or eIF2α (Figure 3A). Identical results were obtained in J558 cells exposed to 2-10 nM SCH727965 alone (Figure 3B). In addition, co-administration of the protein synthesis inhibitor CHX (5μM) with SCH727965 (8 nM) produced a further reduction in XBP-1s expression (Figure 3C). Similar results were observed in Tg/SCH727965-treated 8226 MM cells (Figure 2E). Together, these findings argue against a translational mechanism underlying SCH727965-mediated XBP-1s down-regulation.
Figure 3. Down-regulation of XBP-1s by SCH727965 involves a post-translational mechanism.
(A) U937 cells were stimulated with Tg 2μM alone or in conjunction with SCH727965 (2-10 nM) for 3h after which cells were lysed and and analyzed by Western blot. (B) J558 cells were treated with various concentrations (2-10 nM) of SCH727965 for 3h or with 3 nM SCH727965 for varying exposure intervals (e.g., 1-6h), after which the cells were analyzed by Western blot. (C) J558 cells were treated with SCH727965 (4- 8 nM) alone or together with CHX (5 μg/ml) for 2h, after which levels of XBP-1s were monitored by WB. (D) J558 cells were exposed to 8 nM SCH727965 and 1 μM MG-132 alone or in combination for 2h after which the cell lysed and analyzed by Western blot.
To assess the possible contribution of post-translational events to XBP-1s down-regulation by SCH727965, J558 cells were exposed to SCH727965 (8 nM) in the presence or absence of the proteasome inhibitor MG-132 (1 μM), after which XBP-1s expression was monitored. As shown in Figure 3D, SCH727965 substantially diminished XBP-1s expression, an event that was largely prevented by MG-132 co-administration. Together, these findings are consistent with the notion that a post-translational mechanism (i.e., proteasomal degradation) contributes to SCH727965-mediated down-regulation of XBP-1s expression.
SCH727965 diminishes nuclear accumulation of XBP-1s
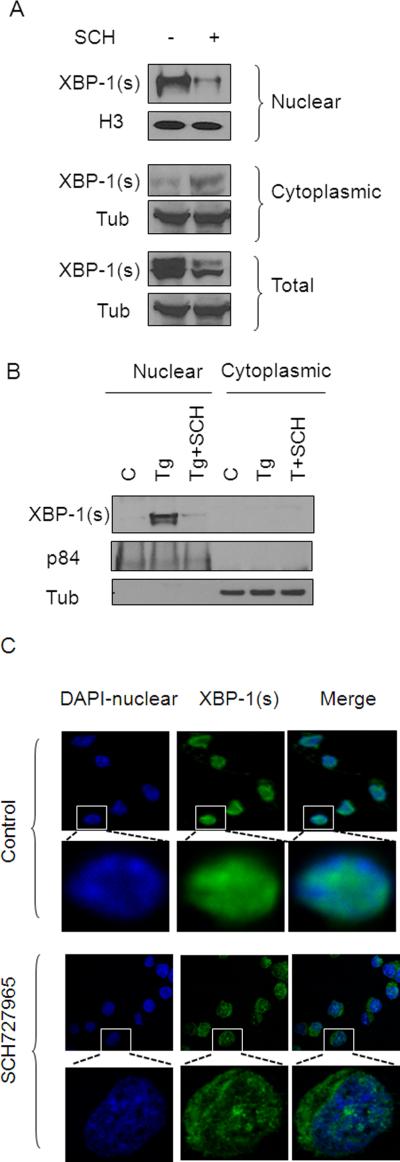
Following cytoplasmic cleavage of XBP-1 by IRE1, XBP-1s translocates to the nucleus where it binds to DNA and drives the transcription of genes involved in the UPR (5), prompting examination of the effects of SCH727967 on XBP-1s nuclear disposition. As shown in Figure 4A, J558 cells exposed to SCH727965 (5 nM; 3h) displayed a dramatic decline in expression of XBP-1s in the nuclear fraction. In contrast, cytoplasmic expression of XBP-1s increased slightly with SCH727965 exposure, although a clear reduction in total XBP-1 expression was observed. Notably, Tg markedly increased XBP-1s nuclear expression in 8226 cells, an effect that was abrogated by SCH727965 co-administration (Figure 4B). Finally, confocal microscopy/immunofluorescence analysis demonstrated that J558 cells displayed clear co-localization of green fluorescence (XBP-1s) and blue fluorescence (nuclear). However, SCH727965-treated cells displayed dramatically reduced nuclear XBP-1s expression but increased cytoplasmic XBP-1s expression (Figure 4C). Virtually identical results were observed in K562 cells exposed to TG (data not shown). These findings argue that SCH727965 attenuates XBP-1s nuclear translocation/accumulation and diminishes the total cellular expression of XBP-1s.
Figure 4. SCH727965 attenuates nuclear XBP-1s accumulation.
(A) J558 cells were exposed to SCH727965 5 nM for 3h, after which cells were harvested. Nuclear and cytoplasmic fractions were separated as described in Material and Methods. H3 was used as a nuclear loading control. (B) 8226 cells were treated with 1μM Tg alone or in combination with SCH727965 (5 nM) for 4h, after which cells were separated using a nuclear extraction kit, with p84 serving as a nuclear marker. (C) J558 cells were exposed to SCH727965 5 nM for 3h, after which cells were harvested and fixed with 4% paraformaldehyde followed by confocal microscopy (magnification 10-100X) to monitor XPB-1s (blue-DAPI: nuclear, green: XBP-1s).
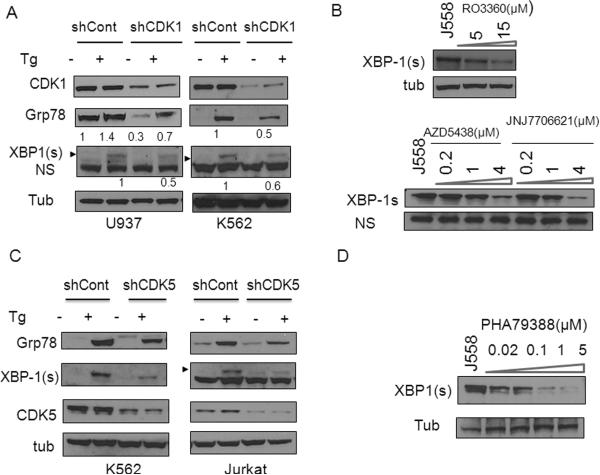
CDK1 or CDK5 interruption plays a functional role in diminishing Grp78/XBP-1s expression and XBP-1s nuclear localization
SCH727965 inhibits CDKs 2, 5, 1, 9 within a similar concentration range (IC50 of 1.0, 1.0, 3.0, and 4.0 nM, respectively) (16). To determine which CDKs might contribute functionally to SCH727965-mediated XBP-1s down regulation, a CDK knockdown strategy was employed. First, CDK1 was knocked down via pooled shRNA plasmids in several cell lines after which responses to ER stress inducers were monitored. As shown in Figure 5A, compared to scrambled sequence controls, shCDK1 U937 cells displayed sharply diminished CDK1 expression. Notably, shCDK1 U937 cells exhibited marked reductions in basal as well as Tg-stimulated Grp78 induction, accompanied by reductions in XBP-1s expression following Tg exposure (Figure 5A). Similarly, K562 cells in which CDK1 was knocked down also displayed pronounced reductions in Grp78 and XPB-1s expression following Tg exposure compared to controls (Figure 5A). Confocal microscopy/immunofluorescence analysis also demonstrated that shCDK1 U937 cells exhibited dramatically reduced nuclear XBP-1s expression following Tg exposure (Supplementary Fig. S2A), recapitulating results obtained with SCH727965 (Figure 4C). In addition, J558 cells treated with the relatively specific CDK1 inhibitors RO3306, AZD5438, or JNJ-7706621 (23-25) for 6h exhibited significantly reduced XBP-1s expression (Figure 5B). Together, these and the preceding shRNA knock-down studies implicate CDK1 in promoting XBP-1s nuclear accumulation and expression as well as Grp78 up-regulation in leukemia and myeloma cells undergoing the UPR.
Figure 5. CDK1 or CDK5 are required for XBP-1s and Grp78 induction.
(A) U937, K562 cells were transfected with shCDK1/pLKO.1 or shControl/pLKO.1 for 48h, after which cells were exposed to Tg (1-5μM) for 3-6h, followed by Western blot analysis. (B) J558 cells were treated with RO3306 (5-15 μM), AZD5438 (0.2-4 μM) or JNJ7706621 (0.2-4 μM) for 6h, after which Western blot analysis was performed to monitor XBP-1s expression. (C) K562 or Jurkat cells were transfected with shCDK5/pLKO.1 or shCont for 48h, followed by exposure to Tg (1.5-5 μM) for 4h, after which cell extracts were subjected to WB analysis using the indicated primary antibodies. (D) J558 cells were treated with PHA793887 (0.02-5 μM) for 6h, after which Western blot analysis was performed to monitor XBP-1s expression.
To determine whether CDK5 might also be involved in UPR regulation, K562 and Jurkat leukemia cells in which CDK5 was knocked down were employed. K562/shCDK5 and Jurkat/shCDK5 cells exhibited marked reductions in basal as well as Tg-stimulated Grp78 induction, accompanied by reductions in XBP-1s expression compared to their control counterparts (Figure 5C). Similar results were obtained in J558 cells (data not shown). Immunofluorescence analysis also demonstrated that shCDK5 K562 cells exhibited sharply reduced nuclear XBP-1s expression following Tg exposure compared to K562/shCont cells (Supplementary Fig. S2B). Furthermore, treatment of J558 cells with PHA793887, an inhibitor of CDK2/5/9 (26) for 6h also displayed diminished XBP-1s expression (Figure 5D). Collectively, these results imply that CDK5, like CDK1, also plays an important role in regulating the IRE1 arm of the UPR. To characterize the possible involvement of CDK2 and 9 in the UPR, cells in which CDK2 or CDK9 were knocked down by shRNA were employed. K562/shCDK9 cells exhibited clear reductions in CDK9 expression, but in sharp contrast to CDK1 or CDK5 knock-down cells, failed to display differences in XBP-1s or Grp78 induction following Tg exposure compared to controls (Supplementary Fig. S3A). Similarly, CDK2 knock-down cells exhibited very modest or no changes in Grp78 and XBP-1s induction following Tg exposure compared to its controls (Supplementary Fig. S3B). Finally, J558 cells treated with a specific CDK4/6 inhibitor (PD332991) (27), or a CDK 9/2/7 inhibitor (SNS-032) (28) did not display changes in Grp78 or XBP-1s expression (data not shown). Together, these findings argue against the possibility that inhibition of CDK2 or CDK9 by SCH727965 is responsible for attenuation of XBP-1s or Grp78 expression.
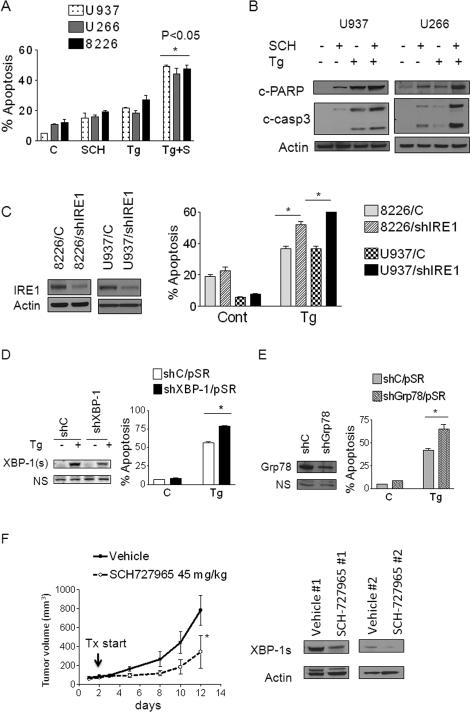
Pharmacologic or genetic disruption of XBP-1s, Grp78, CDK1, or CDK5 increases sensitivity to ER stress inducers
To investigate the functional cytoprotective contribution of the IRE1 arm of the UPR in this setting, effects of blocking the UPR by SCH727965 on the lethality of ER stress inducers (Tg and Tm) were examined. To this end, U937, 8226 and U266 cells were exposed to 1-2 nM SCH727965 in the presence or absence of Tg (5-100nM) for 24h, after which apoptosis was monitored. Exposure to single agents (SCH727965 or Tg) minimally induced apoptosis, whereas combined exposure to SCH727965 and Tg sharply increased apoptosis in all cell lines tested (P < 0.05 versus single agents; Figure 6A). Consistent with these findings, co-treatment with SCH727965 and Tg markedly induced procaspase-3 activation and PARP cleavage (Figure 6B). To test the functional genetic contribution of IRE1, XBP-1s and Grp78 to ER stress inducer-associated lethality, U937 and 8226 cells were transfected with a shIRE1/pLKO.1 plasmid. Both U937/shIRE1 and 8226/shIRE1 cells were significantly more sensitive to Tg-induced apoptosis than their corresponding controls (P < 0.05 in each case; Figure 6C). Similarly, U937/shXBP-1 or U937/shGrp78 knockdown cells were significantly more sensitive to Tg- and Tm-induced apoptosis than controls (P < 0.05; Figure 6D and E). Finally, U937/shCDK1 or U937/shCDK5 cells were also significantly more susceptible to Tg-induced apoptosis than controls (P < 0.05, Supplementary Fig. S4), recapitulating the effects of IRE1, XBP-1, or Grp78 knock-down. Together, these findings argue that disabling components of the IRE1 arm of the UPR by SCH727965 plays a significant functional role in potentiating ER stress-induced lethality.
Figure 6. Inhibition of the UPR or SCH727965 enhances ER stress inducer lethality and suppresses tumor growth and down-regulates XBP-1s expression in vivo.
(A) U937 (dots), U266 (grey) or 8226 (black) cells were exposed to Tg (5nM-100nM) alone or in combination with SCH727965 (1-2 nM) for 24h, after which the percentage of apoptotic cells was determined by 7-AAD uptake. Values represent the means for 3 separate experiments performed in triplicate ± S.D. * = significantly greater than values for Tg alone; P < 0.05. (B) Cells treated as above were prepared and subjected to Western blot analysis using the indicated primary antibodies. (C) U937 and 8226 cells were transfected with shIRE1/pLKO.1 constructs designed against IRE1; A pooled clone from each transfection was selected. These and shCont cells were exposed to Tg (10-50 nM), after which the extent of cell death was monitored by 7-AAD. * = significantly less than values for shControl cells; P < .05. (D) U937 cells were transfected with shXBP-1/pSR or shCont/pSR constructs. Forty eight hours after transfection, cells were treated with Tg 50nM for 24h after which the extent of cell death was monitored by 7-AAD (* = significantly greater than controls; P < .05). (E) U937 cells were transfected with shGrp78/pSR or shCont/pSR constructs. Following transfection, cells were treated with Tg 50nM for 24h, after which the extent of cell death was monitored by 7-AAD (* = significantly greater than controls; P < .05). (F) Athymic NCr-nu/nu mice were subcutaneously inoculated in the right rear flank with 3 × 106 J558 cells. After tumors appeared, treatment was initiated with vehicle or SCH727965 (45 mg/kg, i.p.) for 5 days/week. Tumor size was measured by calipers every other day. * = P<0.05 compared to vehicle (left panel). Xenograft-bearing mice were treated with SCH727965 as above, 10 days after which tumors were excised, lysed, and extracts subjected to Western blot analysis using the indicated antibodies. Blots were subsequently stripped and re-probed with antibodies to actin to ensure equal loading and transfer (right panel).
SCH727965 inhibits tumor growth and down-regulates XBP-1s in vivo
The in vivo anti-tumor activity of SCH727965 was then investigated in athymic nude mice. To this end, 3 × 106 J558 cells were injected in the flanks of mice. After 4 days, when tumors were visible (n=4), mice were treated with SCH727965 45 mg/kg i.p. for 5 days/week. As shown in Figure 6F, right panel, mice treated with SCH727965 displayed a significant reduction in tumor size compared to vehicle treatment (P<0.05). Western blot analysis conducted on excised tumor tissue revealed that SCH727965 also reduced expression of XBP-1s in vivo, consistent with in vitro observations (Figure 6F, left panel).
DISCUSSION
Multiple malignancies are either driven by the UPR or display basal UPR activation, which promotes their survival and/or renders them resistant to chemotherapy (9, 29, 30). The IRE1 arm of the UPR provides neoplastic cells with a mechanism to escape the lethal consequences of proteotoxic stress accompanying high protein turnover, and as a result, has become an important target for therapeutic intervention (31). In particular, the transcription factor XBP-1s is required for plasma cell differentiation and survival, has been implicated in multiple myeloma (MM) pathogenesis, and is highly expressed in MM cells and other cancers, where it is associated with poor survival (9, 32). It therefore represents a logical IRE1 pathway target in MM and potentially other malignancies (9, 33). These considerations have prompted intense efforts to develop XBP-1 inhibitors, including agents that act as IRE1 RNase inhibitors which block XBP-1 splicing e.g STF-083010, MK0186893, MKC-3946 (12-14). Indeed, such agents have been shown to induce MM cell death and to potentiate the activity of anti-myeloma agents that trigger ER stress (12-14). However, the high concentrations (e.g., 10-100 μM) of these agents that are generally required to inhibit XBP-1s expression could limit their potential. In addition, the chaperone protein Grp78 is over-expressed in many cancers and suppresses apoptosis through multiple mechanisms (10). Consequently, in diverse tumor types including, leukemia, lymphoma and epithelial cancers, Grp78 over-expression confers resistance to a wide variety of chemotherapeutic agents (10, 11). However, direct inhibitors of Grp78 are not currently available, and proteins like Grp78 are notoriously difficult to target e.g., by small molecule inhibitors. The present findings suggest that certain CDK inhibitors e.g., SCH727965, may provide an alternative strategy capable of disabling the cytoprotective XBP-1/Grp78 components of the UPR in neoplastic cells. It should be noted that a previous study reported that the CDK inhibitor flavopiridol induced atypical ER stress and autophagic responses in primary chronic lymphoid leukemia (CLL) cell samples (34). Differences between these and the present results may reflect multiple factors, including cell-type specific responses, or, alternatively, the 3-log higher drug concentrations employed in the prior study (34).
The results presented here suggest that in contrast to the actions of IRE endonuclease inhibitors such as STF-083010, which prevent formation of the spliced form of XBP-1 (12-14), CDK inhibitors such as SCH727965 act through a fundamentally different mechanism i.e., prevention of XBP-1s nuclear localization/accumulation and accompanying down-regulation. Specifically, the present findings argue that SCH727965, by inhibiting XBP-1s accumulation and nuclear localization, may in fact increase IRE1 activation, manifested by an increase in IRE1 autophosphorylation, dimerization, and JNK activation (22, 35), responses potentially reflecting a compensatory upstream feedback mechanism. Notably, inhibition of XBP-1s expression induced down-regulation of several established downstream targets such as Erdj4 and Grp78 (7). Results of studies involving protein (CHX) and transcription (actinomycin) inhibitors, as well as RT-PCR, argue against the possibility of transcriptional or translational mechanisms of down-regulation, or interference with XBP-1s splicing. In contrast, proteasome inhibition essentially blocked down-regulation of XBP-1s expression in cells exposed to SCH727965, implicating a post-translational process e.g., proteasomal degradation, in this phenomenon. Based on these findings, a possible explanation for these observations is that prevention of XBP-1s nuclear transport traps this protein in the cytoplasm where it is vulnerable to ubiquitination and proteasomal elimination (36). Additional studies will be required to confirm or refute this hypothesis.
The finding that inhibition of Grp78 and XBP-1s occurred in cells in which CDK1 or CDK5 was knocked down suggests a specific and previously unrecognized role for these CDKs in the IRE1 arm of the UPR. Despite the critical involvement of XBP-1s in UPR responses, current understanding of the regulation of XBP-1s activity is relatively limited. It is known that XBP-1s can be phosphorylated by p38 or through interactions with p85 PI3K, events that enhance its nuclear translocation (37, 38). In the present setting, SCH727965 did not down-regulate phospho-Akt (ser473 or thr308) or phospho-p38 (Nguyen and Grant, unpublished observations), arguing that SCH727965 does not block the UPR through inhibition of the p85 or p38 pathways. Of potential relevance, CDK5 has been reported to mediate ER stress via activation of MEKK1 and calcium channels or regulation of the nuclear translocation of PDX-1(39, 40). In this context, it is conceivable that CDK5, either directly or through interactions with a cooperating protein, promotes XPB-1s nuclear localization.
CDK1 is unique among CDKs in that it is the only CDK sufficient for cell cycle progression in mammalian cells (41). However, several CDK1 functions unrelated to cell cycle regulation have recently been described, including involvement in both homologous recombination- and non-homologous end-joining-related DNA repair as well as phosphorylation of the C/EBPα transcription factor, an event that promotes leukemia cell differentiation (42). It is possible that CDK1 and CDK5 may also promote phosphorylation of XBP-1s and subsequent nuclear localization, as both CDK1 and 5 have shown identical substrate specificities in vitro (43). However, the possibility that CDK1/5 may regulate UPR through modulation of calcium channels cannot be excluded (43). Studies designed to identify the mechanism(s) by which CDK1/5 inhibition diminishes XBP-1s nuclear accumulation, accompanied by Grp78 down-regulation, are currently underway.
In contrast to CDK1/5, knock-down of CDK2 or the transcriptional CDK9 failed to modify XBP-1s or Grp78 expression, arguing against a functional role for these CDKs in SCH727965-mediated attenuation of the UPR. Agents that inhibit CDK9 and repress transcription, including SCH727965, have been shown to down-regulate short-lived anti-apoptotic proteins such as Mcl-1 in various tumor cell types including myeloma and leukemia (44, 45). Moreover, Mcl-1 is known to play a functional role in protecting cells from ER stress (46), raising the possibility that Mcl-1 down-regulation might contribute to potentiation of ER stress-mediated cell death by SCH727965. However, the observation that CDK1 or CDK5 knock-down mimicked the ability of SCH727965 to potentiate Tg and Tm lethality argues that disruption of the IRE1/XBP-1s axis through CDK1 or CDK5 inhibition plays a primary functional role in this phenomenon.
Multiple phase I trials of SCH727965 in both solid tumor as well as hematologic malignancies have been completed or are underway. In a phase I trial in patients with refractory/relapsed CLL, SCH727965 administered at doses of 5-14 mg/m2 as a 2-hr IV infusion on days 1,8, and 15 of a 28-day cycle resulted in a 45% overall response rate, including several responses in patients with poor-prognostic features or who had failed another CDK inhibitor (flavopiridol)(47). In a phase II trial in patients with refractory acute myeloid or lymphoid leukemia in which SCH727965 was administered at a dose of 50 mg/m2 q 21 days, reductions in blasts and predicted pharmacodynamics effects in leukemic cells (e.g., Mcl-1 down-regulation) were observed (48). However, responses were transient, suggesting that more prolonged exposures to this agent e.g., through continuous infusion schedules may be preferable therapeutically. Notably, peak plasma concentrations considerably in excess of those necessary for inhibition of XBP-1s or Grp78 induction (e.g., μM) were achieved in this study, raising the possibility that plasma concentrations capable of inhibiting the IRE1 arm of the UPR may be pharmacologically achievable with alternative schedules.
In summary, the present studies demonstrate that CDK inhibitors such SCH727965, which potently inhibit CDK1 or 5, disrupt the UPR in association with attenuation of XBP-1s nuclear localization and accumulation accompanied by Grp78 down-regulation. These findings also highlight a heretofore unrecognized link between the cell cycle regulatory apparatus and IRE1-related responses to ER stress which offers the potential for therapeutic exploitation. More specifically, the present observations suggest that CDK1 and CDK5 play particularly important roles in activation of the cytoprotective IRE1 arm of the UPR, including induction of Grp78, and that disabling CDK1 or CDK5 either genetically or pharmacologically significantly lowers the threshold for apoptosis in multiple myeloma or leukemia cells exposed to ER stress-inducers. Notably, this process appears to occur through a fundamentally different mechanism from that attributed to endonuclease inhibitors (12-14) i.e., prevention of XBP-1s nuclear localization and accumulation, rather than inhibition of XBP-1s splicing. In the case of SCH727965, the observations that these events occur at concentrations (e.g., low nM) substantially less than those previously required for currently available RNase inhibitors (12-14) and can be recapitulated in vivo, are of potential importance. The significance of these findings is that transformed cells may be more dependent upon a functional UPR to protect them from the lethal effects of protein overload and proteotoxic stress (49). Consequently, concurrently disrupting the activity of CDK1 and CDK5 by agents such as SCH727965 may disable cytoprotective UPR activation, thus rendering neoplastic cells particularly vulnerable to antineoplastic agents eliciting this response (28). In addition, the link between cell cycle regulatory CDKs and the cellular response to ER stress could also provide new therapeutic targets (e.g., CDK1 and CDK5) for the treatment of high-protein turnover disorders in which an activated UPR is required to prevent neoplastic cell death e.g., multiple myeloma and lymphoma (9, 29). Finally, the connection between CDKs and the UPR also provides a theoretical basis for the development of rational combination regimens involving agents which induce the cytoprotective arm of the UPR, including proteasome inhibitors (50), Hsp90 antagonists such as 17-AAG (51), or sorafenib (52). Studies designed to test these possibilities are currently underway.
Supplementary Material
Acknowledgments
Grant support: This study is supported by awards CA93738, CA100866, CA167708, RC2CA148431, and R21CA137823 from the NIH (S. Grant), award 6238-12 from the Leukemia and Lymphoma Society of America (T. Nguyen and S. Grant), and an award from the Multiple Myeloma Research Foundation, Myeloma SPORE award CA142509, and Lymphoma SPORE award 1P50 CA130805 from the NIH (S. Grant). Microscopy was performed at VCU-Department of Neurobiology and Anatomy Microscopy Facility, supported in part by NIH-NINDS center core grant 5P30NS047463.
Abreviations
- CDK
cyclin dependent kinase
- Tg
Thapsigargin
- Tm
Tunicamycin
- UPR
unfolded protein response
Footnotes
Conflict of Interest Statement: The authors whose names are listed above certify that they have no affiliations with or involvement in any organization or entity with any financial interest or non-financial interest in the subject matter or materials discussed in this manuscript.
Authors' Contributions:
Conception and design: Tri Nguyen and Steven Grant
Development of methodology and acquisition of data: Tri Nguyen
Analysis and interpretation of data: Tri Nguyen and Steven Grant
Writing, review, and/or revision of the manuscript: Tri Nguyen and Steven Grant
Study supervision: Steven Grant
Reference List
- 1.Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 2.Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, et al. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318:944–9. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rutkowski DT, Arnold SM, Miller CN, Wu J, Li J, Gunnison KM, et al. Adaptation to ER stress is mediated by differential stabilities of pro-survival and pro-apoptotic mRNAs and proteins. PLoS Biol. 2006;4:e374. doi: 10.1371/journal.pbio.0040374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2:326–32. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 5.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–29. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 6.Yamamoto K, Sato T, Matsui T, Sato M, Okada T, Yoshida H, et al. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev Cell. 2007;13:365–76. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 7.Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23:7448–59. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Acosta-Alvear D, Zhou Y, Blais A, Tsikitis M, Lents NH, Arias C, et al. XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol Cell. 2007;27:53–66. doi: 10.1016/j.molcel.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 9.Carrasco DR, Sukhdeo K, Protopopova M, Sinha R, Enos M, Carrasco DE, et al. The differentiation and stress response factor XBP-1 drives multiple myeloma pathogenesis. Cancer Cell. 2007;11:349–60. doi: 10.1016/j.ccr.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee AS. GRP78 induction in cancer: therapeutic and prognostic implications. Cancer Res. 2007;67:3496–9. doi: 10.1158/0008-5472.CAN-07-0325. [DOI] [PubMed] [Google Scholar]
- 11.Dong D, Stapleton C, Luo B, Xiong S, Ye W, Zhang Y, et al. A critical role for GRP78/BiP in the tumor microenvironment for neovascularization during tumor growth and metastasis. Cancer Res. 2011;71:2848–57. doi: 10.1158/0008-5472.CAN-10-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mimura N, Fulciniti M, Gorgun G, Tai YT, Cirstea D, Santo L, et al. Blockade of XBP1 splicing by inhibition of IRE1alpha is a promising therapeutic option in multiple myeloma. Blood. 2012;119:5772–81. doi: 10.1182/blood-2011-07-366633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Volkmann K, Lucas JL, Vuga D, Wang X, Brumm D, Stiles C, et al. Potent and selective inhibitors of the inositol-requiring enzyme 1 endoribonuclease. J Biol Chem. 2011;286:12743–55. doi: 10.1074/jbc.M110.199737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Papandreou I, Denko NC, Olson M, Van MH, Lust S, Tam A, et al. Identification of an Ire1alpha endonuclease specific inhibitor with cytotoxic activity against human multiple myeloma. Blood. 2011;117:1311–4. doi: 10.1182/blood-2010-08-303099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dai Y, Grant S. Cyclin-dependent kinase inhibitors. Curr Opin Pharmacol. 2003;3:362–70. doi: 10.1016/s1471-4892(03)00079-1. [DOI] [PubMed] [Google Scholar]
- 16.Parry D, Guzi T, Shanahan F, Davis N, Prabhavalkar D, Wiswell D, et al. Dinaciclib (SCH 727965), a novel and potent cyclin-dependent kinase inhibitor. Mol Cancer Ther. 2010;9:2344–53. doi: 10.1158/1535-7163.MCT-10-0324. [DOI] [PubMed] [Google Scholar]
- 17.La RP, Corbin AS, Stoffregen EP, Deininger MW, Druker BJ. Activity of the Bcr-Abl kinase inhibitor PD180970 against clinically relevant Bcr-Abl isoforms that cause resistance to imatinib mesylate (Gleevec, STI571). Cancer Res. 2002;62:7149–53. [PubMed] [Google Scholar]
- 18.Nguyen T, Dai Y, Attkisson E, Kramer L, Jordan N, Nguyen N, et al. HDAC inhibitors potentiate the activity of the BCR/ABL kinase inhibitor KW-2449 in imatinib-sensitive or -resistant BCR/ABL+ leukemia cells in vitro and in vivo. Clin Cancer Res. 2011;17:3219–32. doi: 10.1158/1078-0432.CCR-11-0234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lipson KL, Fonseca SG, Ishigaki S, Nguyen LX, Foss E, Bortell R, et al. Regulation of insulin biosynthesis in pancreatic beta cells by an endoplasmic reticulum-resident protein kinase IRE1. Cell Metab. 2006;4:245–54. doi: 10.1016/j.cmet.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 20.Lee AH, Iwakoshi NN, Anderson KC, Glimcher LH. Proteasome inhibitors disrupt the unfolded protein response in myeloma cells. Proc Natl Acad Sci U S A. 2003;100:9946–51. doi: 10.1073/pnas.1334037100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Papandreou I, Denko NC, Olson M, Van MH, Lust S, Tam A, et al. Identification of an Ire1alpha endonuclease specific inhibitor with cytotoxic activity against human multiple myeloma. Blood. 2011;117:1311–4. doi: 10.1182/blood-2010-08-303099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, et al. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287:664–6. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- 23.Chen S, Chen L, Le NT, Zhao C, Sidduri A, Lou JP, et al. Synthesis and activity of quinolinyl-methylene-thiazolinones as potent and selective cyclin-dependent kinase 1 inhibitors. Bioorg Med Chem Lett. 2007;17:2134–8. doi: 10.1016/j.bmcl.2007.01.081. [DOI] [PubMed] [Google Scholar]
- 24.Byth KF, Thomas A, Hughes G, Forder C, McGregor A, Geh C, et al. AZD5438, a potent oral inhibitor of cyclin-dependent kinases 1, 2, and 9, leads to pharmacodynamic changes and potent antitumor effects in human tumor xenografts. Mol Cancer Ther. 2009;8:1856–66. doi: 10.1158/1535-7163.MCT-08-0836. [DOI] [PubMed] [Google Scholar]
- 25.Emanuel S, Rugg CA, Gruninger RH, Lin R, Fuentes-Pesquera A, Connolly PJ, et al. The in vitro and in vivo effects of JNJ-7706621: a dual inhibitor of cyclin-dependent kinases and aurora kinases. Cancer Res. 2005;65:9038–46. doi: 10.1158/0008-5472.CAN-05-0882. [DOI] [PubMed] [Google Scholar]
- 26.Brasca MG, Albanese C, Alzani R, Amici R, Avanzi N, Ballinari D, et al. Optimization of 6,6-dimethyl pyrrolo[3,4-c]pyrazoles: Identification of PHA-793887, a potent CDK inhibitor suitable for intravenous dosing. Bioorg Med Chem. 2010;18:1844–53. doi: 10.1016/j.bmc.2010.01.042. [DOI] [PubMed] [Google Scholar]
- 27.Leonard JP, LaCasce AS, Smith MR, Noy A, Chirieac LR, Rodig SJ, et al. Selective CDK4/6 inhibition with tumor responses by PD0332991 in patients with mantle cell lymphoma. Blood. 2012;119:4597–607. doi: 10.1182/blood-2011-10-388298. [DOI] [PubMed] [Google Scholar]
- 28.Heath EI, Bible K, Martell RE, Adelman DC, Lorusso PM. A phase 1 study of SNS-032 (formerly BMS-387032), a potent inhibitor of cyclin-dependent kinases 2, 7 and 9 administered as a single oral dose and weekly infusion in patients with metastatic refractory solid tumors. Invest New Drugs. 2008;26:59–65. doi: 10.1007/s10637-007-9090-3. [DOI] [PubMed] [Google Scholar]
- 29.Bagratuni T, Wu P, Gonzalez de CD, Davenport EL, Dickens NJ, Walker BA, et al. XBP1s levels are implicated in the biology and outcome of myeloma mediating different clinical outcomes to thalidomide-based treatments. Blood. 2010;116:250–3. doi: 10.1182/blood-2010-01-263236. [DOI] [PubMed] [Google Scholar]
- 30.Balague O, Mozos A, Martinez D, Hernandez L, Colomo L, Mate JL, et al. Activation of the endoplasmic reticulum stress-associated transcription factor x box-binding protein-1 occurs in a subset of normal germinal-center B cells and in aggressive B-cell lymphomas with prognostic implications. Am J Pathol. 2009;174:2337–46. doi: 10.2353/ajpath.2009.080848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Romero-Ramirez L, Cao H, Nelson D, Hammond E, Lee AH, Yoshida H, et al. XBP1 is essential for survival under hypoxic conditions and is required for tumor growth. Cancer Res. 2004;64:5943–7. doi: 10.1158/0008-5472.CAN-04-1606. [DOI] [PubMed] [Google Scholar]
- 32.Reimold AM, Iwakoshi NN, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, Gravallese EM, et al. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412:300–7. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- 33.Bagratuni T, Wu P, Gonzalez de CD, Davenport EL, Dickens NJ, Walker BA, et al. XBP1s levels are implicated in the biology and outcome of myeloma mediating different clinical outcomes to thalidomide-based treatments. Blood. 2010;116:250–3. doi: 10.1182/blood-2010-01-263236. [DOI] [PubMed] [Google Scholar]
- 34.Mahoney E, Lucas DM, Gupta SV, Wagner AJ, Herman SE, Smith LL, et al. ER stress and autophagy: new discoveries in the mechanism of action and drug resistance of the cyclin-dependent kinase inhibitor flavopiridol. Blood. 2012;120:1262–73. doi: 10.1182/blood-2011-12-400184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shamu CE, Walter P. Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J. 1996;15:3028–39. [PMC free article] [PubMed] [Google Scholar]
- 36.Tirosh B, Iwakoshi NN, Glimcher LH, Ploegh HL. Rapid turnover of unspliced Xbp-1 as a factor that modulates the unfolded protein response. J Biol Chem. 2006;281:5852–60. doi: 10.1074/jbc.M509061200. [DOI] [PubMed] [Google Scholar]
- 37.Lee J, Sun C, Zhou Y, Lee J, Gokalp D, Herrema H, et al. p38 MAPK-mediated regulation of Xbp1s is crucial for glucose homeostasis. Nat Med. 2011;17:1251–60. doi: 10.1038/nm.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park SW, Zhou Y, Lee J, Lu A, Sun C, Chung J, et al. The regulatory subunits of PI3K, p85alpha and p85beta, interact with XBP-1 and increase its nuclear translocation. Nat Med. 2010;16:429–37. doi: 10.1038/nm.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kang MJ, Chung J, Ryoo HD. CDK5 and MEKK1 mediate pro-apoptotic signalling following endoplasmic reticulum stress in an autosomal dominant retinitis pigmentosa model. Nat Cell Biol. 2012;14:409–15. doi: 10.1038/ncb2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saito T, Konno T, Hosokawa T, Asada A, Ishiguro K, Hisanaga S. p25/cyclin-dependent kinase 5 promotes the progression of cell death in nucleus of endoplasmic reticulum-stressed neurons. J Neurochem. 2007;102:133–40. doi: 10.1111/j.1471-4159.2007.04540.x. [DOI] [PubMed] [Google Scholar]
- 41.Santamaria D, Barriere C, Cerqueira A, Hunt S, Tardy C, Newton K, et al. Cdk1 is sufficient to drive the mammalian cell cycle. Nature. 2007;448:811–5. doi: 10.1038/nature06046. [DOI] [PubMed] [Google Scholar]
- 42.Radomska HS, Alberich-Jorda M, Will B, Gonzalez D, Delwel R, Tenen DG. Targeting CDK1 promotes FLT3-activated acute myeloid leukemia differentiation through C/EBPalpha. J Clin Invest. 2012;122:2955–66. doi: 10.1172/JCI43354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dhavan R, Tsai LH. A decade of CDK5. Nat Rev Mol Cell Biol. 2001;2:749–59. doi: 10.1038/35096019. [DOI] [PubMed] [Google Scholar]
- 44.Chen R, Keating MJ, Gandhi V, Plunkett W. Transcription inhibition by flavopiridol: mechanism of chronic lymphocytic leukemia cell death. Blood. 2005;106:2513–9. doi: 10.1182/blood-2005-04-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.MacCallum DE, Melville J, Frame S, Watt K, Anderson S, Gianella-Borradori A, et al. Seliciclib (CYC202, R-Roscovitine) induces cell death in multiple myeloma cells by inhibition of RNA polymerase II-dependent transcription and down-regulation of Mcl-1. Cancer Res. 2005;65:5399–407. doi: 10.1158/0008-5472.CAN-05-0233. [DOI] [PubMed] [Google Scholar]
- 46.Jiang CC, Lucas K, Avery-Kiejda KA, Wade M, deBock CE, Thorne RF, et al. Up-regulation of Mcl-1 is critical for survival of human melanoma cells upon endoplasmic reticulum stress. Cancer Res. 2008;68:6708–17. doi: 10.1158/0008-5472.CAN-08-0349. [DOI] [PubMed] [Google Scholar]
- 47.Johnson AJ, Yeh YY, Smith LL, Wagner AJ, Hessler J, Gupta S, et al. The novel cyclin-dependent kinase inhibitor dinaciclib (SCH727965) promotes apoptosis and abrogates microenvironmental cytokine protection in chronic lymphocytic leukemia cells. Leukemia. 2012;26:2554–7. doi: 10.1038/leu.2012.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gojo I, Sadowska M, Walker A, Feldman EJ, Iyer SP, Baer MR, et al. Clinical and laboratory studies of the novel cyclin-dependent kinase inhibitor dinaciclib (SCH 727965) in acute leukemias. Cancer Chemother Pharmacol. 2013;72:897–908. doi: 10.1007/s00280-013-2249-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luo B, Lee AS. The critical roles of endoplasmic reticulum chaperones and unfolded protein response in tumorigenesis and anticancer therapies. Oncogene. 2012 doi: 10.1038/onc.2012.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Obeng EA, Carlson LM, Gutman DM, Harrington WJ, Jr., Lee KP, Boise LH. Proteasome inhibitors induce a terminal unfolded protein response in multiple myeloma cells. Blood. 2006;107:4907–16. doi: 10.1182/blood-2005-08-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davenport EL, Moore HE, Dunlop AS, Sharp SY, Workman P, Morgan GJ, et al. Heat shock protein inhibition is associated with activation of the unfolded protein response pathway in myeloma plasma cells. Blood. 2007;110:2641–9. doi: 10.1182/blood-2006-11-053728. [DOI] [PubMed] [Google Scholar]
- 52.Rahmani M, Davis EM, Crabtree TR, Habibi JR, Nguyen TK, Dent P, et al. The kinase inhibitor sorafenib induces cell death through a process involving induction of endoplasmic reticulum stress. Mol Cell Biol. 2007;27:5499–513. doi: 10.1128/MCB.01080-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.