Abstract
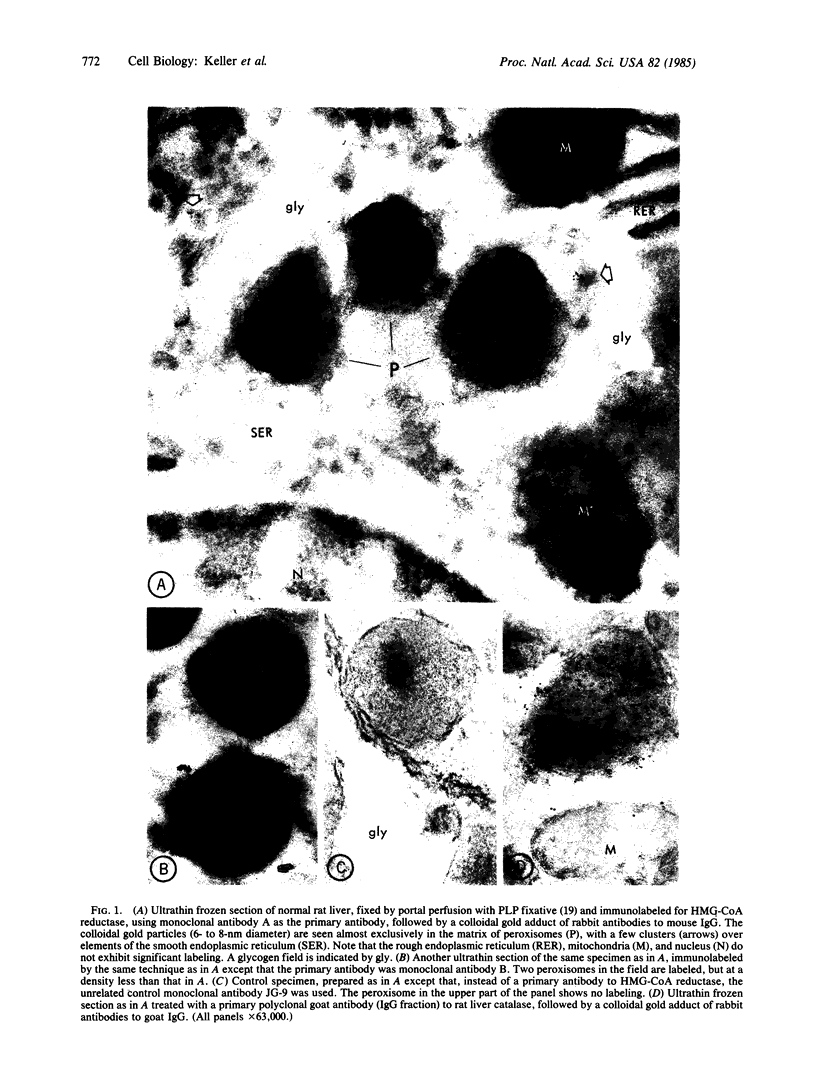
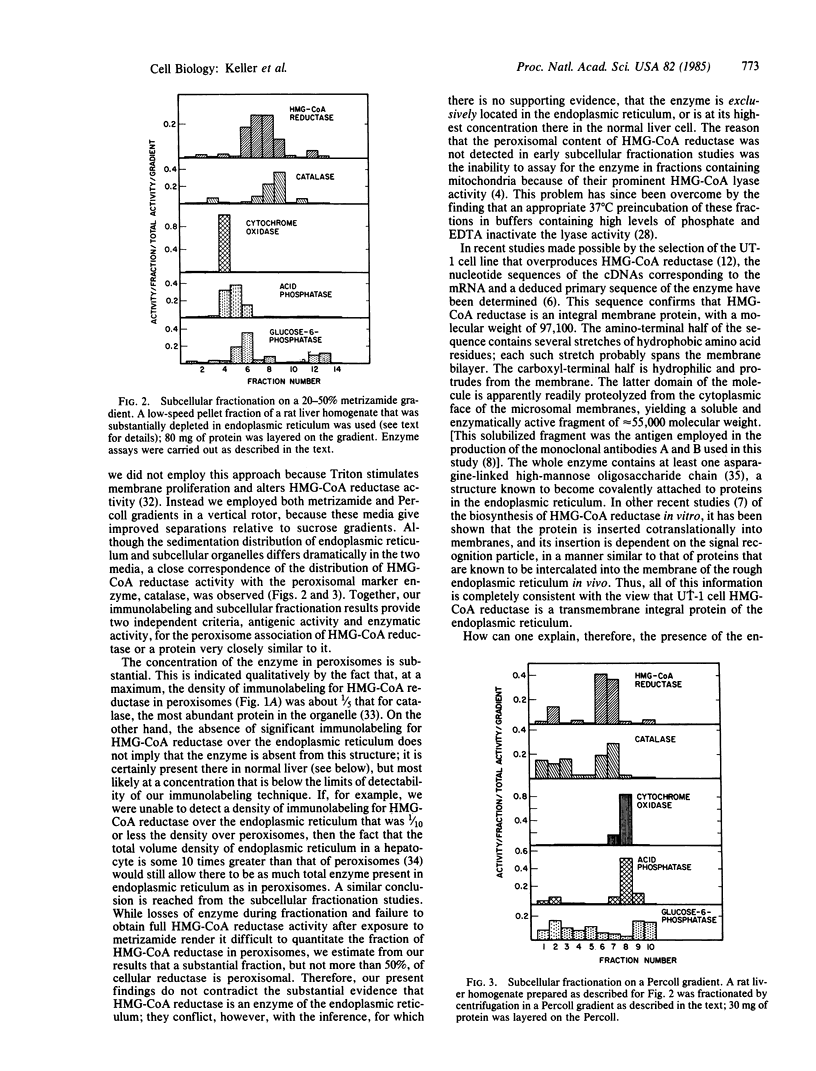
The location inside rat liver parenchymal cells of 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMG-CoA reductase; EC 1.1.1.34), the key regulatory enzyme in cholesterol biosynthesis, has been examined by immunoelectron microscopy and by subcellular fractionation. Although HMG-CoA reductase is generally thought to be exclusively a microsomal enzyme, we find that a substantial portion of cellular HMG-CoA reductase is localized in peroxisomes. Immunoelectron microscopic labeling of ultrathin frozen sections of normal rat liver, using two monoclonal antibodies to purified HMG-CoA reductase, showed that the enzyme is present in the peroxisomes at a higher concentration than at any other site inside the hepatocytes. Subcellular fractionation studies using Percoll and metrizamide gradients demonstrated a close correspondence of peaks of HMG-CoA reductase activity and of catalase activity, again revealing the presence of the reductase enzyme in peroxisomes. HMG-CoA reductase is therefore localized in peroxisomes in addition to being in the microsomal fraction.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BUCHER N. L., OVERATH P., LYNEN F. beta-Hydroxy-beta-methyl-glutaryl coenzyme A reductase, cleavage and condensing enzymes in relation to cholesterol formation in rat liver. Biochim Biophys Acta. 1960 Jun 3;40:491–501. doi: 10.1016/0006-3002(60)91390-1. [DOI] [PubMed] [Google Scholar]
- Baudhuin P. Isolation of rat liver peroxisomes. Methods Enzymol. 1974;31:356–368. doi: 10.1016/0076-6879(74)31039-7. [DOI] [PubMed] [Google Scholar]
- Bronfman M., Inestrosa N. C., Leighton F. Fatty acid oxidation by human liver peroxisomes. Biochem Biophys Res Commun. 1979 Jun 13;88(3):1030–1036. doi: 10.1016/0006-291x(79)91512-2. [DOI] [PubMed] [Google Scholar]
- Brown D. A., Simoni R. D. Biogenesis of 3-hydroxy-3-methylglutaryl-coenzyme A reductase, an integral glycoprotein of the endoplasmic reticulum. Proc Natl Acad Sci U S A. 1984 Mar;81(6):1674–1678. doi: 10.1073/pnas.81.6.1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M. S., Goldstein J. L. Multivalent feedback regulation of HMG CoA reductase, a control mechanism coordinating isoprenoid synthesis and cell growth. J Lipid Res. 1980 Jul;21(5):505–517. [PubMed] [Google Scholar]
- Chin D. J., Gil G., Russell D. W., Liscum L., Luskey K. L., Basu S. K., Okayama H., Berg P., Goldstein J. L., Brown M. S. Nucleotide sequence of 3-hydroxy-3-methyl-glutaryl coenzyme A reductase, a glycoprotein of endoplasmic reticulum. Nature. 1984 Apr 12;308(5960):613–617. doi: 10.1038/308613a0. [DOI] [PubMed] [Google Scholar]
- Clark R. E., Martin G. G., Barton M. C., Shapiro D. J. Production and characterization of monoclonal antibodies to rat liver microsomal 3-hydroxy-3-methylglutaryl-coenzyme A reductase. Proc Natl Acad Sci U S A. 1982 Jun;79(12):3734–3738. doi: 10.1073/pnas.79.12.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DE DUVE C., PRESSMAN B. C., GIANETTO R., WATTIAUX R., APPELMANS F. Tissue fractionation studies. 6. Intracellular distribution patterns of enzymes in rat-liver tissue. Biochem J. 1955 Aug;60(4):604–617. doi: 10.1042/bj0600604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards P. A., Lan S. F., Fogelman A. M. High density lipoproteins and lecithin dispersions increase the activity of 3-hydroxy-3-methylglutaryl coenzyme A reductase by increasing the rate of synthesis and decreasing the rate of degradation of the enzyme. J Biol Chem. 1984 Jul 10;259(13):8190–8194. [PubMed] [Google Scholar]
- Faust J. R., Luskey K. L., Chin D. J., Goldstein J. L., Brown M. S. Regulation of synthesis and degradation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase by low density lipoprotein and 25-hydroxycholesterol in UT-1 cells. Proc Natl Acad Sci U S A. 1982 Sep;79(17):5205–5209. doi: 10.1073/pnas.79.17.5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb Stanley. Submicrosomal localization of hepatic 3-hydroxy-3-methylglutaryl coenzyme a (HMG-CoA) reductase. FEBS Lett. 1972 Aug 1;24(2):153–155. doi: 10.1016/0014-5793(72)80755-5. [DOI] [PubMed] [Google Scholar]
- Greve J. M., Gottlieb D. I. Monoclonal antibodies which alter the morphology of cultured chick myogenic cells. J Cell Biochem. 1982;18(2):221–229. doi: 10.1002/jcb.1982.240180209. [DOI] [PubMed] [Google Scholar]
- Hagey L. R., Krisans S. K. Degradation of cholesterol to propionic acid by rat liver peroxisomes. Biochem Biophys Res Commun. 1982 Aug;107(3):834–841. doi: 10.1016/0006-291x(82)90598-8. [DOI] [PubMed] [Google Scholar]
- Hajra A. K., Bishop J. E. Glycerolipid biosynthesis in peroxisomes via the acyl dihydroxyacetone phosphate pathway. Ann N Y Acad Sci. 1982;386:170–182. doi: 10.1111/j.1749-6632.1982.tb21415.x. [DOI] [PubMed] [Google Scholar]
- Kandutsch A. A., Saucier S. E. Prevention of cyclic and triton-induced increases in hydroxymethylglutaryl coenzyme A reductase and sterol synthesis by puromycin. J Biol Chem. 1969 May 10;244(9):2299–2305. [PubMed] [Google Scholar]
- Keller G. A., Tokuyasu K. T., Dutton A. H., Singer S. J. An improved procedure for immunoelectron microscopy: ultrathin plastic embedding of immunolabeled ultrathin frozen sections. Proc Natl Acad Sci U S A. 1984 Sep;81(18):5744–5747. doi: 10.1073/pnas.81.18.5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarow P. B., Robbi M., Fujiki Y., Wong L. Biogenesis of peroxisomal proteins in vivo and in vitro. Ann N Y Acad Sci. 1982;386:285–300. doi: 10.1111/j.1749-6632.1982.tb21423.x. [DOI] [PubMed] [Google Scholar]
- Leighton F., Poole B., Beaufay H., Baudhuin P., Coffey J. W., Fowler S., De Duve C. The large-scale separation of peroxisomes, mitochondria, and lysosomes from the livers of rats injected with triton WR-1339. Improved isolation procedures, automated analysis, biochemical and morphological properties of fractions. J Cell Biol. 1968 May;37(2):482–513. doi: 10.1083/jcb.37.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum L., Cummings R. D., Anderson R. G., DeMartino G. N., Goldstein J. L., Brown M. S. 3-Hydroxy-3-methylglutaryl-CoA reductase: a transmembrane glycoprotein of the endoplasmic reticulum with N-linked "high-mannose" oligosaccharides. Proc Natl Acad Sci U S A. 1983 Dec;80(23):7165–7169. doi: 10.1073/pnas.80.23.7165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luskey K. L., Chin D. J., MacDonald R. J., Liscum L., Goldstein J. L., Brown M. S. Identification of a cholesterol-regulated 53,000-dalton cytosolic protein in UT-1 cells and cloning of its cDNA. Proc Natl Acad Sci U S A. 1982 Oct;79(20):6210–6214. doi: 10.1073/pnas.79.20.6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean I. W., Nakane P. K. Periodate-lysine-paraformaldehyde fixative. A new fixation for immunoelectron microscopy. J Histochem Cytochem. 1974 Dec;22(12):1077–1083. doi: 10.1177/22.12.1077. [DOI] [PubMed] [Google Scholar]
- Neat C. E., Thomassen M. S., Osmundsen H. Induction of peroxisomal beta-oxidation in rat liver by high-fat diets. Biochem J. 1980 Jan 15;186(1):369–371. doi: 10.1042/bj1860369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikoff A. B., Novikoff P. M. Microperoxisomes and peroxisomes in relation to lipid metabolism. Ann N Y Acad Sci. 1982;386:138–152. doi: 10.1111/j.1749-6632.1982.tb21412.x. [DOI] [PubMed] [Google Scholar]
- Orci L., Brown M. S., Goldstein J. L., Garcia-Segura L. M., Anderson R. G. Increase in membrane cholesterol: a possible trigger for degradation of HMG CoA reductase and crystalloid endoplasmic reticulum in UT-1 cells. Cell. 1984 Apr;36(4):835–845. doi: 10.1016/0092-8674(84)90033-3. [DOI] [PubMed] [Google Scholar]
- Robbi M., Lazarow P. B. Peptide mapping of peroxisomal catalase and its precursor. Comparison to the primary wheat germ translation product. J Biol Chem. 1982 Jan 25;257(2):964–970. [PubMed] [Google Scholar]
- Rodwell V. W., Nordstrom J. L., Mitschelen J. J. Regulation of HMG-CoA reductase. Adv Lipid Res. 1976;14:1–74. doi: 10.1016/b978-0-12-024914-5.50008-5. [DOI] [PubMed] [Google Scholar]
- Shapiro D. J., Imblum R. L., Rodwell V. W. Thin-layer chromatographic assay for HMG-CoA reductase and mevalonic acid. Anal Biochem. 1969 Oct 1;31(1):383–390. doi: 10.1016/0003-2697(69)90279-6. [DOI] [PubMed] [Google Scholar]
- Shapiro D. J., Nordstrom J. L., Mitschelen J. J., Rodwell V. W., Schimke R. T. Micro assay for 3-hydroxy-3-methylglutaryl-CoA reductase in rat liver and in L-cell fibroblasts. Biochim Biophys Acta. 1974 Dec 29;370(2):369–377. doi: 10.1016/0005-2744(74)90098-9. [DOI] [PubMed] [Google Scholar]
- Shapiro D. J., Rodwell V. W. Regulation of hepatic 3-hydroxy-3-methylglutaryl coenzyme A reductase and cholesterol synthesis. J Biol Chem. 1971 May 25;246(10):3210–3216. [PubMed] [Google Scholar]
- Tokuyasu K. T. Immunochemistry on ultrathin frozen sections. Histochem J. 1980 Jul;12(4):381–403. doi: 10.1007/BF01011956. [DOI] [PubMed] [Google Scholar]
- Weibel E. R., Stäubli W., Gnägi H. R., Hess F. A. Correlated morphometric and biochemical studies on the liver cell. I. Morphometric model, stereologic methods, and normal morphometric data for rat liver. J Cell Biol. 1969 Jul;42(1):68–91. doi: 10.1083/jcb.42.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]