Abstract
In addition to redox regulation, protein phosphorylation has gained increasing importance as a regulatory principle in chloroplasts in recent years. However, only very few chloroplast-localized protein kinases have been identified to date. Protein phosphorylation regulates important chloroplast processes such as photosynthesis or transcription. In order to better understand chloroplast function, it is therefore crucial to obtain a complete picture of the chloroplast kinome, which is currently constrained by two effects: first, recent observations showed that the bioinformatics-based prediction of chloroplast-localized protein kinases from available sequence data is strongly biased; and, secondly, protein kinases are of very low abundance, which makes their identification by proteomics approaches extremely difficult. Therefore, the aim of this study was to obtain a complete list of chloroplast-localized protein kinases from different species. Evaluation of protein kinases which were either highly predicted to be chloroplast localized or have been identified in different chloroplast proteomic studies resulted in the confirmation of only three new kinases. Considering also all reports of experimentally verified chloroplast protein kinases to date, compelling evidence was found for a total set of 15 chloroplast-localized protein kinases in different species. This is in contrast to a much higher number that would be expected based on targeting prediction or on the general abundance of protein kinases in relation to the entire proteome. Moreover, it is shown that unusual protein kinases with differing ATP-binding sites or catalytic centres seem to occur frequently within the chloroplast kinome, thus making their identification by mass spectrometry-based approaches even more difficult due to a different annotation.
Keywords: Casein kinase, chloroplast protein kinase, organellar proteomics, photosynthesis, STN7, STN8, subcellular localization, YFP fusion protein
Introduction
Chloroplasts are essential plant organelles of endosymbiotic origin that carry out a wide range of important metabolic pathways such as photosynthesis, and the biosynthesis of amino acids, vitamins, and lipids. These processes require tight regulation and coordination with the metabolic state of the whole plant. To this end, chloroplasts are integrated into the signalling network of the entire cell. In addition to the predominant redox regulation in chloroplasts (Montrichard et al., 2009), reversible protein phosphorylation is a key mechanism for the regulation of cellular processes and for signal transduction in response to environmental changes (Cohen, 2000). Phosphorylation can influence the activity, the subcellular localization, or the stability of target proteins (Stone and Walker, 1995; Cohen, 2000; Schliebner et al., 2008). Plant protein kinases are involved in the regulation of metabolism, cell division, growth, and differentiation, and they mediate cellular responses to biotic and abiotic stress including changing light conditions, altered temperatures, and pathogen invasion (Stone and Walker, 1995; Halford et al., 2004). In eukaryotic genomes it is estimated that 1–3% of all genes encode protein kinases (Stone and Walker, 1995). In the human genome, 518 out of the ~22 200 proteins are annotated as protein kinases (2.3%) (Manning et al., 2002; Orchard et al., 2005), and the nuclear genome of the model plant Arabidopsis thaliana (~27 400 protein-coding genes) encodes ~1050 different protein kinases (3.8%) (Gribskov et al., 2001; Wang et al., 2003; Martin et al., 2009). This slightly higher number in Arabidopsis is probably a result of multiple gene duplications that are generally found in plant genomes (Chevalier and Walker, 2005). No protein kinases are encoded in the chloroplast genome (TAIR; www.arabidopsis.org).
Historically, the first report of protein phosphorylation within chloroplasts dates back to the 1970s, when phosphorylation of light-harvesting complex (LHC) proteins was shown (Bennett, 1977). Stromal protein phosphorylation was discovered in 1983, when pyruvate, orthophosphate dikinase was found to be inactivated by phosphorylation in Zea mays chloroplasts (Ashton and Hatch, 1983). Since that time, more and more targets of protein phosphorylation have been identified in chloroplasts, using different techniques such as incubation of chloroplast protein extracts with radioactively labelled ATP or detection with phosphothreonine-specific antibodies (Laing and Christeller, 1984; Foyer, 1985; Rintamaki et al., 1997). Given that ~2100 proteins are estimated to be imported into the chloroplast based on targeting prediction (Richly and Leister, 2004) and 3.8% of all Arabidopsis proteins encode protein kinases, at least 80 chloroplast protein kinases could be expected (i.e. 3.8% of 2100). However, to date, only a handful of chloroplast-localized protein kinases have been reported in the literature. The most thoroughly described examples are the protein kinases Stt7 and Stl1 from Chlamydomonas reinhardtii and their Arabidopsis orthologues STN7 (At1g68830) and STN8 (At5g01920) (Bellafiore et al., 2005; Bonardi et al., 2005). These kinases were first discovered in a genetic screen in Chlamydomonas aiming at the identification of regulatory factors for photosynthetic acclimation under changing light conditions, a process called state transition (Depege et al., 2003). These kinases are localized within the thylakoid membrane and phosphorylate photosystem II (PSII) core and LHCII proteins resulting in the redistribution of LCHIIs. Analysis of fluorescence recovery after photobleaching revealed that a small portion of chlorophyll (~15%) could move between grana stacks within a 10 min time scale in an STN7-dependent manner (Goral et al., 2010). The phosphorylation of PSII also leads to visible macroscopic changes in the folding of thylakoid membranes. The lack of phosphorylation in the stn7/stn8 double mutant results in an increased size of appressed thylakoid membranes (Fristedt et al., 2009). STN7 was recently shown to provide a link between short-term (state transition) and long-term (nuclear gene expression) acclimation to fluctuating light conditions (Pesaresi et al., 2009). This long-term acclimation also includes a readjustment of distinct metabolic states (Brautigam et al., 2009). All together these processes optimize photosynthesis under fluctuating light conditions, and, accordingly, the phenotype of stn7 mutants becomes particularly obvious under such growth conditions (Brautigam et al., 2009; Tikkanen et al., 2010).
Another well-described chloroplast protein kinase is the chloroplast casein kinase 2 (cpCK2), which was first identified in Sinapis alba. Later, the Arabidopsis homologue (At2g23070) was found to be associated with the plastid RNA polymerase and to phosphorylate parts of the transcription machinery and RNA-binding proteins (Ogrzewalla et al., 2002; Salinas et al., 2006). The plastid sigma factor AtSIG6 is phosphorylated by cpCK2 at different sites. This results in specific and efficient promoter binding of the polymerase, which leads to altered expression of chloroplast genes (Schweer et al., 2010). Recently, the chloroplast sensor kinase CSK (AT1G67840), which is a prokaryotic-like two-component histidine kinase, was shown to couple photosynthesis to chloroplast gene expression via redox signals (Puthiyaveetil et al., 2008) and was identified as an interaction partner of cpCK2 (Puthiyaveetil et al., 2010). Furthermore, studies in Pisum sativum and Spinacia oleracea provided evidence for the presence of protein kinases in the outer envelope of chloroplasts (Soll, 1985, 1988; Soll et al., 1988; Siegenthaler and Bovet, 1993). The importance of phosphorylation in chloroplasts is further highlighted by the results of a recent phosphoproteomic study in Arabidopsis (Reiland et al., 2009). In this study, a total of 353 unique phosphosites was identified in 174 proteins, which were annotated as chloroplast proteins with high confidence. These proteins included LHC proteins, proteins involved in RNA binding and carbohydrate metabolism, photosystem core subunits, and STN7.
In order to understand chloroplast function fully it is crucial to obtain a complete list of protein kinases involved in the regulation of chloroplast processes. Here, the aim is to compile a complete inventory of chloroplast-localized protein kinases according to the current state of knowledge. To this end, the literature was screened for all experimentally verified chloroplast protein kinases, and experimental reports of protein kinases with assigned chloroplast localization were evaluated by analysis of yellow fluorescent protein (YFP) fusion proteins, including also candidates which have been identified only with low confidence in proteomics studies, and candidates selected from bioinformatics-based approaches.
Materials and methods
Plant growth and chloroplast preparations
Arabidopsis thaliana (var. Columbia Col-0) was grown on soil at 22 °C under a 16 h/8 h photoperiod at 150 μmol photons m −2 s −1 for 7 weeks. Chloroplasts were purified as described previously (Seigneurin-Berny et al., 2008) and stored at −80 °C until further use.
Pisum sativum (cultivar Arvika) plants were grown under a 16 h light/8 h dark photoperiod at 21/16 ° (light/dark) at 250 μmol photons m −2 s −1. Chloroplasts were isolated from leaves of 10- to 12-day-old peas as described previously (Waegemann and Soll, 1995).
Preparation of membrane and soluble Arabidopsis chloroplast proteins
Chloroplasts were disrupted by lysis in kinase buffer [20 mM Tricine pH 7.6, 10% glycerol, 0.5% dithiothreitol (DTT)], supplemented with protease inhibitors (complete EDTA free; Roche) and phosphatase inhibitors (Phospho-Stop; Roche). After incubation on ice for 15 min, membranes and soluble components were separated by centrifugation at 60 000 g for 10 min. To remove loosely associated proteins, the membrane fraction was washed with lysis buffer containing 0.8 M NaCl and re-suspended in lysis buffer. The 60 000 g supernatants of the first and second centrifugation were combined, desalted via a PD10 column (GE Healthcare), and concentrated using Vivaspin 500 spin columns (3 kDa cut-off, GE Healthcare). All operations were carried out either on ice or at 4 °.
Protein phosphorylation assays in Arabidopsis
A standard phosphorylation assay (50 μl), using soluble or total membrane proteins, was carried out in lysis buffer supplemented with 2 mM EGTA and 10 mM MgCl2. For the measurement of ATP-dependent phosphorylation, 5 μM cold ATP and 2–5 μCi of [γ-32P]ATP (3000 Ci mmol −1, Perkin Elmer, Waltham, MA, USA) were included in the assay. Control assays for ATP-dependent phosphorylation were carried out using recombinant CPK4 (At4g09570) with histone (S3, Sigma) as substrate. Due to the calcium dependency of CPKs (calcium-dependent protein kinases), the assay included 5 mM CaCl2 instead of EGTA. For the measurement of GTP-dependent phosphorylation the assay contained 5 μM cold GTP and 2–5 μCi of [γ-32P]GTP (6000 Ci mmol −1, Perkin Elmer). Control assays for GTP-dependent phosphorylation were performed using recombinant CK2 (α2, human, Calbiochem) with dephosphorylated casein (from bovine milk, Sigma Aldrich) as substrate. All reactions were carried out for 25 min at room temperature and stopped by the addition of 12 μl of 4× SDS sample buffer.
For the analysis of calcium-dependent phosphorylation in chloroplasts, stromal extracts were incubated with 0.1 μl of [γ-32P]ATP (6000 Ci mmol −1, 10 Ci ml −1; Perkin Elmer), 4 μl of 5× kinase buffer, and 5 mM CaCl2 or 5 mM EGTA in a total volume of 20 μl. The kinase reactions were incubated for 20 min at room temperature and subsequently analysed by SDS–PAGE. The incorporation of γ-32P into proteins was analysed using a Storage Phosphor Screen and a Typhoon Trio Imager by the software ImageQuant (GE Healthcare).
Total leaf protein extraction from pea seedlings
Leaves of 7-day-old P. sativum seedlings were homogenized in extraction buffer [330 mM sorbitol, 20 mM MOPS, 13 mM TRIS, 0.1% bovine serum albumin (BSA), 3 mM MgCl2] using a Waring blender. After filtration through four layers of Miracloth (Merck) and centrifugation for 2 min at 2314 g and 4 °C, the supernatant was buffer exchanged to buffer A (50 mM TRIS pH 7.8, 50 mM NaCl, 10 mM MgCl2) using PD-10 Desalting Columns (GE Healthcare). The final protein extract was stored at 4 °C until further usage.
Stromal protein extraction from pea chloroplasts
Stromal protein extraction from isolated chloroplasts of pea seedlings and subsequent depletion of RuBisCO by size exclusion chromatography was carried out as previously described (Bayer et al., 2011).
Protein phosphorylation assays and inhibitor studies in pea
For the analysis of calcium-dependent phosphorylation, stromal extracts were incubated with 0.1 μl of [γ-32P]ATP (6000 Ci mmol −1, 10 Ci ml −1; Perkin Elmer), 4 μl of 5× kinase buffer, and CaCl2 (0.010–10 mM) or 5 mM MgCl2 in the presence or absence of 5 mM EGTA in a total volume of 20 μl. For protein kinase inhibitor experiments, total pea leaf or stromal protein extracts were mixed with 0.1 μl of [γ-32P]ATP, 4 μl of 5× kinase buffer (100 mM HEPES, 75 mM MgCl2, 1 mM DTT, pH 7.5 with KOH), and 2.5 μl of 800 μM purvalanol B (PurB; in dimethylsulphoxide; Tocris Bioscience) in a total volume of 20 μl. All kinase reactions were incubated for 20 min at room temperature. Subsequently, 8 μl of 4× SDS-PAGE buffer (Laemmli, 1970) were added to each reaction mix and 8 μl were separated on a 12% SDS gel. After Coomassie staining/destaining and drying of the gel, the incorporation of γ-32P into proteins was analysed using a Storage Phosphor Screen and a Typhoon Trio Imager by the software ImageQuant (GE Healthcare).
Subcellular localization analysis of candidate proteins
All candidate protein kinases were cloned in front of YFP in the expression vector pBIN19, expressed in tobacco epidermal cells, and subsequently examined for their subcellular localization by confocal laser scanning microscopy as described previously (Benetka et al., 2008).
Results and Discussion
Selection and localization analysis of candidate protein kinases
First, the aim was to identify novel chloroplast-localized protein kinases by using a bioinformatics-based approach. Therefore, 10 protein kinases with clear prediction for chloroplast localization by different prediction methods (Supplementary Table S1 available at JXB online) were cloned and their subcellular localization as YFP fusion proteins in infiltrated tobacco leaves was investigated. Unexpectedly, a chloroplast localization could not be confirmed for any kinase tested (Supplementary Fig. S1), thus underpinning the previous observation that the prediction of chloroplast targeting is particularly biased for protein kinases, which is in many cases due to interference with N-terminal protein acylation (Mehlmer et al., 2010;Stael et al., 2011). Similar results were previously reported in an analysis of protein kinases, which were also predicted for chloroplast targeting by several prediction programs (Schliebner et al., 2008). Also, in that study, chloroplast localization could only be verified for two out of the nine tested candidates. Combining both studies, in total only two out of 18 protein kinases with a firm prediction for chloroplast localization by different programs could be experimentally confirmed. This result is in strong contrast to the known specificity of ~80% of those targeting prediction programs (Emanuelsson and von Heijne, 2001;Richly and Leister, 2004; Zybailov et al., 2008).
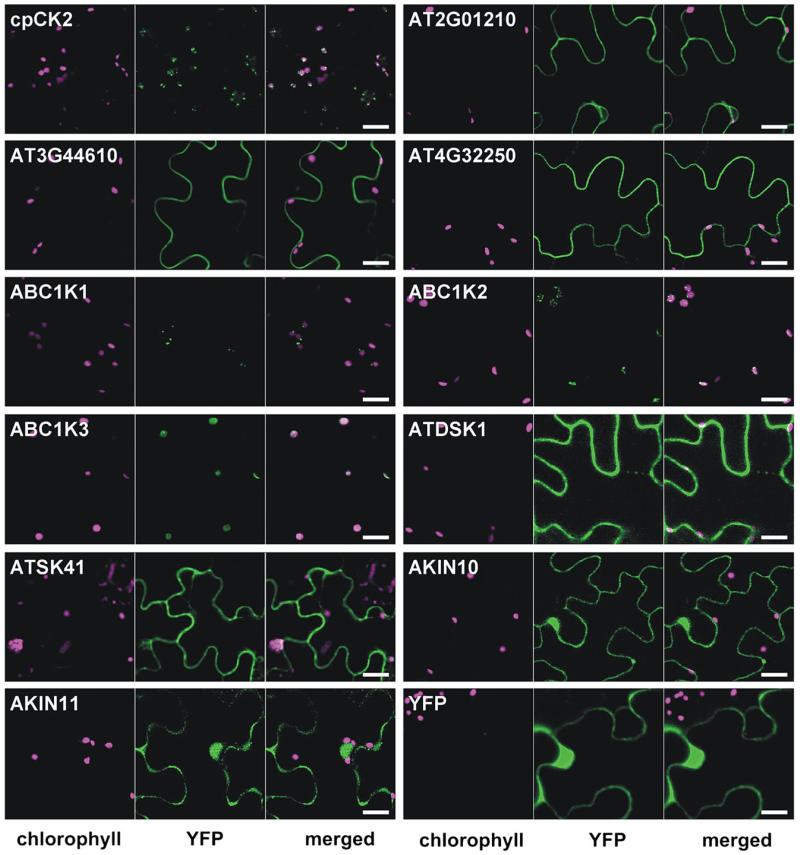
Secondly, available organellar proteomic databases and the literature were searched for experimental evidence of chloroplast-localized protein kinases from Arabidopsis and other plant species, and verification of the localization of the Arabidopsis proteins was sought by YFP fusion analysis. cpCK2 was used as positive control and the following 10 protein kinases were selected: At2g01210, At3g44610, At4g32250, ABC1K1 (At1g71810), ABC1K2 (At1g79600), ABC1K3 (At4g31390), AtDSK1 (At3g13690), AtSK41 (At1g09840), AKIN10 (At3g01090), and AKIN11 (At3g29160) (Fig. 1). At2g01210 and At4g32250 were assigned to chloroplasts in mass spectrometric studies (Dunkley et al., 2006; Zybailov et al., 2008; Ferro et al., 2010), and At3g44610 is listed in the plastid database plprot (Kleffmann et al., 2006). In the Plant Proteome Database (PPDB) six members of the ABC1 family are annotated as putative plastid protein kinases (ABC1K1–ABC1K6) in addition to the known chloroplast protein kinases STN7, STN8, and cpCK2 (Sun et al., 2008). However, according to the protein domain database PROSITE (www.expasy.org/prosite), ABC1K4 (At5g05200) and ABC1K5 (At5g64940) do not contain a protein kinase domain and were therefore excluded from further studies. The ABC1 kinases were identified in proteomic studies of plastoglobules (Ytterberg et al., 2006; Zybailov et al., 2008). These are lipoprotein particles inside chloroplasts, which function in the storage and synthesis of vitamin E, lipids, and quinones (Vidi et al., 2006). In addition to the protein kinase domain, these kinases contain an ABC1 domain, which was first described in the yeast mitochondrial ABC1 protein in the regulation of the bc1 complex (Bousquet et al., 1991).
Fig. 1.
Localization analysis of selected protein kinases. Tobacco leaves infiltrated with genes of interest fused in front of YFP in the plant expression plasmid pBIN19 were analysed by confocal laser scanning microscopy 2 d after infiltration. Chlorophyll autofluorescence (magenta) is shown in the first channel and the YFP signal (green) in the second channel. The third channel shows the merged image. Scale bar=20 μm.
Further reports of chloroplast protein kinases from other organisms include MsK4 from Medicago sativa and NtDSK1 from Nicotiana tabacum. MsK4 was found to localize to starch granules and to be involved in the regulation of carbohydrate metabolism in response to salt stress (Kempa et al., 2007). NtDSK1 is a dual-specificity protein kinase and its expression is regulated in response to light, suggesting a role in chloroplast light signalling (Cho et al., 2001). The closest Arabidopsis homologues of MsK4 and NtDSK1, AtSK41 and AtDSK1, respectively, were analysed. Further, the SNF1-related protein kinases AKIN10 and AKIN11, which were recently reported to be chloroplast localized (Fragoso et al., 2009), were also analysed. Finally, YFP alone was included as a negative control.
Chloroplast localization, indicated by an overlap of the YFP signal and the chlorophyll autofluorescence, could only be confirmed for the positive control cpCK2 and the three ABC1 family protein kinases. In contrast, At2g01210, At3g44610, and At4g32250 seemed to localize to the plasma membrane, and AtDSK1 and AtSK41 to the plasma membrane and the cytoplasm. AKIN10 and AKIN11 were found in the cytoplasm as well as in the nucleus (Fig. 1), which is in perfect agreement with their proposed function as central regulators of energy metabolism in response to starvation and stress (Baena-Gonzalez et al., 2007), and with the localization of their substrates (Baena-Gonzalez et al., 2007; Jossier et al., 2009). The negative control YFP was observed in the nucleus and in the cytoplasm.
A current survey of chloroplast-localized protein kinases
To summarize the current state of knowledge on chloroplast-localized protein kinases, a revised survey was created by integrating the data obtained in this study and data extracted from the current literature. The search was extended to all chloroplast-containing organisms so far investigated (Table 1). It is important to note that the chloroplast localization of a protein kinase was only considered to be true if compelling experimental evidence such as immunolocalization or fluorescent protein fusion studies, or chloroplast import assays were found. Sole identification in a chloroplast proteomic study was not accepted as a proof of localization, due to the high risk of detecting contaminations with the sensitive mass spectrometry instruments (Baginsky, 2009).
Table 1.
Survey of chloroplast-localized protein kinases with conventional and unusual ATP binding or active sites
| Name | AGIcode (or GenBank) | Organism(s) | PROSITE kinase domain | Reference(s) | ||
|---|---|---|---|---|---|---|
| Chloroplast localized | Conventional | Stt7/STN7 | AT1G68830 | Chlamydomonas; Arabidopsis | Complete | Depege et al. (2003); Bellafiore et al. (2005); Bonardi et al. (2005) |
| Stl1/STN8 | AT5G01920 | Chlamydomonas; Arabidopsis | Complete | Depege et al. (2003); Bonardi et al. (2005) | ||
| cpCK2 | AT2G23070 | Sinapis alba; Arabidopsis | Complete | Ogrzewalla et al. (2002); Salinas et al. (2006) | ||
| TAK1 | AT4G02630 | Arabidopsis | Complete | Snyders and Kohorn (1999) | ||
| CIPK13 | AT2G34180 | Arabidopsis | Complete | Schliebner et al. (2008) | ||
| At1g51170 | AT1G51170 | Arabidopsis | Complete | Schliebner et al. (2008) | ||
| MsK4 | AF432335.1 | Medicago sativa | Complete | Kempa et al. (2007) | ||
| NtDSK1 | AF106957.1 | Nicotiana tabacum | Complete | Cho et al. (2001) | ||
| Unusual | ABC1K1 | AT1G71810 | Arabidopsis | Unusual ATP site | Ytterberg et al. (2006); Zybailov et al. (2008); this study | |
| ABC1K2 | AT1G79600 | Arabidopsis | Unusual ATP site | Vidi et al. (2006); Ytterberg et al. (2006); Zybailov et al. (2008); this study | ||
| ABC1K3 | AT4G31390 | Arabidopsis | Unusual ATP site | Ytterberg et al. (2006); Zybailov et al. (2008); this study | ||
| AtRP1 | AT4G21210 | Zea mays; Arabidopsis | Absent | Burnell and Chastain (2006) | ||
| CSK | AT1G67840 | Arabidopsis | Absent | Puthiyaveetil et al. (2008) | ||
| NDPK2 | AT5G63310 | Arabidopsis | Absent | Bolter etal. (2007); Reiland et al. (2009) | ||
| PPK | AT5G16810 | Arabidopsis | Single D absent | Bayer etal. (2011) | ||
| Localization ambiguous | Conventional | AKIN10 | AT3G01090 | Arabidopsis | Complete | Fragoso et al. (2009); this study |
| AKIN11 | AT3G29160 | Arabidopsis | Complete | Fragoso et al. (2009); this study | ||
| MKK4 | AT1G51660 | Arabidopsis | Complete | Samuel et al. (2008) | ||
| AtSK41 | AT1G09840 | Arabidopsis | Complete | This study | ||
| AtDSK1 | AT3G13690 | Arabidopsis | Complete | This study |
In a similar survey from 2008 (Schliebner et al., 2008), the seven protein kinases STN7, STN8, cpCK2, AtRP1 (At4g21210), TAK1 (At4g02630), At1g51170, and CIPK13 (At2g34180) were listed. AtRP1 was classified as a protein phosphatase, exhibiting both protein kinase and phosphatase activity on pyruvate, orthophosphate dikinase (Chastain et al., 2008). TAK1 was originally purified from thylakoids using a specific antiserum (Snyders and Kohorn, 1999), and its chloroplast localization has also been confirmed by chloroplast import assays in an independent study (Bonardi, 2006). TAK1 is part of a family of protein kinases together with TAK2 (At1g01540) and TAK3 (At4g01330), which both have initially been suggested to localize to thylakoids due to sequence homology with TAK1 (Snyders and Kohorn, 1999). However, in the meantime, TAK2 and TAK3 have been found localized at the plasma membrane according to the PPDB database. The protein kinases At1g51170 and CIPK13 were both identified due to the presence of a predicted chloroplast targeting peptide, and their localization was verified by red fluorescent protein fusion analysis (Schliebner et al., 2008). Whereas nothing is known about At1g51170, CIPK13 belongs to the well-characterized family of calcineurin B-like protein (CBL)-interacting protein kinases (CIPKs) (Batistic and Kudla, 2009).
Based on YFP analyses carried out in this study, the ABC1 protein kinases ABC1K1, ABC1K2, and ABC1K3 could now be added to the complement of chloroplast-localized protein kinases. It should be noted, however, that ABC1K6 (At3g24190), which exhibits high sequence homology (e-value 3e −113) with the other kinases of the ABC1 kinase family, is most probably chloroplast localized as well, as it has been identified with high confidence in a chloroplast proteomic study (Zybailov et al., 2008). Furthermore, the chloroplast-localized nucleoside diphosphate kinase 2 (NDPK2; At5g63310), which has been shown to undergo autophosphorylation (Shen et al., 2006), was included. NDPK2 had originally been implicated in phytochrome-mediated signalling in the cytoplasm (Choi et al., 1999), and in salt stress and H2O2 signalling involving SOS2 and two mitogen-activated protein kinases (MPK3 and MPK6) (Moon et al., 2003;Verslues et al., 2007). However, recently, NDPK2 was clearly shown to be localized in chloroplasts by green fluorescent protein analysis, import assay, and immunoblotting (Bolter et al., 2007). Moreover, the protein kinases MsK4 and NtDSK1, the chloroplast sensor kinase CSK, and the protein kinase PPK (At5g16810), which has recently been identified in a directed proteomic study (Bayer et al., 2011), could be included (Table 1).
In contrast, contradictory evidence was obtained for the localization of the protein kinases AKIN10 and AKIN11, and therefore their localization was considered as ambiguous. Similarly, the Arabidopsis MAPKK MKK4 (At1g51660) was shown to be targeted to chloroplasts by an in vitro import assay (Samuel et al., 2008). However, this is in contradiction to the nuclear and cytoplasmic localization observed in a previous study (Koroleva et al., 2005) and to its function in the cytoplasmic- and nuclear-localized MAPK signalling cascade. Therefore, the chloroplast localization of MKK4 is also contested. Finally, AtSK41 and AtDSK1, the closest Arabidopsis homologues of Medicago MsK4 and tobacco NtDSK1, respectively, could not be verified inside the chloroplast by YFP fusion analysis. However, MsK4 appeared to be localized in chloroplasts only when starch was present, which might explain the difference. It could also be that AtSK41 is not the real Arabidopsis orthologue of MsK4, because it belongs to the family of GSK3/shaggy-like protein kinases consisting of at least 10 members with overall high sequence homology (Jonak and Hirt, 2002; Wang et al., 2003). In contrast, NtDSK1 seems to be a tobacco-specific protein kinase, because Arabidopsis AtDSK1 shares only 41% sequence identity, and this is most probably due to the well-conserved protein kinase domain. Summarizing, experimental evidence was found for 15 chloroplast-localized protein kinases from all organisms, of which 13 orthologues could be assigned in Arabidopsis.
Ca2+-dependent protein kinase activity in chloroplasts
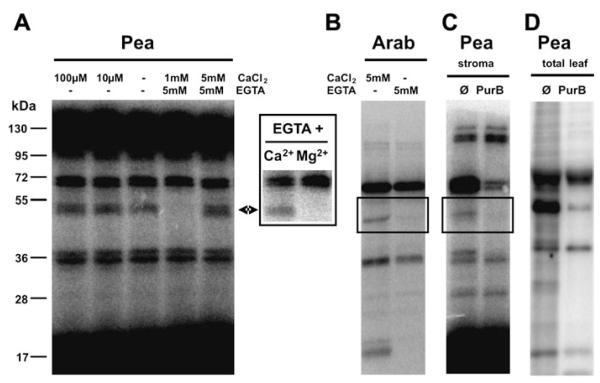
The presence of CIPK13 in chloroplasts would provide a first mechanism to decode Ca2+ signals, which are known to occur inside chloroplasts (Sai and Johnson, 2002). CIPKs require CBLs as regulatory proteins that interact with CIPKs upon calcium binding via internal EF-hand motifs (Batistic et al., 2008; Batistic and Kudla, 2009). However, none of the 10 CBLs known in Arabidopsis has been reported or predicted to be chloroplast localized. Nevertheless, it was possible to demonstrate calcium-dependent protein kinase activity in pea chloroplasts by incubation of stromal extracts with radioactively labelled ATP in the presence or absence of free calcium (Fig. 2A). Increasing calcium levels did not change the phosphorylation pattern much as compared with the control without addition of Ca2+ or EGTA (middle lane in Fig. 2A). Only the intensity of an ~50 kDa band increased slightly. This weak effect is most probably due to the presence of residual amounts of Ca2+ in the stromal extracts. In contrast, the full depletion of calcium by EGTA resulted in the complete loss of phosphorylation of this 50 kDa band. Moreover, phosphorylation could only be restored by addition of calcium but not by addition of magnesium (see inset box next to Fig. 2A), thus indicating a true calcium-dependent phosphorylation event. Importantly, this effect is not limited to pea because a similar Ca2+-dependently phosphorylated band of ~50 kDa was also visible in Arabidopsis chloroplast extracts (Fig. 2B).
Fig. 2.
Protein kinase activity in chloroplasts: Ca2+ dependency and the effect of the protein kinase inhibitor purvalanol B (PurB). (A) Ca2+-dependent phosphorylation of RuBisCO-depleted stromal proteins from pea chloroplasts. The extracted proteins were incubated with radioactively labelled ATP in the absence or presence of CaCl2 and/or EGTA. The insert shows a protein kinase assay in the presence of 5 mM EGTA and 5 mM CaCl2 or MgCl2, respectively. (B) Ca2+-dependent phosphorylation of stromal proteins from Arabidopsis chloroplasts. (C and D) The effect of the protein kinase inhibitor PurB on pea protein kinases. A RuBisCO-depleted stromal extract and a total leaf extract were incubated with radioactively labelled ATP in the absence or presence of 100 μM PurB. Ø=control reaction without inhibitor.
In previous attempts to identify chloroplast-localized protein kinases an affinity purification was used on the immobilized canonical protein kinase inhibitor PurB (Bayer et al., 2011). Interestingly, PurB was also able to inhibit the phosphorylation of the 50 kDa band (Fig. 2C), indicating that this activity is caused by a canonical protein kinase and might be attributed to CIPK13. Therefore, future experiments should include the careful localization analysis of all CBLs and the analysis of the calcium-dependent phospho-rylation pattern in chloroplasts of CIPK13 knock-out lines.
Unusual protein kinases in chloroplasts
When different protein kinase inhibitors were tested for their ability to inhibit protein kinase activities in stromal extracts it was noticed that addition of PurB to RuBisCO-depleted stromal protein extracts from pea had a very weak effect besides the inhibition of the 50 kDa band (Fig. 2C). In contrast, PurB, which has been shown to interact with >80 protein kinases from total human cell extracts (Wissing et al., 2007), proved to be the most active protein kinase inhibitor of plant protein kinases in a whole leaf extract (Fig. 2D). This observation suggested that unusual protein kinases might be present in chloroplasts.
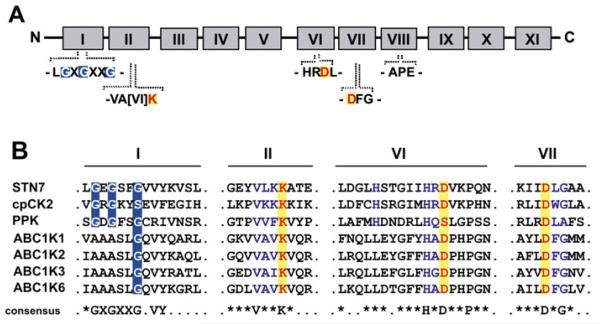
The conserved protein kinase catalytic domains range from 250 to 300 amino acids and have been divided into 11 major subdomains (I–XI) as depicted in Fig. 3A (Hanks et al., 1988). Many of those subdomains directly participate in ATP binding and phosphotransfer like the so-called ‘phosphate gripper’ consensus GXGXXG, which is also found in many other nucleotide-binding proteins (Saraste et al., 1990;Knowles, 1991). A multiple sequence alignment of the subdomains I and II and VI and VII, which harbour critical amino acids for ATP binding and phosphotransfer, shows that chloroplast protein kinases can be divided into two categories: conventional and unusual protein kinases (Fig. 3B). Conventional protein kinases, such as STN7/8 or cpCK2, contain a conserved protein kinase domain consisting of a complete ATP-binding region as shown for subdomains I and II, and an active site signature according to PROSITE as shown for subdomains VI and VII. In contrast, the unusual protein kinase PPK lacks a characteristic aspartate residue in subdomain VI, which usually constitutes the kinase active centre (Fig. 3B). Moreover, AtRP1, NDPK2, and CSK lack the complete protein kinase domain, but all were shown to retain protein kinase activity (Shen et al., 2006; Chastain et al., 2008;Puthiyaveetil et al., 2008). Furthermore, the alignment of subdomain I of all four ABC1 kinases with the conventional serine/threonine-specific protein kinases STN7 and cpCK2 revealed that despite an overall high conservation, two crucial glycine residues within the ATP-binding site are missing (Fig. 3B). This differing structure of the ATP-binding pocket of ABC1 kinases in comparison with conventional protein kinases could indicate a different mode of nucleotide binding.
Fig. 3.
Conserved protein kinase domains and their presence in chloroplast-localized protein kinases. (A) Illustration of the 11 highly conserved protein kinase domains according to Hanks et al. (1988), highlighting the ATP-binding motifs in subdomain I and II and essential catalytic residues in subdomains II, VI, and VII. (B) Alignment of the ‘classical’ protein kinases STN7 and cpCK2 with the unusual kinases PPK and the ABC1 kinases. The ATP-binding sites are highlighted in subdomain I and the essential catalytic residues are indicated in subdomains II, VI, and VII, respectively. A consensus sequence for homologous regions is displayed below.
The mystery of chloroplast-localized protein kinases
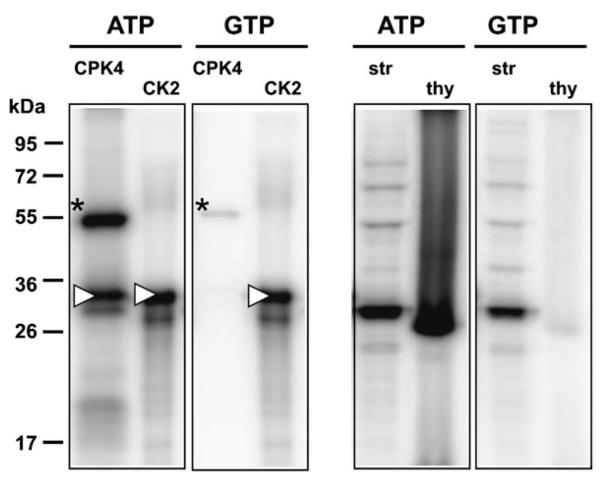
Based on phosphoproteomic studies and experiments with phospho-specific antibodies and radioactively labelled ATP, there is unequivocal evidence for extensive phosphorylation of chloroplast proteins in the stroma as well as at the thylakoids (Laing and Christeller, 1984; Foyer, 1985; Rintamaki et al., 1997; Reiland et al., 2009; Tikkanen et al., 2010). However, while at least 80 chloroplast-localized protein kinases could be expected in Arabidopsis based on targeting predictions, to date only 13 (1.2%) out of all ~1050 Arabidopsis protein kinases can be assigned to the chloroplast with high confidence. This clearly shows that targeting prediction fails in the case of protein kinases and that they seem to be under-represented in chloroplasts. It rather appears that only a relatively small number of protein kinases is responsible for the vast majority of chloroplast phosphorylation events. This would be in accordance with recent results of a chloroplast phosphoproteomic study, which showed that cpCK2 phosphorylation motifs are strongly over-represented among 174 identified chloroplast phosphoproteins (Reiland et al., 2009). This suggests that cpCK2 functions as a central regulator of chloroplast processes. To test this hypothesis experimentally, use was made of the unique feature of CK2 to use GTP as co-substrate as efficiently as ATP (Niefind et al., 1999). The phosphorylation patterns of stromal extracts and thylakoids were compared after the removal of nucleotides by gel filtration and subsequent incubation with either ATP or GTP (Fig. 4). The calcium-dependent protein kinase CPK4 served as control for an ATP-dependent kinase, and commercial CK2 served as control for cpCK2 activity. It was found that indeed, the activity of CPK4 towards histone as substrate was completely dependent on ATP, whereas CK2 did phosphorylate casein equally efficiently in the presence of both ATP and GTP (Fig. 4, left panels). Looking at the chloroplast samples it was found that the phosphorylation patterns in stromal extracts were very similar, thus indicating that cpCK2 presents the dominating protein kinase activity in the stroma (Fig. 4, right panels). In the thylakoid samples, the LHCs (~29 kDa) presented the main phosphorylation targets, and their phosphorylation was strictly dependent on ATP as expected for the phosphorylation by STN7/8. In line with these results, the analysis of phosphorylation patterns in wild-type and stn7stn8 double knock-out mutants revealed that all major phosphorylation events in isolated thylakoid membranes could be attributed to STN7 and STN8 (Tikkanen et al., 2010).
Fig. 4.
The effect of using ATP versus GTP as co-substrate for phosphorylation assays. Protein kinase assays were performed as described in the Materials and methods using purified protein kinases and their substrates (left panel) or chloroplast extracts (right panel) with ATP or GTP as co-substrate. The calcium-dependent protein kinase CPK4 and histone as substrate served as control for a strictly ATP-dependent kinase, and commercial CK2 and casein as substrate were used as a positive control. The autophosphorylation of CPK4 is indicated by an asterisk and the substrate phosphorylation of histone and casein is indicated by a white triangle. Gel-filtrated stromal extracts (str) incubated with ATP and GTP are shown in lanes 5 and 7, respectively and the phosphorylation of thylakoid membranes (thy) by stromal extracts in the presence of either ATP or GTP is shown in lanes 6 and 8, respectively.
In contrast, out of all 217 Arabidopsis protein phosphatases, already 10 (4.6%) are known to be chloroplast localized (Schliebner et al., 2008; Pribil et al., 2010). Therefore, they seem to be over-represented in chloroplasts compared with kinases, implicating an important regulatory role for chloroplast phosphatases.
Furthermore, considering the ABC1 family protein kinases, AtRP1, NDPK2, CSK, and PPK and the unexpected results from protein kinase inhibitor experiments, evidence for unusual chloroplast protein kinases accumulates. Seven out of the 13 experimentally verified chloroplast protein kinases in Arabidopsis contain unusual kinase domains, thus suggesting that unusual protein kinases are responsible for many of the remaining protein phosphorylation events occurring inside the chloroplast.
Conclusions
Chloroplast phosphorylation has already been known for >30 years, and the importance of chloroplast protein kinases has already been demonstrated, for example in the case of STN7, STN8, or cpCK2. Considering the experimental reports of extensive phosphorylation and based on targeting prediction, numerous protein kinases are generally assumed to be present inside chloroplasts. However, in contrast to this expectation, the complement of chloroplast protein kinases seems to be much smaller than initially anticipated. It seems that STN7, STN8, and cpCK2 represent the major protein kinase activities in chloroplasts. In addition to those three, a number of other protein kinases, including unusual protein kinases such as the ABC1 protein kinases or PPK, most probably have more specialized functions, which need to be elucidated in the future. For this reason, the identification of novel chloroplast protein kinases cannot rely solely on directed approaches involving mass spectrometry, as this depends on the correct functional annotation of proteins. It is therefore suggested rather to use known chloroplast protein kinase substrates in order to identify their interacting protein kinases as has been done for the identification of a protein kinase that phosphorylates chloroplast precursor proteins (Martin et al., 2006).
Supplementary Material
Acknowledgements
This work has been funded by the Austrian GEN-AU program in the ERA-PG project CROPP (Project no. 818514), the Austrian Science Foundation FWF (P19825-B12), and by the EU in the Marie-Curie ITN COSI (ITN 2008 GA 215-174). The authors thank Helga Waltenberger for technical assistance, Gustav Ammerer and all members of the mass spectrometry facilities of the MFPL for their good collaboration, Felix Kessler (University of Neuchatel) for discussion about ABC kinases, and Giulio Superti-Furga and Uwe Rix (CeMM, Vienna) for providing protein kinase inhibitors. The purified recombinant CPK4 was a kind gift of Dr Norbert Mehlmer (LMU Munich).
Abbreviations
- AGI
Arabidopsis gene identifier
- CPK
calcium-dependent protein kinase
- PurB
purvalanol B
- RuBisCO
ribulose-1,5-bisphosphate carboxylase/oxygenase
- YFP
yellow fluorescent protein
Footnotes
Supplementary data are available at JXB online.
Figure S1. Localization analysis of predicted chloroplast-localized protein kinases.
Table S1. Overview of predicted chloroplast-localized protein kinases.
References
- Ashton AR, Hatch MD. Regulation of C4 photosynthesis: regulation of pyruvate, Pi dikinase by ADP-dependent phosphorylation and dephosphorylation. Biochemical and Biophysical Research Communications. 1983;115:53–60. doi: 10.1016/0006-291x(83)90967-1. [DOI] [PubMed] [Google Scholar]
- Baena-Gonzalez E, Rolland F, Thevelein JM, Sheen J. A central integrator of transcription networks in plant stress and energy signalling. Nature. 2007;448:938–942. doi: 10.1038/nature06069. [DOI] [PubMed] [Google Scholar]
- Baginsky S. Plant proteomics: concepts, applications, and novel strategies for data interpretation. Mass Spectrometry Reviews. 2009;28:93–120. doi: 10.1002/mas.20183. [DOI] [PubMed] [Google Scholar]
- Batistic O, Kudla J. Plant calcineurin B-like proteins and their interacting protein kinases. Biochimica et Biophysica Acta. 2009;1793:985–992. doi: 10.1016/j.bbamcr.2008.10.006. [DOI] [PubMed] [Google Scholar]
- Batistic O, Sorek N, Schultke S, Yalovsky S, Kudla J. Dual fatty acyl modification determines the localization and plasma membrane targeting of CBL/CIPK Ca2+ signaling complexes in Arabidopsis. The Plant Cell. 2008;20:1346–1362. doi: 10.1105/tpc.108.058123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer RG, Stael S, Csaszar E, Teige M. Mining the soluble chloroplast proteome by affinity chromatography. Proteomics. 2011;11:1287–1299. doi: 10.1002/pmic.201000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellafiore S, Barneche F, Peltier G, Rochaix JD. State transitions and light adaptation require chloroplast thylakoid protein kinase STN7. Nature. 2005;433:892–895. doi: 10.1038/nature03286. [DOI] [PubMed] [Google Scholar]
- Benetka W, Mehlmer N, Maurer-Stroh S, Sammer M, Koranda M, Neumuller R, Betschinger J, Knoblich JA, Teige M, Eisenhaber F. Experimental testing of predicted myristoylation targets involved in asymmetric cell division and calcium-dependent signalling. Cell Cycle. 2008;7:3709–3719. doi: 10.4161/cc.7.23.7176. [DOI] [PubMed] [Google Scholar]
- Bennett J. Phosphorylation of chloroplast membrane polypeptides. Nature. 1977;269:344–346. [Google Scholar]
- Bolter B, Sharma R, Soll J. Localisation of Arabidopsis NDPK2—revisited. Planta. 2007;226:1059–1065. doi: 10.1007/s00425-007-0549-4. [DOI] [PubMed] [Google Scholar]
- Bonardi V. Molecular-genetic characterization of thylakoid protein phosphorylation in Arabidopsis thaliana. Ludwig-Maximilians-Universität München; Munich: 2006. PhD thesis. [Google Scholar]
- Bonardi V, Pesaresi P, Becker T, Schleiff E, Wagner R, Pfannschmidt T, Jahns P, Leister D. Photosystem II core phosphorylation and photosynthetic acclimation require two different protein kinases. Nature. 2005;437:1179–1182. doi: 10.1038/nature04016. [DOI] [PubMed] [Google Scholar]
- Bousquet I, Dujardin G, Slonimski PP. ABC1, a novel yeast nuclear gene has a dual function in mitochondria: it suppresses a cytochrome b mRNA translation defect and is essential for the electron transfer in the bc 1 complex. EMBO Journal. 1991;10:2023–2031. doi: 10.1002/j.1460-2075.1991.tb07732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brautigam K, Dietzel L, Kleine T, et al. Dynamic plastid redox signals integrate gene expression and metabolism to induce distinct metabolic states in photosynthetic acclimation in Arabidopsis. The Plant Cell. 2009;21:2715–2732. doi: 10.1105/tpc.108.062018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnell JN, Chastain CJ. Cloning and expression of maize-leaf pyruvate, Pi dikinase regulatory protein gene. Biochemical and Biophysical Research Communications. 2006;345:675–680. doi: 10.1016/j.bbrc.2006.04.150. [DOI] [PubMed] [Google Scholar]
- Chastain CJ, Xu W, Parsley K, Sarath G, Hibberd JM, Chollet R. The pyruvate, orthophosphate dikinase regulatory proteins of Arabidopsis possess a novel, unprecedented Ser/Thr protein kinase primary structure. The Plant Journal. 2008;53:854–863. doi: 10.1111/j.1365-313X.2007.03366.x. [DOI] [PubMed] [Google Scholar]
- Chevalier D, Walker JC. Functional genomics of protein kinases in plants. Briefings in Functional Genomics and Proteomics. 2005;3:362–371. doi: 10.1093/bfgp/3.4.362. [DOI] [PubMed] [Google Scholar]
- Cho HS, Yoon GM, Lee SS, Kim YA, Hwang I, Choi D, Pai HS. A novel dual-specificity protein kinase targeted to the chloroplast in tobacco. FEBS Letters. 2001;497:124–130. doi: 10.1016/s0014-5793(01)02439-5. [DOI] [PubMed] [Google Scholar]
- Choi G, Yi H, Lee J, Kwon YK, Soh MS, Shin B, Luka Z, Hahn TR, Song PS. Phytochrome signalling is mediated through nucleoside diphosphate kinase 2. Nature. 1999;401:610–613. doi: 10.1038/44176. [DOI] [PubMed] [Google Scholar]
- Cohen P. The regulation of protein function by multisite phosphorylation—a 25 year update. Trends in Biochemical Sciences. 2000;25:596–601. doi: 10.1016/s0968-0004(00)01712-6. [DOI] [PubMed] [Google Scholar]
- Depege N, Bellafiore S, Rochaix JD. Role of chloroplast protein kinase Stt7 in LHCII phosphorylation and state transition in Chlamydomonas. Science. 2003;299:1572–1575. doi: 10.1126/science.1081397. [DOI] [PubMed] [Google Scholar]
- Dunkley TP, Hester S, Shadforth IP, et al. Mapping the Arabidopsis organelle proteome. Proceedings of the National Academy of Sciences, USA. 2006;103:6518–6523. doi: 10.1073/pnas.0506958103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, von Heijne G. Prediction of organellar targeting signals. Biochimica et Biophysica Acta. 2001;1541:114–119. doi: 10.1016/s0167-4889(01)00145-8. [DOI] [PubMed] [Google Scholar]
- Ferro M, Brugiere S, Salvi D, et al. AT_CHLORO: s comprehensive chloroplast proteome database with subplastidial localization and curated information on envelope proteins. Molecular and Cellular Proteomics. 2010;9:1063–1084. doi: 10.1074/mcp.M900325-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH. Stromal protein phosphorylation in spinach (Spinacia oleracea) chloroplasts. Biochemical Journal. 1985;231:97–103. doi: 10.1042/bj2310097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragoso S, Espindola L, Paez-Valencia J, Gamboa A, Camacho Y, Martinez-Barajas E, Coello P. SnRK1 isoforms AKIN10 and AKIN11 are differentially regulated in Arabidopsis plants under phosphate starvation. Plant Physiology. 2009;149:1906–1916. doi: 10.1104/pp.108.133298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fristedt R, Willig A, Granath P, Crevecoeur M, Rochaix JD, Vener AV. Phosphorylation of photosystem II controls functional macroscopic folding of photosynthetic membranes in Arabidopsis. The Plant Cell. 2009;21:3950–3964. doi: 10.1105/tpc.109.069435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goral TK, Johnson MP, Brain AP, Kirchhoff H, Ruban AV, Mullineaux CW. Visualizing the mobility and distribution of chlorophyll proteins in higher plant thylakoid membranes: effects of photoinhibition and protein phosphorylation. The Plant Journal. 2010;62:948–959. doi: 10.1111/j.0960-7412.2010.04207.x. [DOI] [PubMed] [Google Scholar]
- Gribskov M, Fana F, Harper J, Hope DA, Harmon AC, Smith DW, Tax FE, Zhang G. PlantsP: a functional genomics database for plant phosphorylation. Nucleic Acids Research. 2001;29:111–113. doi: 10.1093/nar/29.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford NG, Hey S, Jhurreea D, Laurie S, McKibbin RS, Zhang Y, Paul MJ. Highly conserved protein kinases involved in the regulation of carbon and amino acid metabolism. Journal of Experimental Botany. 2004;55:35–42. doi: 10.1093/jxb/erh019. [DOI] [PubMed] [Google Scholar]
- Hanks SK, Quinn AM, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- Jonak C, Hirt H. Glycogen synthase kinase 3/SHAGGY-like kinases in plants: an emerging family with novel functions. Trends in Plant Sciences. 2002;7:457–461. doi: 10.1016/s1360-1385(02)02331-2. [DOI] [PubMed] [Google Scholar]
- Jossier M, Bouly JP, Meimoun P, Arjmand A, Lessard P, Hawley S, Grahame Hardie D, Thomas M. SnRK1 (SNF1-related kinase 1) has a central role in sugar and ABA signalling in Arabidopsis thaliana. The Plant Journal. 2009;59:316–328. doi: 10.1111/j.1365-313X.2009.03871.x. [DOI] [PubMed] [Google Scholar]
- Kempa S, Rozhon W, Samaj J, et al. A plastid-localized glycogen synthase kinase 3 modulates stress tolerance and carbohydrate metabolism. The Plant Journal. 2007;49:1076–1090. doi: 10.1111/j.1365-313X.2006.03025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleffmann T, Hirsch-Hoffmann M, Gruissem W, Baginsky S. plprot: a comprehensive proteome database for different plastid types. Plant and Cell Physiology. 2006;47:432–436. doi: 10.1093/pcp/pcj005. [DOI] [PubMed] [Google Scholar]
- Knowles JR. Enzyme catalysis: not different, just better. Nature. 1991;350:121–124. doi: 10.1038/350121a0. [DOI] [PubMed] [Google Scholar]
- Koroleva OA, Tomlinson ML, Leader D, Shaw P, Doonan JH. High-throughput protein localization in Arabidopsis using Agrobacterium-mediated transient expression of GFP-ORF fusions. The Plant Journal. 2005;41:162–174. doi: 10.1111/j.1365-313X.2004.02281.x. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laing WA, Christeller JT. Chloroplast phosphoproteins: distribution of phosphoproteins within spinach chloroplasts. Plant Science Letters. 1984;36:99–104. [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- Martin DM, Miranda-Saavedra D, Barton GJ. Kinomer v. 1.0: a database of systematically classified eukaryotic protein kinases. Nucleic Acids Research. 2009;37:D244–D250. doi: 10.1093/nar/gkn834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin T, Sharma R, Sippel C, Waegemann K, Soll J, Vothknecht UC. A protein kinase family in Arabidopsis phosphorylates chloroplast precursor proteins. Journal of Biological Chemistry. 2006;281:40216–40223. doi: 10.1074/jbc.M606580200. [DOI] [PubMed] [Google Scholar]
- Mehlmer N, Wurzinger B, Stael S, Hofmann-Rodrigues D, Csaszar E, Pfister B, Bayer R, Teige M. The Ca2+-dependent protein kinase CPK3 is required for MAPK-independent salt-stress acclimation in Arabidopsis. The Plant Journal. 2010;63:484–498. doi: 10.1111/j.1365-313X.2010.04257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montrichard F, Alkhalfioui F, Yano H, Vensel WH, Hurkman WJ, Buchanan BB. Thioredoxin targets in plants: the first 30 years. Journal of Proteomics. 2009;72:452–474. doi: 10.1016/j.jprot.2008.12.002. [DOI] [PubMed] [Google Scholar]
- Moon H, Lee B, Choi G, et al. NDP kinase 2 interacts with two oxidative stress-activated MAPKs to regulate cellular redox state and enhances multiple stress tolerance in transgenic plants. Proceedings of the National Academy of Sciences, USA. 2003;100:358–363. doi: 10.1073/pnas.252641899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niefind K, Putter M, Guerra B, Issinger OG, Schomburg D. GTP plus water mimic ATP in the active site of protein kinase CK2. Nature Structural Biology. 1999;6:1100–1103. doi: 10.1038/70033. [DOI] [PubMed] [Google Scholar]
- Ogrzewalla K, Piotrowski M, Reinbothe S, Link G. The plastid transcription kinase from mustard (Sinapis alba L.). A nuclear-encoded CK2-type chloroplast enzyme with redox-sensitive function. European Journal of Biochemistry. 2002;269:3329–3337. [PubMed] [Google Scholar]
- Orchard S, Hermjakob H, Apweiler R. Annotating the human proteome. Molecular and Cellular Proteomics. 2005;4:435–440. doi: 10.1074/mcp.R500003-MCP200. [DOI] [PubMed] [Google Scholar]
- Pesaresi P, Hertle A, Pribil M, et al. Arabidopsis STN7 kinase provides a link between short- and long-term photosynthetic acclimation. The Plant Cell. 2009;21:2402–2423. doi: 10.1105/tpc.108.064964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pribil M, Pesaresi P, Hertle A, Barbato R, Leister D. Role of plastid protein phosphatase TAP38 in LHCII dephosphorylation and thylakoid electron flow. PLoS Biology. 2010;8:e1000288. doi: 10.1371/journal.pbio.1000288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthiyaveetil S, Ibrahim IM, Jelicic B, Tomasic A, Fulgosi H, Allen JF. Transcriptional control of photosynthesis genes: the evolutionarily conserved regulatory mechanism in plastid genome function. Genome Biology and Evolution. 2010;2:888–896. doi: 10.1093/gbe/evq073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthiyaveetil S, Kavanagh TA, Cain P, Sullivan JA, Newell CA, Gray JC, Robinson C, van der Giezen M, Rogers MB, Allen JF. The ancestral symbiont sensor kinase CSK links photosynthesis with gene expression in chloroplasts. Proceedings of the National Academy of Sciences, USA. 2008;105:10061–10066. doi: 10.1073/pnas.0803928105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiland S, Messerli G, Baerenfaller K, Gerrits B, Endler A, Grossmann J, Gruissem W, Baginsky S. Large-scale Arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks. Plant Physiology. 2009;150:889–903. doi: 10.1104/pp.109.138677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richly E, Leister D. An improved prediction of chloroplast proteins reveals diversities and commonalities in the chloroplast proteomes of Arabidopsis and rice. Gene. 2004;329:11–16. doi: 10.1016/j.gene.2004.01.008. [DOI] [PubMed] [Google Scholar]
- Rintamaki E, Salonen M, Suoranta UM, Carlberg I, Andersson B, Aro EM. Phosphorylation of light-harvesting complex II and photosystem II core proteins shows different irradiance-dependent regulation in vivo. Application of phosphothreonine antibodies to analysis of thylakoid phosphoproteins. Journal of Biological Chemistry. 1997;272:30476–30482. doi: 10.1074/jbc.272.48.30476. [DOI] [PubMed] [Google Scholar]
- Sai J, Johnson CH. Dark-stimulated calcium ion fluxes in the chloroplast stroma and cytosol. The Plant Cell. 2002;14:1279–1291. doi: 10.1105/tpc.000653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas P, Fuentes D, Vidal E, Jordana X, Echeverria M, Holuigue L. An extensive survey of CK2 alpha and beta subunits in Arabidopsis: multiple isoforms exhibit differential subcellular localization. Plant and Cell Physiology. 2006;47:1295–1308. doi: 10.1093/pcp/pcj100. [DOI] [PubMed] [Google Scholar]
- Samuel MA, Chaal BK, Lampard G, Green BR, Ellis BE. Surviving the passage: non-canonical stromal targeting of an Arabidopsis mitogen-activated protein kinase kinase. Plant Signaling Behavior. 2008;3:6–12. doi: 10.4161/psb.3.1.4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste M, Sibbald PR, Wittinghofer A. The P-loop—a common motif in ATP- and GTP-binding proteins. Trends in Biochemical Sciences. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- Schliebner I, Pribil M, Zuhlke J, Dietzmann A, Leister D. A survey of chloroplast protein kinases and phosphatases in. Arabidopsis thaliana. Current Genomics. 2008;9:184–190. doi: 10.2174/138920208784340740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweer J, Turkeri H, Link B, Link G. AtSIG6, a plastid sigma factor from Arabidopsis, reveals functional impact of cpCK2 phosphorylation. The Plant Journal. 2010;62:192–202. doi: 10.1111/j.1365-313X.2010.04138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seigneurin-Berny D, Salvi D, Joyard J, Rolland N. Purification of intact chloroplasts from Arabidopsis and spinach leaves by isopycnic centrifugation. Current Protocols in Cell Biology. 2008 doi: 10.1002/0471143030.cb0330s40. Chapter 3, Unit 3 30. [DOI] [PubMed] [Google Scholar]
- Shen Y, Kim JI, Song PS. Autophosphorylation of Arabidopsis nucleoside diphosphate kinase 2 occurs only on its active histidine residue. Biochemistry. 2006;45:1946–1949. doi: 10.1021/bi051868a. [DOI] [PubMed] [Google Scholar]
- Siegenthaler PA, Bovet L. A unique protein-kinase activity is responsible for the phosphorylation of the 26- and 14-kDa proteins but not of the 67-kDa protein in the chloroplast envelope membranes of spinach. Planta. 1993;190:231–240. [Google Scholar]
- Snyders S, Kohorn BD. TAKs, thylakoid membrane protein kinases associated with energy transduction. Journal of Biological Chemistry. 1999;274:9137–9140. doi: 10.1074/jbc.274.14.9137. [DOI] [PubMed] [Google Scholar]
- Soll J. Phosphoproteins and protein-kinase activity in isolated envelopes of pea (Pisum sativum L.) chloroplasts. Planta. 1985;166:394–400. doi: 10.1007/BF00401178. [DOI] [PubMed] [Google Scholar]
- Soll J. Purification and characterization of a chloroplast outer-envelope-bound, ATP-dependent protein kinase. Plant Physiology. 1988;87:898–903. doi: 10.1104/pp.87.4.898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soll J, Fischer I, Keegstra K. A guanosine 5′-triphosphate-dependent protein kinase is localized in the outer envelope membrane of pea chloroplasts. Planta. 1988;176:488–496. doi: 10.1007/BF00397655. [DOI] [PubMed] [Google Scholar]
- Stael S, Bayer RG, Mehlmer N, Teige M. Protein N-acylation overrides differing targeting signals. FEBS Letters. 2011;585:517–522. doi: 10.1016/j.febslet.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone JM, Walker JC. Plant protein kinase families and signal transduction. Plant Physiology. 1995;108:451–457. doi: 10.1104/pp.108.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Zybailov B, Majeran W, Friso G, Olinares PD, van Wijk KJ. PPDB, the Plant Proteomics Database at Cornell. Nucleic Acids Research. 2008;37:D969–D974. doi: 10.1093/nar/gkn654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikkanen M, Grieco M, Kangasjarvi S, Aro EM. Thylakoid protein phosphorylation in higher plant chloroplasts optimizes electron transfer under fluctuating light. Plant Physiology. 2010;152:723–735. doi: 10.1104/pp.109.150250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues PE, Batelli G, Grillo S, Agius F, Kim YS, Zhu J, Agarwal M, Katiyar-Agarwal S, Zhu JK. Interaction of SOS2 with nucleoside diphosphate kinase 2 and catalases reveals a point of connection between salt stress and H2O2 signaling in Arabidopsis thaliana. Molecular and Cellular Biology. 2007;27:7771–7780. doi: 10.1128/MCB.00429-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidi PA, Kanwischer M, Baginsky S, Austin JR, Csucs G, Dormann P, Kessler F, Brehelin C. Tocopherol cyclase (VTE1) localization and vitamin E accumulation in chloroplast plastoglobule lipoprotein particles. Journal of Biological Chemistry. 2006;281:11225–11234. doi: 10.1074/jbc.M511939200. [DOI] [PubMed] [Google Scholar]
- Waegemann K, Soll J. Characterization and isolation of the chloroplast protein import machinery. Methods in Cell Biology. 1995;50:255–267. doi: 10.1016/s0091-679x(08)61035-3. [DOI] [PubMed] [Google Scholar]
- Wang D, Harper JF, Gribskov M. Systematic trans-genomic comparison of protein kinases between Arabidopsis and. Saccharomyces cerevisiae. Plant Physiology. 2003;132:2152–2165. doi: 10.1104/pp.103.021485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissing J, Jansch L, Nimtz M, Dieterich G, Hornberger R, Keri G, Wehland J, Daub H. Proteomics analysis of protein kinases by target class-selective prefractionation and tandem mass spectrometry. Molecular and Cellular Proteomics. 2007;6:537–547. doi: 10.1074/mcp.T600062-MCP200. [DOI] [PubMed] [Google Scholar]
- Ytterberg AJ, Peltier JB, van Wijk KJ. Protein profiling of plastoglobules in chloroplasts and chromoplasts. A surprising site for differential accumulation of metabolic enzymes. Plant Physiology. 2006;140:984–997. doi: 10.1104/pp.105.076083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zybailov B, Rutschow H, Friso G, Rudella A, Emanuelsson O, Sun Q, van Wijk KJ. Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS One. 2008;3:e1994. doi: 10.1371/journal.pone.0001994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.