Abstract
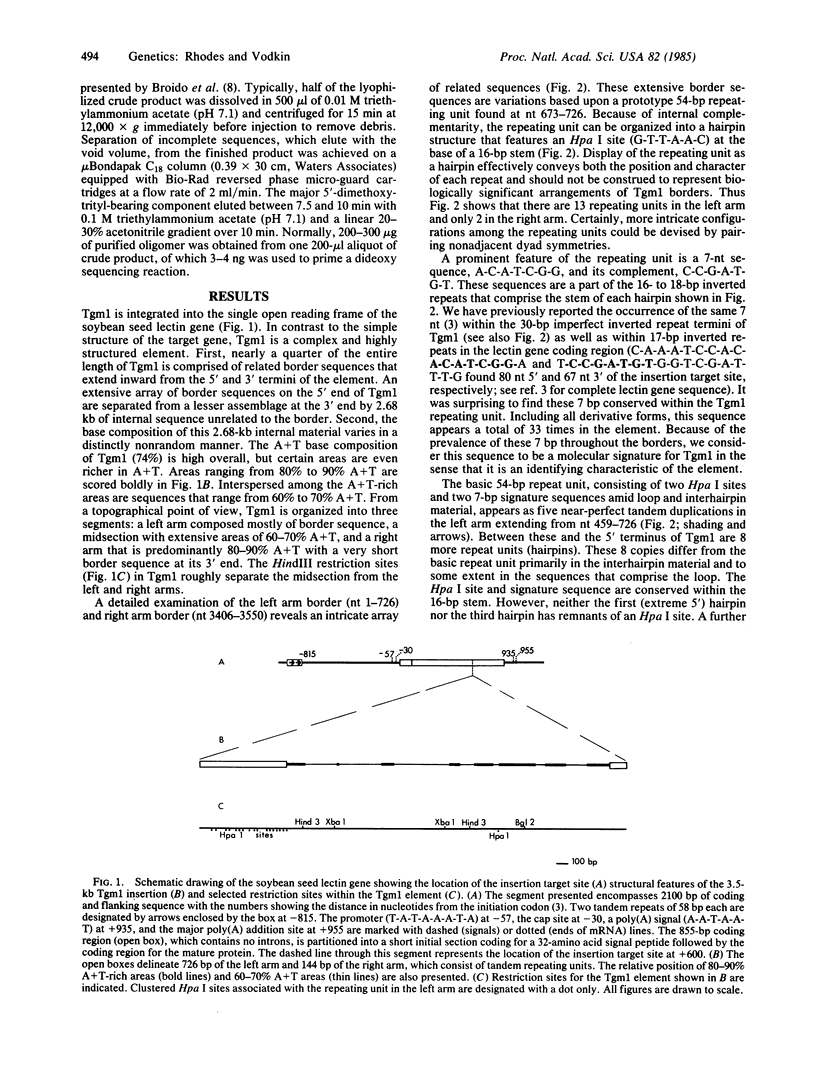
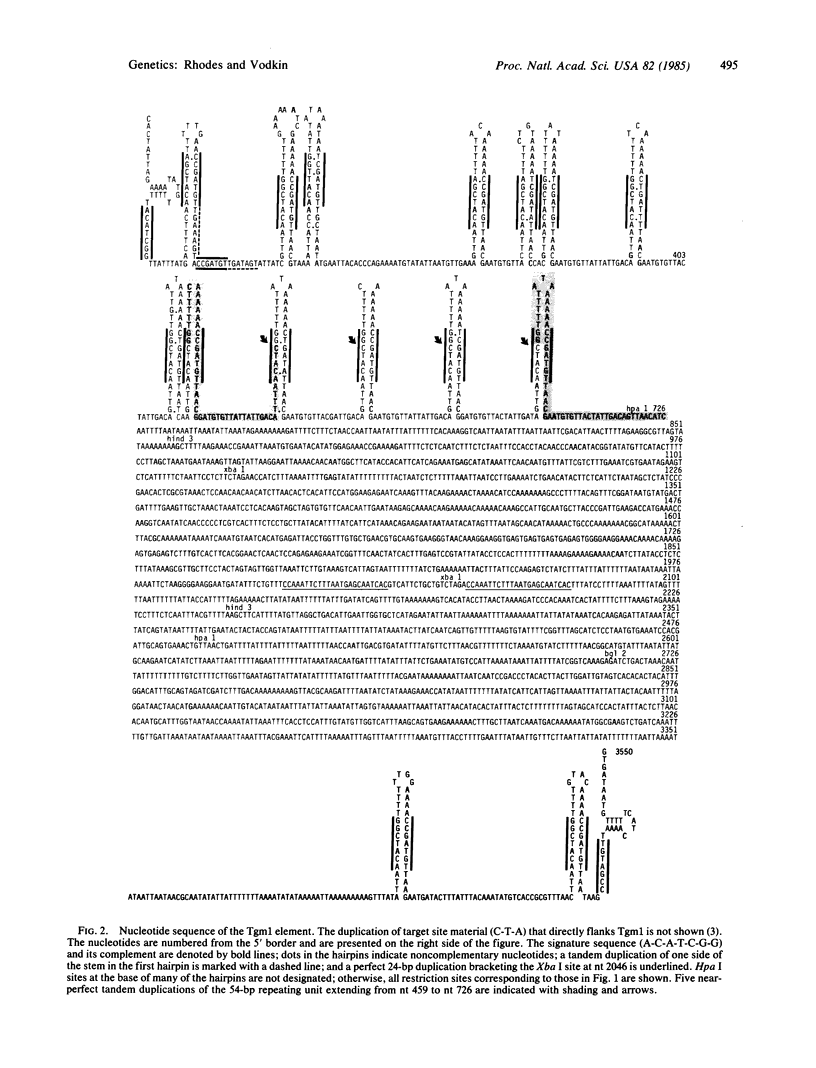
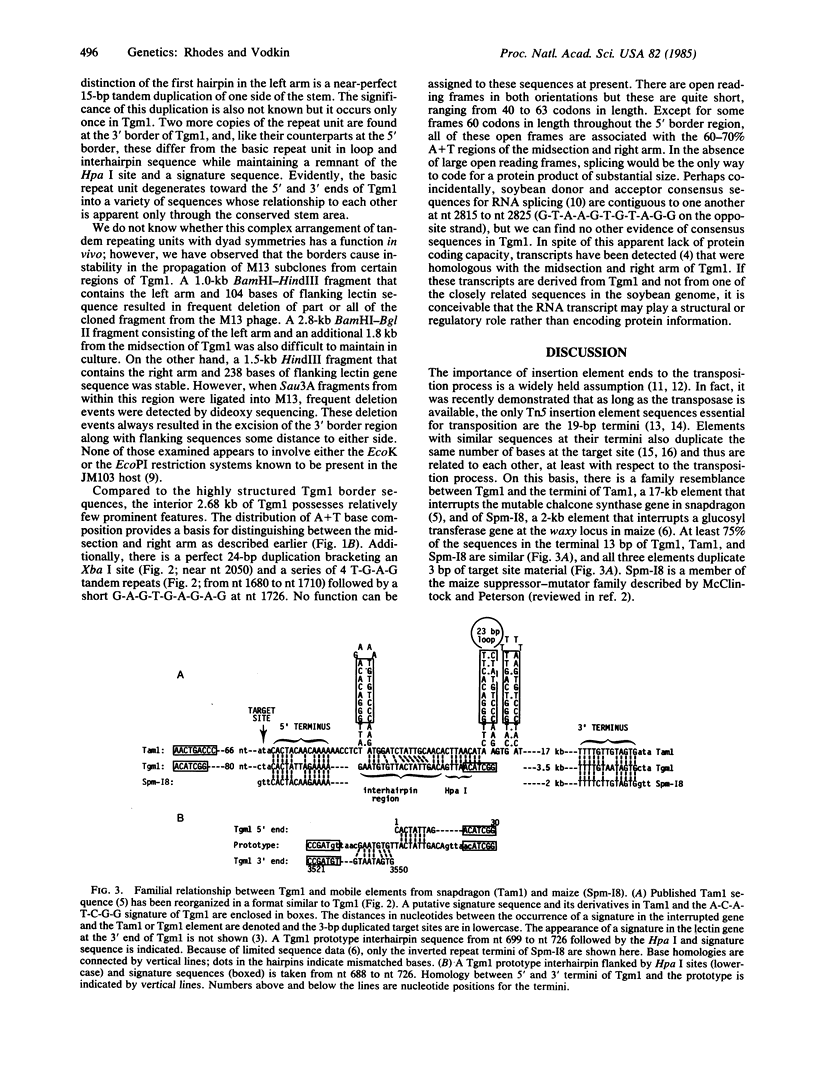
The recessive allele for soybean seed lectin results from the insertion of a DNA segment (designated Tgm1) into the coding region of the gene. The termini of Tgm1 display structural features characteristic of a transposable element. The complete sequence of Tgm1 contains 3550 base pairs (bp) and can be divided into three regions (left arm, mid-section, and right arm). No large open reading frames were found, but an extensive, highly structured border with homology to the lectin gene was revealed. The left border (726 bp) comprising most of the left arm and extreme right border (144 bp) of the right arm consist of various forms of a basic 54-bp repeating unit. This 54-bp unit is comprised of a stem-loop structure and interhairpin sequence that occurs 13 times in the left arm and 2 times in the right arm of Tgm1. Progressively degenerate forms of this repeating unit appear toward the termini of Tgm1, but the dyad symmetry remains highly conserved. Seven nucleotides (A-C-A-T-C-G-G and its complement) maintained within the stem also appear as a subset of inverted repeats found at nearly equal distances from the target site in the lectin gene. Together with the inverted repeat termini and a duplication in the left arm, this 7-bp sequence occurs a total of 33 times in Tgm1. We infer that the dyad symmetries containing this sequence are involved in target gene selection. The repeating unit format of Tgm1 describes a distinct class of eukaryotic elements that includes representatives known to be mobile in snapdragon and maize.
Keywords: soybean seed lectin, transposable element, DNA sequence
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bonas U., Sommer H., Saedler H. The 17-kb Tam1 element of Antirrhinum majus induces a 3-bp duplication upon integration into the chalcone synthase gene. EMBO J. 1984 May;3(5):1015–1019. doi: 10.1002/j.1460-2075.1984.tb01921.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broido M. S., Zon G., James T. L. Complete assignment of the non-exchangeable proton nmr resonances of [d-(GGAATTCC)]2 using two-dimensional nuclear overhauser effect spectra. Biochem Biophys Res Commun. 1984 Mar 15;119(2):663–670. doi: 10.1016/s0006-291x(84)80301-0. [DOI] [PubMed] [Google Scholar]
- Calos M. P., Miller J. H. Transposable elements. Cell. 1980 Jul;20(3):579–595. doi: 10.1016/0092-8674(80)90305-0. [DOI] [PubMed] [Google Scholar]
- Goldberg R. B., Hoschek G., Vodkin L. O. An insertion sequence blocks the expression of a soybean lectin gene. Cell. 1983 Jun;33(2):465–475. doi: 10.1016/0092-8674(83)90428-2. [DOI] [PubMed] [Google Scholar]
- Johnson R. C., Reznikoff W. S. DNA sequences at the ends of transposon Tn5 required for transposition. Nature. 1983 Jul 21;304(5923):280–282. doi: 10.1038/304280a0. [DOI] [PubMed] [Google Scholar]
- Larson R., Messing J. Apple II software for M13 shotgun DNA sequencing. Nucleic Acids Res. 1982 Jan 11;10(1):39–49. doi: 10.1093/nar/10.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCLINTOCK B. Chromosome organization and genic expression. Cold Spring Harb Symp Quant Biol. 1951;16:13–47. doi: 10.1101/sqb.1951.016.01.004. [DOI] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paro R., Goldberg M. L., Gehring W. J. Molecular analysis of large transposable elements carrying the white locus of Drosophila melanogaster. EMBO J. 1983;2(6):853–860. doi: 10.1002/j.1460-2075.1983.tb01513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter S. S. DNA sequence of a foldback transposable element in Drosophila. Nature. 1982 May 20;297(5863):201–204. doi: 10.1038/297201a0. [DOI] [PubMed] [Google Scholar]
- Reed R. R., Young R. A., Steitz J. A., Grindley N. D., Guyer M. S. Transposition of the Escherichia coli insertion element gamma generates a five-base-pair repeat. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4882–4886. doi: 10.1073/pnas.76.10.4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasakawa C., Carle G. F., Berg D. E. Sequences essential for transposition at the termini of IS50. Proc Natl Acad Sci U S A. 1983 Dec;80(23):7293–7297. doi: 10.1073/pnas.80.23.7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-Sommer Z., Gierl A., Klösgen R. B., Wienand U., Peterson P. A., Saedler H. The Spm (En) transposable element controls the excision of a 2-kb DNA insert at the wx allele of Zea mays. EMBO J. 1984 May;3(5):1021–1028. doi: 10.1002/j.1460-2075.1984.tb01922.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek M., DasSarma S., RajBhandary U. L., Khorana H. G. A transposable element from Halobacterium halobium which inactivates the bacteriorhodopsin gene. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7268–7272. doi: 10.1073/pnas.79.23.7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodkin L. O., Rhodes P. R., Goldberg R. B. cA lectin gene insertion has the structural features of a transposable element. Cell. 1983 Oct;34(3):1023–1031. doi: 10.1016/0092-8674(83)90560-3. [DOI] [PubMed] [Google Scholar]



