Abstract
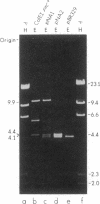
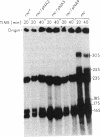
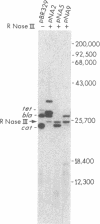
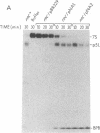
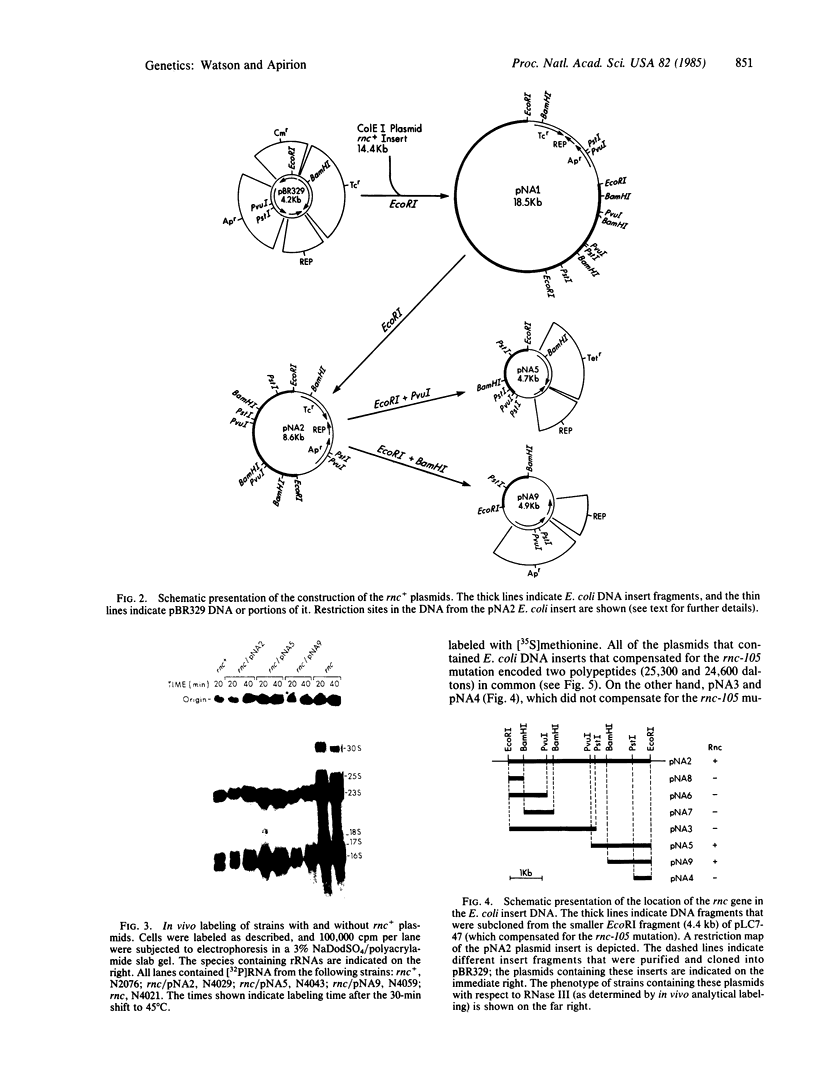
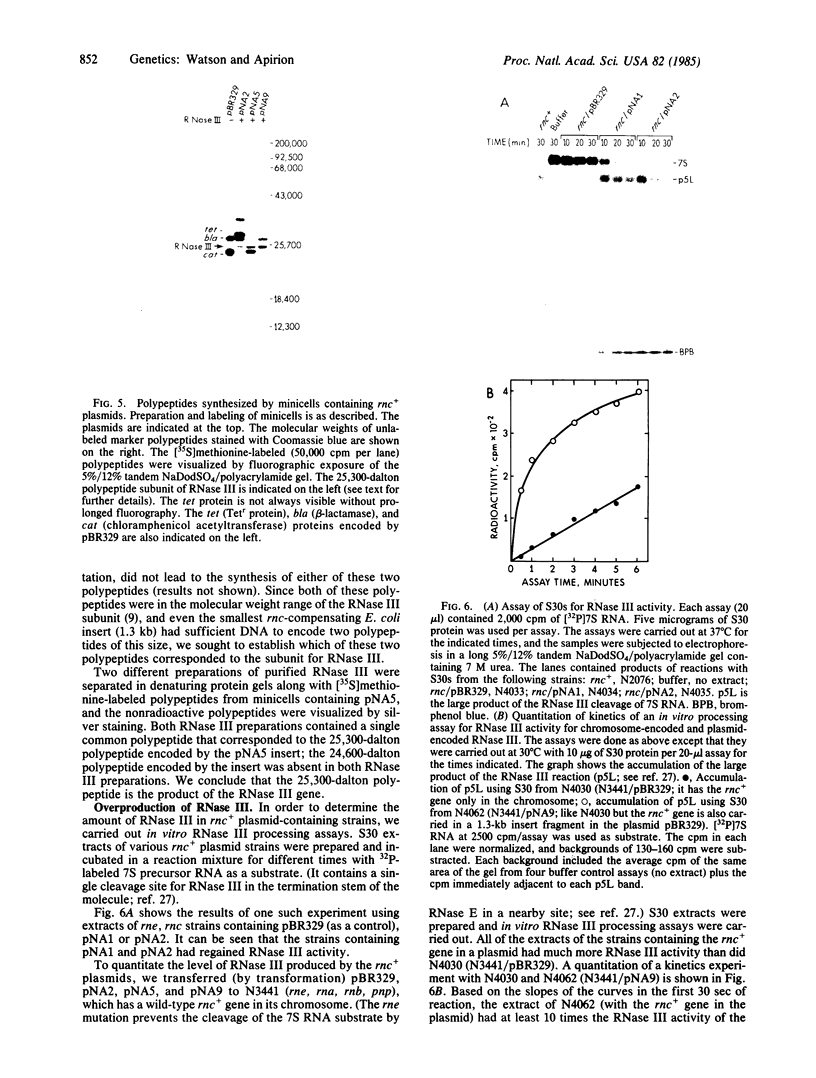
A ColE1 plasmid from the Clarke and Carbon collection [Clarke, L. & Carbon, J. (1976) Cell 9, 91-99] that contains a 14.4-kilobase Escherichia coli DNA insert complements the rnc-105 mutation, which destroys the activity of the RNA-processing enzyme RNase III. This insert and smaller restriction endonuclease fragments derived from it were cloned into the plasmid pBR329. A number of these recombinant plasmids complemented the rnc-105 mutation in a recA genetic background. The smallest cloned fragment that compensated for the rnc-105 mutation was 1.3 kilobase in size. This fragment led to the synthesis of two polypeptides. One of these polypeptides was 25,300 daltons and corresponded in size to the subunit of RNase III. Fragments cloned in opposite orientations led to synthesis of RNase III, indicating that the cloned fragments contained an endogenous promoter. Extracts of an rnc+ E. coli strain containing an rnc+ plasmid had at least 10 times more RNase III activity than did an analogous strain containing the pBR329 plasmid.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abelson J. RNA processing and the intervening sequence problem. Annu Rev Biochem. 1979;48:1035–1069. doi: 10.1146/annurev.bi.48.070179.005131. [DOI] [PubMed] [Google Scholar]
- Apirion D. Altered ribosomes in a suppressor strain of Escherichia coli. J Mol Biol. 1966 Apr;16(2):285–301. doi: 10.1016/s0022-2836(66)80173-0. [DOI] [PubMed] [Google Scholar]
- Apirion D. Isolation, genetic mapping and some characterization of a mutation in Escherichia coli that affects the processing of ribonuleic acid. Genetics. 1978 Dec;90(4):659–671. doi: 10.1093/genetics/90.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apirion D., Watson N. Mapping and characterization of a mutation in Escherichia coli that reduces the level of ribonuclease III specific for double-stranded ribonucleic acid. J Bacteriol. 1975 Oct;124(1):317–324. doi: 10.1128/jb.124.1.317-324.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 7. Microbiol Rev. 1983 Jun;47(2):180–230. doi: 10.1128/mr.47.2.180-230.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain J. P. Fluorographic detection of radioactivity in polyacrylamide gels with the water-soluble fluor, sodium salicylate. Anal Biochem. 1979 Sep 15;98(1):132–135. doi: 10.1016/0003-2697(79)90716-4. [DOI] [PubMed] [Google Scholar]
- Clarke L., Carbon J. A colony bank containing synthetic Col El hybrid plasmids representative of the entire E. coli genome. Cell. 1976 Sep;9(1):91–99. doi: 10.1016/0092-8674(76)90055-6. [DOI] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covarrubias L., Bolivar F. Construction and characterization of new cloning vehicles. VI. Plasmid pBR329, a new derivative of pBR328 lacking the 482-base-pair inverted duplication. Gene. 1982 Jan;17(1):79–89. doi: 10.1016/0378-1119(82)90103-2. [DOI] [PubMed] [Google Scholar]
- Csonka L. N., Clark A. J. Construction of an Hfr strain useful for transferring recA mutations between Escherichia coli strains. J Bacteriol. 1980 Jul;143(1):529–530. doi: 10.1128/jb.143.1.529-530.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Date T., Wickner W. Isolation of the Escherichia coli leader peptidase gene and effects of leader peptidase overproduction in vivo. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6106–6110. doi: 10.1073/pnas.78.10.6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn J. J. RNase III cleavage of single-stranded RNA. Effect of ionic strength on the fideltiy of cleavage. J Biol Chem. 1976 Jun 25;251(12):3807–3814. [PubMed] [Google Scholar]
- Ehresmann B., Imbault P., Weil J. H. Spectrophotometric determination of protein concentration in cell extracts containing tRNA's and rRNA's. Anal Biochem. 1973 Aug;54(2):454–463. doi: 10.1016/0003-2697(73)90374-6. [DOI] [PubMed] [Google Scholar]
- Frazer A. C., Curtiss R., 3rd Production, properties and utility of bacterial minicells. Curr Top Microbiol Immunol. 1975;69:1–84. doi: 10.1007/978-3-642-50112-8_1. [DOI] [PubMed] [Google Scholar]
- Gegenheimer P., Apirion D. Processing of procaryotic ribonucleic acid. Microbiol Rev. 1981 Dec;45(4):502–541. doi: 10.1128/mr.45.4.502-541.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegenheimer P., Watson N., Apirion D. Multiple pathways for primary processing of ribosomal RNA in Escherichia coli. J Biol Chem. 1977 May 10;252(9):3064–3073. [PubMed] [Google Scholar]
- Gesteland R. F. Isolation and characterization of ribonuclease I mutants of Escherichia coli. J Mol Biol. 1966 Mar;16(1):67–84. doi: 10.1016/s0022-2836(66)80263-2. [DOI] [PubMed] [Google Scholar]
- Goldblum K., Apririon D. Inactivation of the ribonucleic acid-processing enzyme ribonuclease E blocks cell division. J Bacteriol. 1981 Apr;146(1):128–132. doi: 10.1128/jb.146.1.128-132.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindler P., Keil T. U., Hofschneider P. H. Isolation and characterization of a ribonuclease 3 deficient mutant of Escherichia coli. Mol Gen Genet. 1973 Oct 16;126(1):53–59. doi: 10.1007/BF00333481. [DOI] [PubMed] [Google Scholar]
- Lennette E. T., Apirion D. Genetic analysis of an Escherichia coli syndrome. J Bacteriol. 1971 Dec;108(3):1322–1328. doi: 10.1128/jb.108.3.1322-1328.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra T. K., Apirion D. RNase E, an RNA processing enzyme from Escherichia coli. J Biol Chem. 1979 Nov 10;254(21):11154–11159. [PubMed] [Google Scholar]
- Motamedi H., Lee K., Nichols L., Schmidt F. J. An RNA species involved in Escherichia coli ribonuclease P activity. Gene cloning and effect on transfer RnA synthesis in vivo. J Mol Biol. 1982 Dec 15;162(3):535–550. doi: 10.1016/0022-2836(82)90387-4. [DOI] [PubMed] [Google Scholar]
- Ray A., Apirion D. Cloning the gene for ribonuclease E, an RNA processing enzyme. Gene. 1980 Dec;12(1-2):87–94. doi: 10.1016/0378-1119(80)90018-9. [DOI] [PubMed] [Google Scholar]
- Reed R. E., Baer M. F., Guerrier-Takada C., Donis-Keller H., Altman S. Nucleotide sequence of the gene encoding the RNA subunit (M1 RNA) of ribonuclease P from Escherichia coli. Cell. 1982 Sep;30(2):627–636. doi: 10.1016/0092-8674(82)90259-8. [DOI] [PubMed] [Google Scholar]
- Robertson H. D., Webster R. E., Zinder N. D. Purification and properties of ribonuclease III from Escherichia coli. J Biol Chem. 1968 Jan 10;243(1):82–91. [PubMed] [Google Scholar]
- Roozen K. J., Fenwick R. G., Jr, Curtiss R., 3rd Synthesis of ribonucleic acid and protein in plasmid-containing minicells of Escherichia coli K-12. J Bacteriol. 1971 Jul;107(1):21–33. doi: 10.1128/jb.107.1.21-33.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy M. K., Apirion D. Purification and properties of ribonuclease E, an RNA-processing enzyme from Escherichia coli. Biochim Biophys Acta. 1983 Sep 28;747(3):200–208. doi: 10.1016/0167-4838(83)90098-5. [DOI] [PubMed] [Google Scholar]
- Silver P., Wickner W. Genetic mapping of the Escherichia coli leader (signal) peptidase gene (lep): a new approach for determining the map position of a cloned gene. J Bacteriol. 1983 May;154(2):569–572. doi: 10.1128/jb.154.2.569-572.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W. Genetic mapping of a mutation that causes ribonucleases III deficiency in Escherichia coli. J Bacteriol. 1975 Oct;124(1):307–316. doi: 10.1128/jb.124.1.307-316.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeberényi J., Apirion D. Synthesis and processing of 5 S rRNA from an rrnB minigene in a plasmid. Biochim Biophys Acta. 1984 Oct 5;783(1):15–25. doi: 10.1016/0167-4781(84)90073-3. [DOI] [PubMed] [Google Scholar]
- Szeberényi J., Roy M. K., Apirion D. 7 S RNA: a single site substrate for the RNA processing enzyme ribonuclease E of Escherichia coli. Biochim Biophys Acta. 1983 Aug 2;740(3):282–290. doi: 10.1016/0167-4781(83)90137-9. [DOI] [PubMed] [Google Scholar]
- Szeberényi J., Roy M. K., Vaidya H. C., Apirion D. 7S RNA, containing 5S ribosomal RNA and the termination stem, is a specific substrate for the two RNA processing enzymes RNase III and RNase E. Biochemistry. 1984 Jun 19;23(13):2952–2957. doi: 10.1021/bi00308a016. [DOI] [PubMed] [Google Scholar]
- Thuring R. W., Sanders J. P., Borst P. A freeze-squeeze method for recovering long DNA from agarose gels. Anal Biochem. 1975 May 26;66(1):213–220. doi: 10.1016/0003-2697(75)90739-3. [DOI] [PubMed] [Google Scholar]
- Zwizinski C., Wickner W. Purification and characterization of leader (signal) peptidase from Escherichia coli. J Biol Chem. 1980 Aug 25;255(16):7973–7977. [PubMed] [Google Scholar]