Abstract
We analysed effects of abscisic acid (ABA, a negative regulatory hormone), alone and in combination with positive or neutral hormones, including salicylic acid (SA), on rice growth and expression of cell cycle-related genes. ABA significantly inhibited shoot growth and induced expression of OsKRP4, OsKRP5, and OsKRP6. A yeast two-hybrid assay showed that OsKRP4, OsKRP5, and OsKRP6 interacted with OsCDKA;1 and/or OsCDKA;2. When SA was simultaneously supplied with ABA, the antagonistic effect of SA completely blocked ABA inhibition. SA also blocked ABA inhibition of DNA replication and thymidine incorporation in the shoot apical meristem. These results suggest that ABA arrests cell cycle progression by inducing expression of OsKRP4, OsKRP5, and OsKRP6, which inhibit the G1/S transition, and that SA antagonizes ABA by blocking expression of OsKRP genes.
Progression of the cell cycle is primarily controlled by universally conserved molecular machinery in which a family of Ser/Thr protein kinases, known as cyclin-dependent kinases (CDKs), plays a central role1,2,3. A large number of CDKs have been identified in plants4. Eight classes of CDKs were defined in Arabidopsis based on phylogenetic analyses5,6, and eight groups of CDKs similar to those in Arabidopsis were identified in the rice genome4; of these, the CDKA and CDKB groups have been well studied. CDKA is closely related to yeast Cdc2/Cdc28 and to human CDK1, CDK2, and CDK37. Although levels of CDKA transcripts and proteins are quite stable throughout the cell cycle8, their expression levels are altered by plant hormones. For instance, expression of the Arabidopsis CDKA;1 gene is increased twofold by individual treatment with either auxin or cytokinin, and increases threefold by simultaneous treatment with these hormones9. CDKB is a plant-specific CDK that accumulates in a manner dependent on the cell cycle phase, reaching a maximum level at the G2/M transition8,10,11. Knockdown of the CDKB2;1 gene induces polyploidy in rice, indicating an essential role of CDKB in regulating the G2/M transition12. CDKB is also affected by plant hormones; in barley, the CDKB1 gene is downregulated by the plant stress hormone abscisic acid (ABA)13. The CDKB gene is thought to be expressed primarily in dividing cells, while the CDKA gene is expressed in both dividing and differentiated cells10.
CDK activity is negatively regulated by binding of the INHIBITOR OF CDK/KIP-RELATED PROTEIN (ICK/KRP)14,15,16,17. ICK/KRP induces arrest or delay of the cell cycle in response to intra- or extracellular signals18. Expression of ICK/KRP genes is dependent on the cell cycle phase. In Arabidopsis, KRP3 and KRP5 genes are expressed at high levels in the S phase, KRP4 expression peaks in the early G2 phase, KRP1 expression increases up to the G2/M boundary, and KRP6 expression peaks at the G1/S boundary6. KRP genes are also differentially expressed in plant organs. In rice, KRP1 is expressed in mature leaves and germinating seeds, KRP3 is expressed in the syncytial endosperm, KRP4 and KRP5 are expressed in the shoot apex, and KRP5 and KRP6 are expressed in young panicles17,19. ABA has been shown to affect the expression of ICK/KRP genes. Expression of the alfalfa KRPMt gene and the Arabidopsis KRP1 gene is induced by ABA16,20. Arabidopsis KRP1 was shown to interact with CDKA;1, suggesting that ABA arrests the cell cycle at the G1/S boundary via this interaction16.
The activity and substrate specificity of CDKs are dependent upon their association with cyclins3. In plants, A-, B-, and D-type cyclins are thought to play a major role in cell cycle control21. The A- and B-type cyclins are expressed from the S to the M phase and control DNA replication, the G2/M transition, and mitosis; the D-type cyclin is thought to be a sensor of external signals and to play an essential role in cell cycle progression and in the re-entry of quiescent cells into the cell cycle22. Plant cyclin D (CycD) has been classified into the following six groups based on similarities in amino acid sequences: CycD1, CycD2,/CycD4, CycD3, CycD5, CycD6, and CycD723. Riou-Khamlichi et al.24 reported that cytokinin activation of the Arabidopsis cell cycle is primarily mediated by transcriptional regulation of CycD3 at the G1-S boundary.
CDKs, KRPs, and CyCs are known to be regulated by plant hormones. However, to our knowledge, no studies have investigated the synergistic or antagonistic effects of combinations of plant hormones, other than auxin-cytokinin, on the cell cycle. Since levels of plant hormones are differentially altered according to growth stage and environmental conditions25, hormones are thought to regulate cell cycle progression via additive or antagonistic interactions. Thus, elucidation of the combined effects of plant hormones is important for understanding the mechanisms of cell cycle regulation in response to the environment and plant developmental stage. Here, we analysed the combined effects of the negative regulatory hormone ABA and of positive or neutral hormones, including indole-3-acetic acid (IAA), 6-benzylaminopurine (BAP), gibberellic acid (GA3), and salicylic acid (SA), on plant growth and expression of CDK, KRP, and CycD genes in rice. We found that SA antagonized the inhibitory effects of ABA on shoot growth and cell cycle progression in rice seedlings.
Results
Effect of exogenous hormones on shoot growth in young rice seedlings
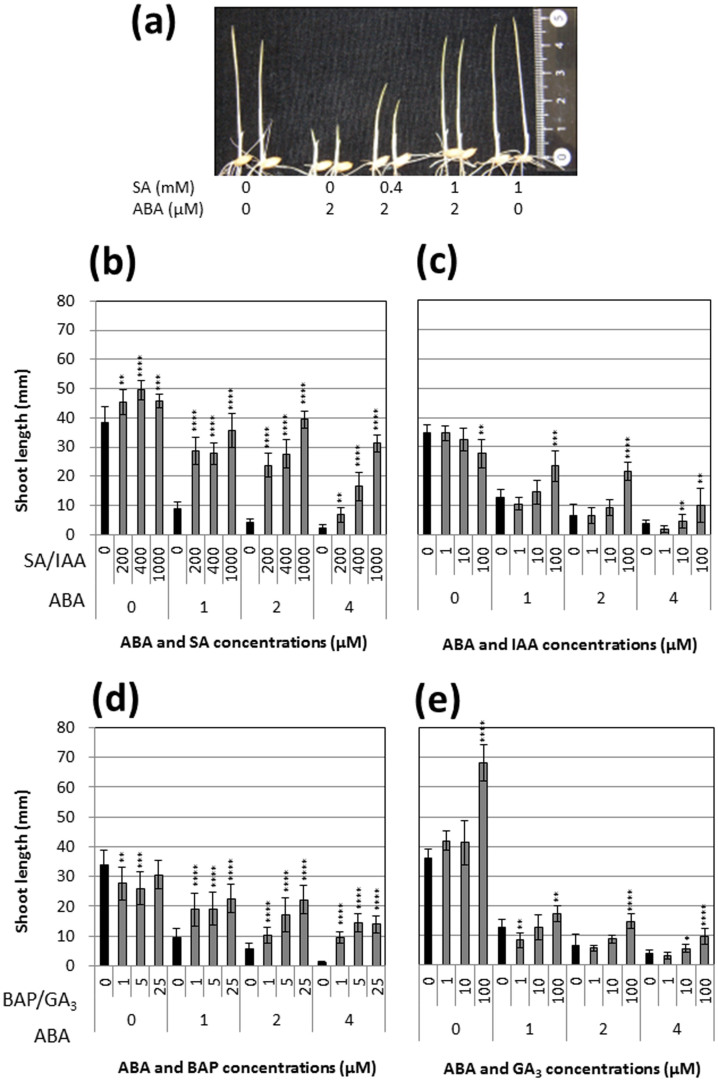
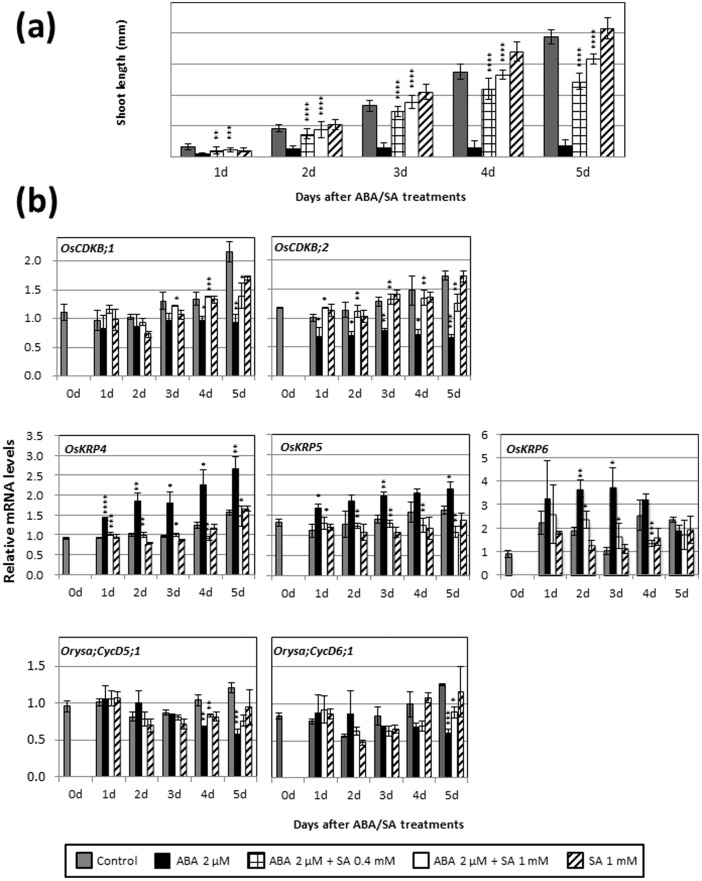
Shoot growth was markedly inhibited by exogenous ABA (Fig. 1a–e), while exogenous SA resulted in moderate promotion of shoot growth (Fig. 1a,b). Treatment with IAA and BAP inhibited shoot growth (Fig. 1c,d); in contrast, exogenous GA3 significantly promoted shoot growth (Fig. 1e). When treated with SA in addition to 1 μM ABA, inhibition of shoot growth by ABA was greatly suppressed (Fig. 1b). Inhibition caused by treatment with 2 or 4 μM ABA was reduced in a dose-dependent manner in seedlings treated with various concentrations of SA (Fig. 1a,b). Treatment with IAA and BAP also reduced ABA inhibition, but to a lesser extent than treatment with SA (Fig. 1c,d). GA3 showed an antagonistic effect against ABA but its effect was very small compared with the effects of SA, IAA, and BAP (Fig. 1e). Thus, the antagonistic effect of the hormones and concentrations tested against ABA was greatest for SA.
Figure 1. Effects of plant hormone treatment on shoot growth in young rice seedlings.
Abscisic acid (ABA) treatment was combined with salicylic acid (SA) (a,b), indole acetic acid (IAA) (c), 6-benzylaminopurine (BAP) (d), or gibberellic acid (GA3) (e). Error bars show SD (n = 9). Asterisks indicate a significant difference from the ABA-only treatment (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; Student's t-test).
Effects of exogenous hormones on expression of cell cycle-related genes
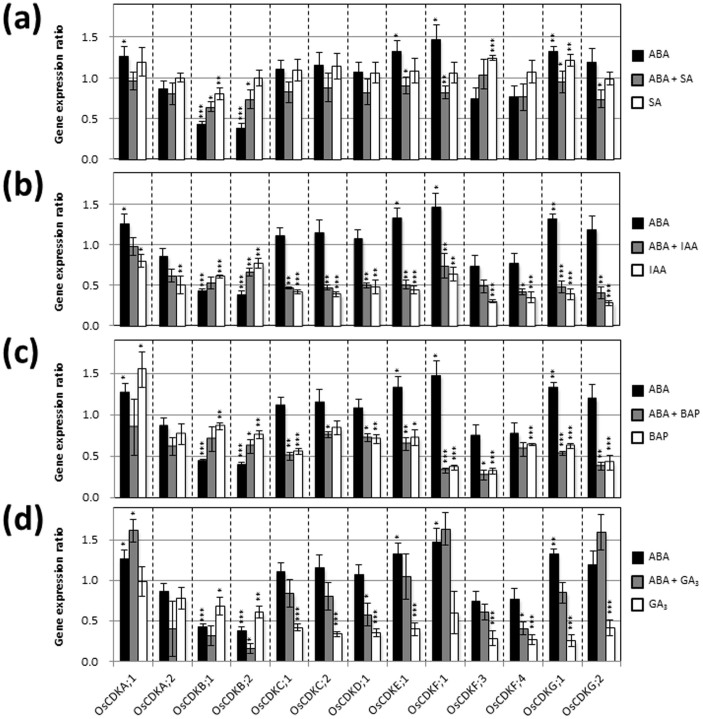
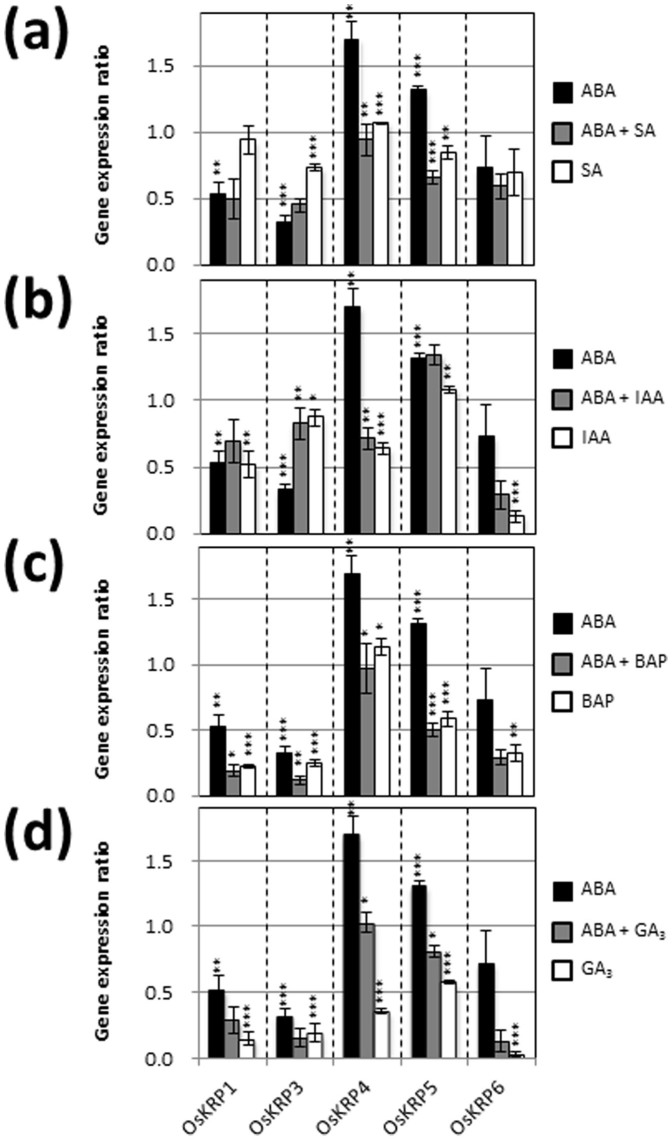
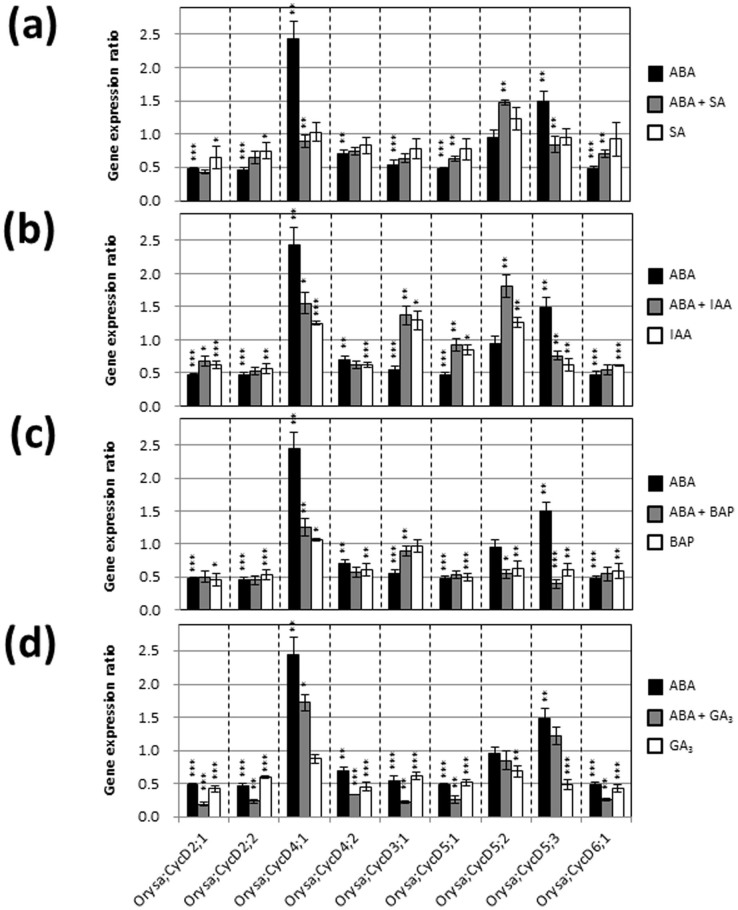
Expression of genes that code for CDKs, KRPs, and CycDs, which are central to cell cycle regulation, was investigated in young rice seedlings treated with various plant hormones. Real-time PCR analysis indicated that exogenous ABA significantly suppressed the expression of OsCDKB;1, OsCDKB;2, Orysa;CycD2;1, Orysa;CycD2;2, Orysa;CycD3;1, Orysa;CycD4;2, Orysa;CycD5;1, and Orysa;CycD6;1, and induced the expression of OsCDKA;1, OsCDKF;1, OsCDKG;1, OsKRP4, OsKRP5, Orysa;CycD4;1, and Orysa;Cyc D5;3 (Figs. 2,3,4 and Supplementary Table S1). Exogenous SA also affected the expression of some genes encoding CDKs, KRPs, and CycDs; however, these effects were relatively small compared to those of IAA, BAP, and GA3 (Figs. 2,3,4). When SA, IAA, BAP, and GA3 were individually combined with ABA treatment, substantial negative effects of ABA on expression of genes encoding CDKs and CycDs, and substantial positive effects of ABA on expression of genes encoding KRPs were reversed (SA, 6 genes; IAA, 5 genes; BAP, 4 genes; and GA3, 2 genes) (Figs. 2,3,4).
Figure 2. Gene expression ratios of OsCDKs after treatment for 5 d with various plant hormones.
Asterisks above ABA bars indicate a significant difference from the control (hormone-untreated) mean. Asterisks above ABA + SA bars indicate a significant difference from the ABA-only treatment. Error bars show SD (n = 3). Asterisks above SA bars indicate a significant difference from the control mean. *P < 0.05, ** P < 0.01, *** P < 0.001; Student's t-test.
Figure 3. Gene expression ratios of OsKRPs after treatment for 5 d with various plant hormones.
Error bars show SD (n = 3). Asterisks indicate significant differences as described in Fig. 2.
Figure 4. Gene expression ratios of Orysa;CycDs after treatment for 5 d with various plant hormones.
Error bars show SD (n = 3). Asterisks indicate significant differences as described in Fig. 2.
While the antagonistic effect of SA against ABA was greatest among the hormones and concentrations tested in this study, the effects of SA alone on gene expression were smallest among the hormones investigated. Therefore, we further examined the antagonistic effects of SA against ABA suppression of shoot growth and expression of genes encoding CDKs (OsCDKsB;1 and OsCDKB;2) and CycDs (CycD5;1 and CycD6;1) (Figs. 2 and 4), and ABA induction of genes encoding KRPs (OsKRP4 and OsKRP5) (Fig. 3). Because the microarray data showed that OsKRP6 was induced by 6 h of ABA treatment (Supplementary Fig. S1), this gene was also included in the analysis.
Shoot growth was inhibited by 1 d of treatment with ABA, but this inhibition was blocked by 1 d treatment with SA (Fig. 5a). ABA substantially and consistently reduced the OsCDKB;2 transcript level from 1 to 5 d after treatment (Fig. 5b). In contrast, expression of OsCDKB;1 was not substantially reduced by ABA during the first 2 d, but decreased later (Fig. 5b). When SA was combined with ABA application, OsCDKB;2 transcript levels remained similar to those of the controls from 1 to 4 d after hormone treatment (Fig. 5b). These results suggest that changes in expression of OsCDKB;2 involved ABA inhibition of shoot growth and its release by SA. Expression of OsKRP4 and OsKRP5 in ABA-treated seedlings gradually increased from 1 to 5 d; OsKRP6 expression increased from 2 to 3 d and decreased to the control level at 5 d (Fig. 5b). In contrast, when SA was combined with ABA, transcript levels of OsKRP4, OsKRP5, and OsKRP6 remained similar to those of the control throughout the 5-d hormone treatment (Fig. 5b). These results suggest that OsKRP4, OsKRP5, and OsKRP6 participate in ABA inhibition and that SA blocks this inhibition by suppressing induction of these genes by ABA. To determine whether ABA has a direct effect on these genes, microarray analysis was conducted using seedlings treated with ABA for 6 h. Expression of OsKRP4 and OsKRP6 increased significantly (P = 0.0042 and 0.0048; Student's t-test, respectively) while expression of OsKRP3 and OsKRP5 did not change significantly and expression of OsKRP1 was significantly reduced (P = 0.016; Student's t-test, (Supplementary Fig. S1). These findings suggest that OsKRP4 and OsKRP6 were directly upregulated by ABA. Further studies are necessary to determine whether ABA acts directly on these KRP genes. Expression of Orysa;CycD5;1 and Orysa;CycD6;1 did not change significantly after 1–3 d of ABA treatment, but declined significantly after 4–5 d of ABA treatment (Fig. 5b). When SA was combined with ABA, expression of Orysa;CycD5;1 and Orysa;Cyc6;1 was reversed at 4 or 5 d after hormone treatment (Fig. 5b), suggesting that these genes were not involved in ABA inhibition of shoot growth and its release by SA (effects that were observed during the first 3 d of hormone treatment, Fig. 5a).
Figure 5. Changes in shoot growth (a) and expression of OsCDKB, OsKRP, and Orysa;CycD genes (b) in shoots of young rice seedlings after treatment with ABA and SA.
(a) Error bars show SD (n = 9). Asterisks indicate a significant difference from the ABA-only treatment. (b) Error bars show SD (n = 3). Asterisks indicate a significant difference between the ABA-only and ABA + SA treatments. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; Student's t-test.
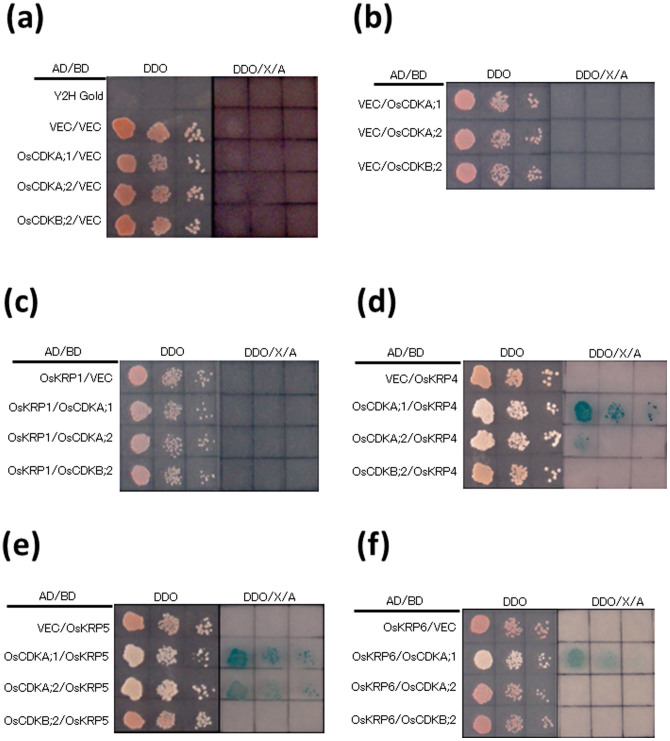
Interactions between OsKRPs and OsCDKs
Interactions between OsKRP1, OsKRP4, OsKRP5, OsKRP6, and OsCDKs were investigated using a yeast two-hybrid (Y2H) system. OsiICK6 has been reported to interact with OsCDKA in indica rice26, and OsiICK6 corresponds to OsKRP4 in japonica. We observed that both OsKRP4 and OsKRP5 interacted with OsCDKA;1 and OsCDKA;2, and that OsKRP6 interacted with OsCDKA;1; these KRPs did not interact with OsCDKB;2 (Fig. 6d–f). No interaction was found between OsKRP1 and OsCDKs (Fig. 6c). These results suggest that OsKRP4, OsKRP5, and OsKRP6 inhibit the functioning of OsCDKA;1 and/or OsCDKA;2. Since we observed induction of OsKRP4, OsKRP5, and OsKRP6 by ABA (Fig. 5b), ABA was thought to negatively regulate the cell cycle via inhibition of OsCDKA;1 and/or OsCDKA;2.
Figure 6. Interactions of OsKRPs with different OsCDKs.
(a) AD-CDKs and BD-VEC, (b) AD-VEC and BD-CDKs, (c) AD-OsKRP1 and BD-CDKs, (d) AD-OsKRP4 and BD-CDKs, (e) AD-OsKRP5 and BD-CDKs, (f) AD-OsKRP6 and BD-CDKs. Columns in each panel represent serial dilutions.
Effects of ABA and SA on DNA replication and thymidine incorporation in the shoot apex
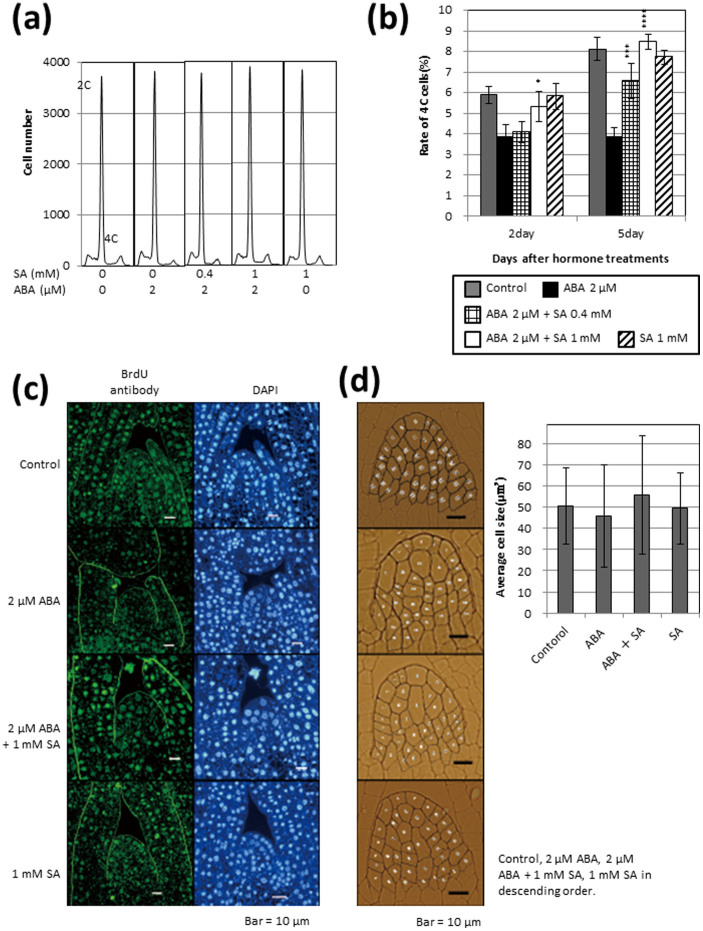
To investigate individual effects of ABA and antagonistic effects of SA against ABA on cell cycle progression in the shoot apex, ploidy levels were analysed by flow cytometry (Fig. 7a,b and Supplementary Table S2). Reduced numbers of cells with 4C DNA content were observed in ABA-treated seedlings. When SA was combined with ABA, the reduction in 4C cells was prevented in a concentration-dependent manner. These results indicate that ABA prevents cell cycle progression before DNA synthesis (S phase) and that SA blocks this inhibition.
Figure 7. Effects of ABA and SA on DNA replication (a,b) and thymidine incorporation (c) in the shoot apex of rice seedlings.
(d) Average cell size in hormone-untreated and hormone-treated tissues. Nuclear DNA copy numbers were analysed by flow cytometry. (a) Representative flow cytometric data for 2C and 4C cells in basal shoot tissues including the shoot apex. (b) Ratio of 4C cells in hormone-untreated and hormone-treated tissues. Five biological replicates were performed. Error bars show SD (n = 5). (d) Error bars show SD (n = 40). Asterisks indicate significant differences between the ABA-only and ABA + SA treatments (*P < 0.05, ***P < 0.001, ****P < 0.001; Student's t-test).
DNA synthesis in the shoot apical meristem was investigated using a thymidine incorporation assay by indirect immunofluorescence labelling with anti-BrdU antibody and DAPI staining (Fig. 7c). The intensity of blue-white fluorescence under UV excitation in DAPI-stained sections was similar in ABA-treated and control seedlings. In contrast, the intensity of green-yellow fluorescence of Alexa FluorR 488 under blue-light excitation in BrdU-labelled sections was markedly reduced in ABA-treated seedlings. This reduction was completely blocked when SA was combined with ABA. These results suggest that ABA inhibits cell nuclear DNA synthesis and that SA blocks this inhibition. No significant size differences were found between control and hormone-treated cells (Fig. 7d), suggesting that ABA-induced inhibition of shoot growth and the reversal of this inhibition by SA occur through cell division rather than cell elongation.
Discussion
We found that expression of OsKRP4, OsKRP5, and OsKRP6 was induced by exogenous ABA (Fig. 5b). By investigating the interactions between KRPs and CDKs using a Y2H system, we found that OsKRP4, OsKRP5, and OsKRP6 interact with OsCDKA;1 and/or OsCDKA;2 (Fig. 6), suggesting that induction of the OsKRP genes leads to inhibition of OsCDKA;1 and/or OsCDKA;2. Inhibition of the cell cycle by ABA was investigated in detail using synchronized tobacco BY-2 cells, and ABA was found to arrest the cell cycle at the G1/S transition but not to affect subsequent phases27. In Arabidopsis, ABA induces expression of the KRP1 gene, which inhibits CDKA;1, a gene that promotes the G1/S transition, causing cells to remain in the G1 phase16. Therefore, it is conceivable that ABA prevents cell cycle progression at the G1/S transition by inducing genes for KRP4, KRP5, and KRP6 that interact with OsCDKA;1 and/or OsCDKA;2.
It is possible that increased levels of OsKRP4, OsKRP5, and OsKRP6 caused inhibition of OsCDKA;1, OsCDKF;1, and OsCDKG;1, which were induced by ABA treatment; however, OsCDKF;1 and OsCDKG;1 have not been demonstrated to interact with those KRPs. Expression of OsKRP1 and OsKRP3 was reduced by ABA (Fig. 3); while reduced expression of these proteins may affect cell cycle progression, the effect is thought to be small because OsKRP1 was not observed to interact with OsCDKAs. In addition, the transcript levels of OsKRP1 and OsKRP3 were much lower than the total sum of KRP4, KRP5, and KRP6 transcripts (Supplementary Fig. S1).
We also found that ABA suppresses the expression of OsCDKB;1 and OsCDKB;2 (Fig. 2 and Fig. 5b). ABA reduced the expression of CDKB1 and CYCB1, which are specifically expressed at the G2/M boundary in barley13. It has also been reported that polyploid cells resulting from endomitosis were present in CDKB2;1-knockdown rice, indicating an important role for OsCDKB2;1 during mitosis12. Therefore, ABA may also inhibit the G2/M transition in rice by suppressing expression of OsCDKB genes.
Expression of Orysa;CycD2;1, Orysa;CycD2;2, Orysa;CycD3;1, Orysa;CycD4;2, Orysa;CycD5;1, and Orysa;CycD6;1 decreased substantially after 4–5 d of ABA treatment (Fig. 4), but negative effects of ABA were not observed during the first 3 d of treatment (Fig. 5b and Supplementary Fig. S2). This suggests that these CycD genes are not associated with ABA-induced inhibition of shoot growth, which occurred during the first 2 d after ABA treatment (Fig. 5a).
Individual effects of auxin, cytokinin, and GA3 on cell cycle-related genes have been reported previously28. Expression of CDKA;1 was increased twofold by auxin treatment in Arabidopsis9; however, expression of OsCDKA;1 and OsCDKA;2 was suppressed by IAA treatment in our study (Fig. 2). An inhibitor of mevalonic acid (a cytokinin precursor) synthesis arrested the cell cycle at the G2 phase in tobacco BY-2 cells, and zeatin released this arrest, suggesting that cytokinin is essential for cell cycle progression29. The expression of CDKA;1 was reported to increase twofold by cytokinin treatment in Arabidopsis, indicating that cytokinin promotes the G1/S transition30. However, expression of OsCDKA;1 was not affected by BAP, and the expression of other CDK genes was significantly suppressed by BAP treatment in our study (Fig. 2). GA3 was reported to induce expression of CDKB2;1, which occurs specifically at the G2 phase31,32, while in our study, expression of OsCDKBs was significantly suppressed by GA3 (Fig. 2). These discrepancies among results are thought to be a result of differences in growth stage. We used 5-day-old seedlings, while 11- to 13-week-old rice plants were analysed by Fabian et al.32. Suppression of the KRP2 gene by IAA was reported in Arabidopsis33; we also observed IAA- (as well as BAP- and GA3-) induced suppression of OsKRPs (Fig. 3).
Cytokinin signals have been reported to regulate CycDs24, but no CycD genes were induced by BAP in our study (Fig. 4). Treatment with BAP alone also failed to promote shoot growth (Fig. 1d); we thus did not observe positive effects of cytokinins on growth of young rice seedlings at the concentrations tested in this study.
We evaluated the combined effects of plant hormones on shoot growth and expression of CDK, KRP, and CycD genes. Although all of the investigated hormones reduced ABA-induced inhibition of shoot growth, SA was the most effective antagonist against ABA (Fig. 1). Moreover, SA was more effective than IAA and BAP at blocking ABA suppression of OsCDKB;1 and OsCDKB;2, and GA3 promoted ABA suppression of OsCDKB;2 (Fig. 2). SA, BAP, and GA3 strongly suppressed ABA induction of OsKRP4 and OsKRP5, while IAA repressed induction of OsKRP4 only (Fig. 3). Thus, SA antagonizes the effects of ABA most strongly, but the effect of SA alone is relatively small compared to the effect of hormone combinations. Consequently, we attempted to determine the inhibitory mechanisms of SA on ABA in more detail.
The effects of KRPs on plant growth have been investigated by overexpressing KRP genes or by examining mutants that overaccumulate these proteins. Rice plants overexpressing OsKRP4 had short culms and significantly reduced seed fertility26, and overexpression of OsKRP1 inhibited the development of embryos and pollen grains34. Reduced cell numbers and small, aberrant leaves were observed in Arabidopsis plants overexpressing KRP218. In these plants, the endoreduplication cycle was inhibited, resulting in increased numbers of cells with 2C DNA and reduced numbers with 4C and 8C DNA. Development of male and female gametes was inhibited by cell cycle arrest caused by overaccumulation of KRP6 in anthers and embryo sacs in Arabidopsis mutants deficient in the gene encoding RHF E3 ligase, which is required for targeted degradation of KRP6 by proteasomes35. Thus, overexpression of KRP genes or overaccumulation of KRPs significantly inhibits plant growth and development of reproductive organs. Since we found that ABA induced expression of OsKRP4, OsKRP5, and OsKRP6 (Fig. 5), it is conceivable that ABA inhibits shoot growth in rice seedlings via an increase in OsKRPs coupled with a decrease in OsCDKBs.
ABA was previously reported to inhibit mitosis and thymidine incorporation in root meristems of sunflower36 and to diminish bromodeoxyuridine incorporation into Arabidopsis root meristems37. Our data also suggest that ABA inhibits DNA replication and reduces thymidine incorporation in shoot apical meristems of young rice seedlings (Fig. 7). Reduction of the 4C population and thymidine incorporation into the ABA-treated shoot apex indicate that ABA delays the G1/S transition, probably by inducing expression of OsKRP4, OsKRP5, and OsKRP6 (Fig. 3) because OsKRP4, OsKRP5, and OsKRP6 interacted with OsCDKA;1 and/or OsCDKA;2, which promote the G1/S transition (Fig. 6). Interestingly, SA completely blocked ABA inhibition of DNA replication and thymidine incorporation (Fig. 7). SA suppressed ABA-induced expression of OsKRP4, OsKRP5, and OsKRP6 (Fig. 5b), thus antagonizing ABA inhibition of cell cycle progression. However, changes in the expression of cell cycle-related genes observed in this study may have been caused by blockage of the cell cycle at the G1/S transition. Further studies using mutants for cell cycle genes are necessary to ascertain whether these changes in expression were a cause or result of interference at the G1/S transition.
Although SA is known to be a key endogenous signalling molecule that mediates systemic acquired resistance38, an antagonistic interaction is also known to occur between ABA and SA. ABA inhibits pathways up- and downstream of SA signalling; conversely, SA inhibits both ABA synthesis and ABA signalling39. Inhibition of ABA synthesis cannot explain the antagonistic effect of SA shown here, because we supplied exogenous ABA to rice seedlings. Therefore, it is conceivable that SA inhibition of ABA signalling leads to suppression of the inhibitory action of ABA on shoot growth and cell cycle progression. However, there may be other mechanisms for the antagonistic effect of SA on ABA. A future study will be initiated to investigate the effects of SA on the transport, degradation, or inactivation of ABA, in addition to its effects on ABA signalling.
Methods
Plant materials and growth conditions
Seeds of the Oborozuki cultivar of rice (Oryza sativa japonica group) were washed with sterilized water and soaked in water for 2 d at 28°C in the dark. After soaking, germinated seeds were grown under hydroponic conditions as follows. Nine seeds were placed on a plastic grid (approximately 40 × 40 mm) and floated in a plastic cup containing 100 mL distilled water. Floated seeds were grown for 3 d at 25°C in the dark and were then transferred to a new cup containing 100 mL distilled water with various concentrations and combinations of plant hormones (0–4 μM ABA, 0–1000 μM SA, 0–100 μM IAA, 0–25 μM BAP, and 0–100 μM GA3). Hydroponic culture was performed in a growth chamber in continuous darkness at 25°C for 5 d. After incubation, shoot lengths were measured.
RNA extraction, cDNA synthesis, and quantitative real-time PCR
Seven KRP genes, 15 CDK genes, and 13 D-type cyclin genes have been identified in the rice genome4,26. Among these, complementary DNA (cDNA) clones encoding OsKRP2, OsCDKC;3, OsCDKF;2, Orysa;CycD1;1, Orysa;CycD1;2, Orysa;CycD2;3, and Orysa;CycD5;4 have not been identified, suggesting that expression of these genes is extremely low. In addition, OsKRP7, which lacks both a CDK- and CYCD-binding box is thought to be a pseudogene4. We analysed KRP, CDK, and CycD genes, with the exception of those just mentioned. Shoot samples were collected daily from seedlings grown with or without hormones (2 μM ABA; 1 mM SA; 100 μM IAA; 25 μM BAP; 100 μM GA3; 2 μM ABA + 1 mM SA; 2 μM ABA + 100 μM IAA; 2 μM ABA + 25 μM BAP; or 2 μM ABA + 100 μM GA3) for 5 d in the dark. Tissue samples were frozen with liquid N2 and stored at −80°C until use. Total RNA was extracted from 100 to 200 mg of each sample using FastPrep Green RNA Isolation solution (MP Biomedicals, USA). RNA samples were treated with DNase I (Takara, Japan) to remove genomic DNA. cDNA was synthesized from 0.5-μg samples of RNA using a PrimeScript RT Master Mix kit (Takara, Japan). The levels of expression were examined by quantitative real-time PCR with a LightCycler Carousel-based system (Roche, Switzerland) using a LightCycler TaqMan Master kit (Roche). The gene-specific primer sets and universal TaqMan probes were designed using information from the Roche Assay Design Center (available through http://www.roche-applied-science.com.) The primer sequences and corresponding universal probes are listed in Supplementary Table S3. Relative mRNA abundance was normalized against the level of ubiquitin. The analyses were performed using three biological replicates. Gene expression ratios were presented as the ratio of expression levels in hormone-treated seedlings to those in untreated seedlings.
Yeast two-hybrid experiments
Y2H assays were performed using the Matchmaker Gold Yeast Two-Hybrid System (Clontech, USA). The open-reading frames of OsCDKA;1, OsCDKA;2, OsCDKB;2, OsKRP1, OsKRP4, OsKRP5, and OsKRP6 were amplified by PCR with the first cDNA cloning primers, and the first PCR products were then amplified by PCR with the second primers (see Supplementary Table S3). PCR fragments of OsCDKA;1, OsCDKA;2, OsCDKB;2, OsKRP1, OsKRP4, OsKRP5, and OsKRP6 were fused into pGADT7-AD and/or pGBKT7-BD (Clontech) using an In-Fusion HD Cloning Kit (Clontech). The final constructs were confirmed by sequencing. Different combinations of bait and prey constructs were transformed into Y2HGold (Clontech), and their ability to grow on leucine- and tryptophan-deficient minimal media (DDO) supplemented with X-α-Gal and Aureobasidin A (DDO/X/A) was assayed at 30°C.
Cell flow cytometric analysis
Nuclear DNA copy numbers were analysed by flow cytometry. Rice seedlings were hydroponically cultured for 3 d as described above (Plant materials and growth conditions) and then grown with or without various plant hormones (2 μM ABA; 2 μM ABA + 0.4 mM SA; 2 μM ABA + 1 mM SA; or 1 mM SA) for 2 d or 5 d. Five shoot tissue samples (0.5 cm) including the shoot apex were chopped into small pieces for 30 s with a razor blade in nuclei extraction buffer provided by the manufacturer (Partec North America) and filtered through a 30-μm nylon mesh tube to isolate plant nuclei. The filtered solution was suspended in a 4-fold volume of DNA-staining buffer and incubated for 30 s. The stained nuclei solution was analysed using a flow cytometer (Partec PA, Tokyo) (n = 6,000–20,000 nuclei per sample). The frequencies of 2C and 4C nuclei were calculated as [2C nuclei/(2C nuclei + 4C nuclei)] × 100, and [4C nuclei/(2C nuclei + 4C nuclei)] × 100, respectively.
Thymidine incorporation assay
A thymidine incorporation assay was used to determine the extent of DNA synthesis in response to plant hormones using a thymidine analogue, 5-bromo-2′-deoxyuridine (BrdU). Rice seedlings were hydroponically cultured for 3 d as described above and then grown with plant hormones (2 μM ABA; 2 μM ABA + 1 mM SA; or 1 mM SA) and 150 μM BrdU in the presence of 6 μM 5-fluorodeoxyuridine for 2 d at 25°C. The BrdU-labelled samples were fixed with 4% (w/v) paraformaldehyde in 0.1 M phosphate buffer (pH 7.0) for 16 h at 4°C. After fixation, samples were washed with 1× PBS containing 0.2 M sucrose, dehydrated in a graded ethanol series, and embedded in Technovit 7100 resin (Kulzer, Wehrheim, Germany). Then, 0.5-μm-thick sections were cut using an ultramicrotome (Ultracut N, Reichert-Nissei, Tokyo) and mounted on microscope slides. DAPI staining and BrdU labelling were carried out as described by Asahina et al.40. Alexa Fluor 488-conjugated anti-BrdU (Invitrogen) was used to detect anti-BrdU antibody.
Measurement of cell size
Microscopy images were prepared as described above (Thymidine incorporation assay) and analysed by ImageJ software (version 1.4; http://rsbweb.nih.gov/ij/). After setting the image scale, 40 cells per sample were manually outlined and measured.
Microarray experiments
Shoot samples were collected from seedlings grown with or without 2 μM ABA for 6 h. Total RNA was extracted as described above (RNA extraction, cDNA synthesis, and quantitative real-time PCR). The integrity of each RNA sample was examined using lab-on-a-chip technology with the RNA 6000 Nano LabChip kit and a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Microarrays were performed using a Low-Input Quick Amp Labeling Kit (Agilent) and a rice 4 × 44 K custom oligoDNA microarray (Agilent) according to the manufacturer's instructions. Hybridization microarray slides were scanned with a Microarray Scanner and the resulting images were analysed using Feature Extraction software (Agilent), applying standard normalization procedures. The analyses were performed using three biological replicates. Probes used for microarray analysis are listed in Supplementary Table S4.
Author Contributions
Y.S. and A.M. contributed to the study design. A.M. conducted the study and analysed the data. Y.S. and A.M. wrote and edited the manuscript.
Supplementary Material
Supplementary information
Acknowledgments
We thank Drs. Hideki Kato, Akira Endo, Koichi Shinoda, and Tetsuo Maoka for suggestions regarding the experiments, and Noriko Goto for technical assistance. This work was supported by the Science and Technology Research Promotion Program for Agriculture, Forestry, Fisheries and Food Industry of Japan.
References
- Pines J. Cyclins and cyclin-dependent kinases - a biochemical view. Biochem J. 308, 697–711 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan D. Principles of CDK regulation. Nature 374, 131–134 (1995). [DOI] [PubMed] [Google Scholar]
- Morgan D. Cyclin-dependent kinases: Engines, clocks, and microprocessors. Annu. Rev. Cell Dev. Biol. 13, 261–291 (1997). [DOI] [PubMed] [Google Scholar]
- Guo J., Song J., Wang F. & Zhang X. S. Genome-wide identification and expression analysis of rice cell cycle genes. Plant Mol. Biol. 64, 349–360 (2007). [DOI] [PubMed] [Google Scholar]
- Vandepoele K. et al. Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14, 903–916 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges d. e. Jager S. M., Gruissem W. & Murray J. A. Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J. 41, 546–566 (2005). [DOI] [PubMed] [Google Scholar]
- Hirt H. et al. Complementation of a yeast-cell cycle mutant by an alfalfa cDNA-encoding a protein-kinase homologous to p34cdc2. Proc. Natl. Acad. Sci. USA 88, 1636–1640 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magyar Z. et al. Cell cycle phase specificity of putative cyclin-dependent kinase variants in synchronized alfalfa cells. Plant Cell 9, 223–235 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemerly A. et al. Cdc2a expression in Arabidopsis is linked with competence for cell division. Plant Cell 5, 1711–1723 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda M., Umeda-Hara C., Yamaguchi M., Hashimoto J. & Uchimiya H. Differential expression of genes for cyclin-dependent protein kinases in rice plants. Plant Physiol. 119, 31–40 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen S. et al. Requirement of B2-type cyclin-dependent kinases for meristem integrity in Arabidopsis thaliana. Plant Cell 20, 88–100 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M. et al. CDKB2 is involved in mitosis and DNA damage response in rice. Plant J. 69, 967–977 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendreau E. Cayla T. & Corbineau F. S phase of the cell cycle: a key phase for the regulation of thermodormancy in barley grain. J. Exp. Bot. 63, 5535–5543 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr C. & Roberts J. CDK inhibitors: positive and negative regulators of G1-phase progression. Gene. Dev. 13, 1501–1512 (1999). [DOI] [PubMed] [Google Scholar]
- De Veylder L. et al. Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13, 1653–1667 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Qi Q., Schorr P., Cutler A. J., Crosby W. L. & Fowke L. C. ICK1, a cyclin-dependent protein kinase inhibitor from Arabidopsis thaliana interacts with both Cdc2a and CycD3, and its expression is induced by abscisic acid. Plant J. 15, 501–510 (1998). [DOI] [PubMed] [Google Scholar]
- Torres Acosta A., Fowke L. C. & Wang H. Analyses of phylogeny, evolution, conserved sequences and genome-wide expression of the ICK/KRP family of plant CDK inhibitors. Ann. Bot. 107, 1141–1157 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkest A. et al. The cyclin-dependent kinase inhibitor KRP2 controls the onset of the endoreduplication cycle during Arabidopsis leaf development through inhibition of mitotic CDKA;1 kinase complexes. Plant Cell 17, 1723–1736 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani M., Naganuma T., Tsutsumi K. & Saitoh Y. The syncytium-specific expression of the Orysa;KRP3 CDK inhibitor: implication of its involvement in the cell cycle control in the rice (Oryza sativa L.) syncytial endosperm. J. Exp. Bot. 61, 791–798 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettko-Szandtner A. et al. Activation of an alfalfa cyclin-dependent kinase inhibitor by calmodulin-like domain protein kinase. Plant J. 46, 111–123 (2006). [DOI] [PubMed] [Google Scholar]
- de Jager S. M., Maughan S., Dewitte W., Scofield S. & Murray J. A. The developmental context of cell-cycle control in plants. Semin. Cell Dev. Biol. 16, 385–396 (2005) [DOI] [PubMed] [Google Scholar]
- Konoa A. et al. The Arabidopsis D-type cyclin CYCD4 controls cell division in the stomatal lineage of the hypocotyl epidermis. Plant Cell 19, 1265–1277 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G. et al. Genome-wide analysis of the cyclin family in Arabidopsis and comparative phylogenetic analysis of plant cyclin-like proteins. Plant Physiol. 135, 1084–1099 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riou-Khamlich C., Huntley R., Jacqmard A. & Murray J. A. H. Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science 283, 1541–1544 (1999). [DOI] [PubMed] [Google Scholar]
- Richard C., Lescot M., Inze D. & Veylder L. D. Effect of auxin, cytokinin, and sucrose on cell cycle gene expression in Arabidopsis thaliana cell suspension cultures. Plant Cell Tiss. Org. 69, 167–176 (2002). [Google Scholar]
- Yang R. et al. Analyses of two rice (Oryza sativa) cyclin-dependent kinase inhibitors and effects of transgenic expression of OsiICK6 on plant growth and development. Ann. Bot. 107, 1087–1101 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiatek A., Lenjou M., Van Bockstaele D., Inzé D. & Van Onckelen H. Differential effect of jasmonic acid and abscisic acid on cell cycle progression in tobacco BY-2 cells. Plant Physiol. 128, 201–211 (2002). [PMC free article] [PubMed] [Google Scholar]
- Komaki S. & Sugimoto K. Control of the plant cell cycle by developmental and environmental cues. Plant Cell Physiol. 53, 953–964 (2012). [DOI] [PubMed] [Google Scholar]
- Laureys F. et al. Zeatin is indispensable for the G2-M transition in tobacco BY-2 cells. FEBS Lett. 426, 29–32 (1998). [DOI] [PubMed] [Google Scholar]
- Riou-Khamlichi C., Huntley R., Jacqmard A. & Murray J. A. H. Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science 283, 1541–1544 (1999). [DOI] [PubMed] [Google Scholar]
- Sauter M., Mekhedov S. L. & Kende H. Gibberellin promotes histone h1 kinase-activity and the expression of cdc2 and cyclin genes during the induction of rapid growth in deep-water rice internodes. Plant J. 7, 623–632 (1995). [DOI] [PubMed] [Google Scholar]
- Fabian T., Lorbiecke R., Umeda M. & Sauter M. The cell cycle genes cycA1;1 and cdc2Os-3 are coordinately regulated by gibberellin in planta. Planta 211, 376–383 (2000). [DOI] [PubMed] [Google Scholar]
- Sanz L. et al. The Arabidopsis D-type cyclin CYCD2;1 and the inhibitor ICK2/KRP2 modulate auxin-induced lateral root formation. Plant Cell 23, 641–660 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroco R. et al. The cyclin-dependent kinase inhibitor orysa; KRP1 plays an important role in seed development of rice. Plant Physiol. 142, 1053–1064 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. et al. Targeted degradation of the cyclin-dependent kinase inhibitor ICK4/KRP6 by RING-type E3 ligases is essential for mitotic cell cycle progression during Arabidopsis gametogenesis. Plant Cell 20, 1538–1554 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson J. M., Hubick K. T., Yeung E. C. & Reid D. M. Developmental responses to drought and abscisic-acid in sunflower roots. 2. Mitotic-activity. J. Exp. Bot. 41, 339–350 (1990). [Google Scholar]
- Leung J. et al. Arabidopsis ABA response gene ABI1 - features of a calcium-modulated protein phosphatase. Science 264, 1448–1452 (1994). [DOI] [PubMed] [Google Scholar]
- Umemura K. et al. Contribution of salicylic acid glucosyltransferase, OsSGT1, to chemically induced disease resistance in rice plants. Plant J. 57, 463–472 (2009). [DOI] [PubMed] [Google Scholar]
- Yasuda M. et al. Antagonistic interaction between systemic acquired resistance and the abscisic acid-mediated abiotic stress response in Arabidopsis. Plant Cell 20, 1678–1692 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asahina M. et al. Gibberellin produced in the cotyledon is required for cell division during tissue reunion in the cortex of cut cucumber and tomato hypocotyls. Plant Physiol. 129, 201–210 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information