Abstract
Many oomycete and fungal plant pathogens are obligate biotrophs, which extract nutrients only from living plant tissue and cannot grow apart from their hosts. Although these pathogens cause significant crop losses, little is known about the molecular basis or evolution of obligate biotrophy. Here, we report the genome sequence of the oomycete Hyaloperonospora arabidopsidis (Hpa), an obligate biotroph and natural pathogen of Arabidopsis thaliana. In comparison to genomes of related, hemi-biotrophic Phytophthora species, the Hpa genome exhibits dramatic reductions in genes encoding: 1) RXLR effectors and other secreted pathogenicity proteins; 2) enzymes for assimilation of inorganic nitrogen and sulphur; 3) proteins associated with zoospore formation and motility. These attributes comprise a genomic signature of evolution towards obligate biotrophy.
The oomycete Hyaloperonospora arabidopsidis (Hpa, formerly Peronospora parasitica or Hyaloperonospora parasitica) is a natural pathogen of Arabidopsis thaliana and a model for dissection of A. thaliana pathogen response networks (1, 2). Hpa belongs to a group of “downy mildew” pathogens, comprising over 800 species that cause disease on hundreds of plant species (3). Downy mildew pathogens are related to other destructive oomycete plant pathogens (e.g., Phytophthora species) (4, 5). Oomycetes belong to the kingdom Straminopila, which includes brown algae and diatoms. Although oomycetes and fungi share morphological and ecological similarities, they evolved independently to colonize plants.
Hpa hyphae grow between plant cells and establish feeding structures called haustoria, which have also evolved in fungal pathogens (2, 6). Downy mildews are obligately biotrophic and cannot be cultured apart from their hosts. In contrast, Phytophthora species are hemibiotrophic; an initial phase of biotrophic growth is followed by a necrotrophic phase. Molecular phylogenies show that downy mildews arose from a paraphyletic, Phytophthora-like, hemibiotrophic ancestor (4, 5, 7). Thus, insight into the genomic basis and evolution of obligate biotrophy can be obtained through comparison of the Hpa genome to the recently sequenced genomes of Phytophthora species (8, 9).
Genome analysis was performed on the Hpa Emoy2 isolate (1), using DNA from asexual spores (10). Sanger shotgun sequencing at 9.5-fold coverage, combined with 97 sequenced BAC inserts, yielded an assembly of 77.8 Mb. Illumina sequencing at 46-fold coverage yielded 3.8 Mb of additional sequence, which was integrated into the Sanger assembly to form the 81.6 Mb final version (v8.3.2). 42% of the Hpa genome is comprised of repetitive elements (Table S1). Analysis of Sanger and Illumina read depth suggests that v8.3 contains approximately 12.7 Mb composed of tandem repeats compressed into reduced copies in the assembly, indicative of a genome size of around 100 Mb (Fig. S1). The CEGMA pipeline, combined with manual examination, identified Hpa orthologs of 95% of the 248 conserved single copy eukaryotic genes (Fig. S2). Moreover, 94% of 31,759 EST reads aligned to the assembly, indicating that the draft genome assembly encompasses a very high percentage of the Hpa gene space.
14,543 genes were computationally predicted in v8.3, of which 80% are supported by ESTs and/or Illumina cDNA tags. This predicted gene content is similar to P. ramorum (65 Mb, 15,743 genes) and lower than P. sojae (95 Mb, 19,027 genes) (9) or P. infestans (240 MB, 17,887 genes) (8). 6882 predicted genes in Hpa had no identifiable ortholog in sequenced Phytophthora species, or similarity to known proteins, and as such represent potentially lineage-specific genes. Some of these genes may play roles that are specific to biotrophy. For example, a novel family of secreted small, cysteine-rich proteins exists in Hpa (PPAT12/24-like; Table 1)(11).
Table 1.
Copy numbers of annotated Hpa genes for hydrolases, PAMPS and effectors, compared to Phytophthora genomes.
| Gene product | H. arabidopsidis | P. sojae | P. ramorum |
|---|---|---|---|
| Extracellular Proteases | 18 | 47 | 48 |
| Glycosyl hydrolases | >60 | 125 | 114 |
| Endoglucanases (EGL12) | 3 | 10 | 8 |
| Polygalacturonases | 3 | 25 | 16 |
| Pectin methyl esterases | 3 | 19 | 15 |
| Cutinases | 2 | 16 | 4 |
| Chitinases | 1 | 5 | 2 |
| Elicitins | 1 | 18 | 17 |
| Elicitin-like | 14 | 39 | 31 |
| CBEL and CBEL-like | 2 | 13 | 15 |
| RXLR | 134 | 396 | 374 |
| NLP | 10 | 29 | 40 |
| Crinklers | 20 | 40 | 8 |
| PPAT12/24-like | 8 | 0 | 0 |
Pathogenicity genes were compared among Hpa and Phytophthora species, revealing that families encoding host-targeted, degradative enzymes (secreted proteinases, cell wall-degrading enzymes) are reduced in Hpa (Table 1). Two notable examples are the family 12 endoglucanases (EGL12) and pectin methyl esterases (Pect). Phylogenetic analyses delineated several EGL12 and Pect gene clades containing genes from P. sojae and P. ramorum but not Hpa (Figs. S6 & S7). Because Hpa and P. sojae likely share a sister group relationship relative to P. ramorum (4, 5), it is probable that a number of EGL12 and Pect genes were lost from Hpa following divergence of the lineage leading to Hpa and P. sojae. Hydrolytic enzymes that target the host cell wall can release cell wall fragments that elicit host defenses. It is conceivable that in evolving a biotrophic lifestyle, Hpa has lost most of the secreted hydrolytic enzymes that were present in a hemi-biotrophic ancestor.
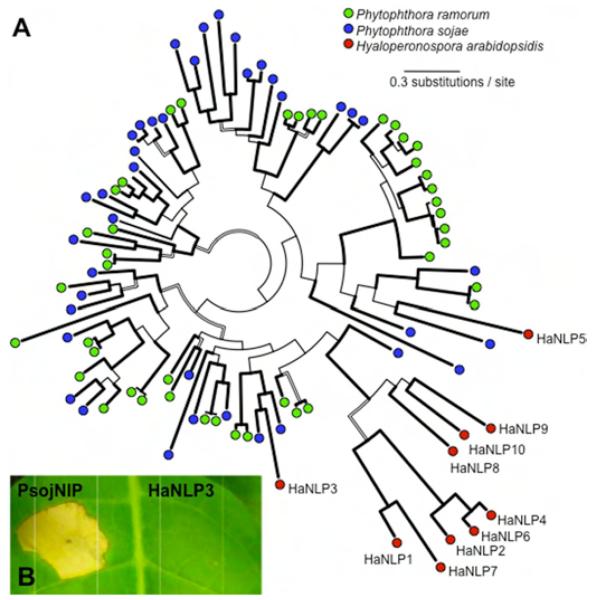
Similarly, gene families encoding necrosis and ethylene-inducing (Nep1)-like proteins (NLPs) are significantly reduced in Hpa, compared to P. sojae and P. ramorum. NLPs in Phytophthora and Pythium can trigger plant cell death and defenses (12), which has been implicated in the transition from biotrophy to necrotrophy. Only three of the 13 oomycete NLP clades contain genes from Hpa. However, one clade contains an expanded family that is specific to Hpa (Fig. 1A). All ten HaNLP genes are supported by transcriptional data. Of these, HaNLP3 is most closely related to the PsojNIP and PiNPP1.1 proteins, but it did not induce necrosis in Nicotiana tabacum (Fig. 1B). These results suggest that downy mildew NLP genes may have evolved a different function than in Phytophthora. Copy number reduction was also evident for genes encoding known pathogen-associated molecular patterns (PAMPs) such as sterol-binding elicitins (13) and carbohydrate-binding CBEL genes (14) (Table 1). These examples further suggest that selection for “stealth” (avoidance of host defences) was a major force during downy mildew evolution.
Fig. 1.
Diversity, evolutionary history, and functional analysis of oomycete necrosis and ethylene-inducing (Nep1)-like proteins (NLPs). (A) Phylogeny of oomycete NLPs. A consensus tree from the Bayesian Inference is shown. Thick lines indicate high support in Minimum Evolution (> 90), Maximum Likelihood (> 90) and Bayesian inference (> 0.95). Hollow lines indicate branches highly supported in at least two analyses. Branches with high support in less than two analyses are represented by thin lines. (B) An Hpa NLP ortholog does not induce necrosis in plant leaves. NLP genes were transiently expressed in Nicotiana tabacum by agroinfiltration.
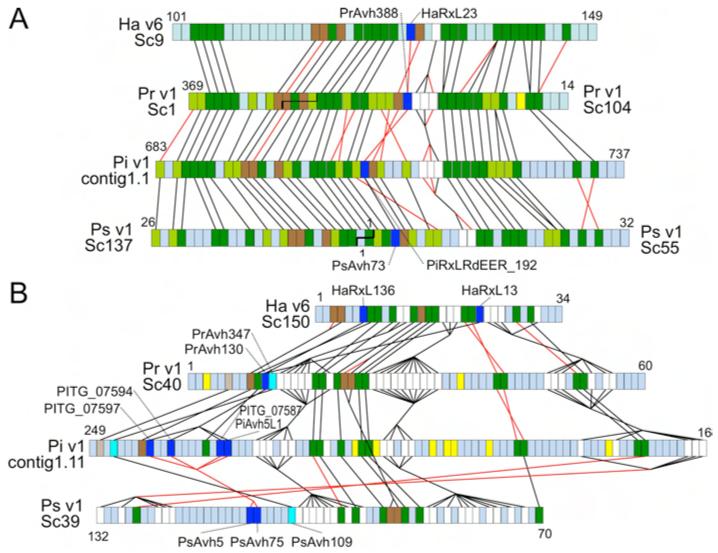
Phytophthora genomes encode hundreds of potential effector proteins (9, 15, 16) with RXLR cell entry motifs (16, 17) that likely function to suppress host defenses (18, 19). The Hpa genome contained 134 high-confidence effector gene candidates (HaRxL genes), including the known effector genes Atr1 and Atr13 (20, 21), significantly fewer than in the Phytophthora genomes (9, 15). SNPs arising from heterozygosity in v8.3 occurred at 5-fold higher rate in RXLR effector candidates (1 per ~500bp) than in other genes (1 per ~2500bp). Only 36% of the high-confidence Hpa effectors had significant matches in any Phytophthora genome (sequence similarity > 30%) consistent with strong divergent selection on RXLR effector genes (15, 22). Moreover, Hpa effectors generally were not located in syntenic locations relative to Phytophthora genomes, except for three families of effectors, which have unusually high levels of sequence conservation (Fig. 2).
Fig. 2.
Synteny of conserved RXLR effectors. (A) Region around HaRxL23, spanning scaffold_9:467737- 739923 (v6) and supercontig16:325445- 40488 (v8.3.2) (B) Region around HaRxL13 and HaRxL136, spanning scaffold_150: 3503-183330 (v6) and supercontig35:456072-268293 (v8.3.2). Colored boxes show order of gene models. Non-coding DNA is not represented. Dark green, orthologs; light green, orthologs found only in Phytophthora; dark brown, syntenic paralogs; light brown, syntenic orthologs found only in Phytophthora; white, syntenic gene families; dark blue, syntenic conserved RXLR effectors; cyan, syntenic RXLR effectors conserved only in Phytophthora; yellow, RXLR effectors not syntenic or conserved; blue-gray, other genes not conserved or syntenic. Black lines join syntenic genes with the same orientation; red lines join genes with reversed orientations. Staggered black lines in (A) show scaffold joins predicted from the synteny analysis. HaRxL23, HaRxL13 and HaRxL136 have 47%, 38% and 40% amino acid identity, respectively, with their most similar Phytophthora ortholog, within the normally hypervariable C terminus.
As obligate biotrophs, downy mildews may have lost some metabolic pathways. We identified several potential metabolic defects in Hpa compared to P. sojae, and P. ramorum (Fig. S9). For example, genes for nitrate and nitrite reductases, a nitrate transporter and sulfite reductase were missing (Fig. S10; Table S3), which is also a feature of the genomes of obligately parasitic rust and powdery mildew fungi (23, 24). Hpa also lacks genes required for synthesis of arachidonic acid and polyamine oxidases.
Flagellated zoospores are produced by many oomycetes (25). Contrastingly, several downy mildew lineages germinate by extending infective germ tubes from non-motile conidiospores, although evidence exists for a rare zoosporic stage in some otherwise conidial downy mildews (26, 27). To conclusively determine whether spore motility has been lost from the Hpa lineage, we searched the Hpa genome for 90 flagella-associated genes using Chlamydomonas sequences and their Phytophthora orthologs (28). No matches were detected in Hpa for any of these. Similarly, many Phytophthora adhesion-related genes are reduced in number or absent from Hpa, consistent with the lack of adherent cysts that normally develop from zoospores during infection.
In summary, analysis of Hpa gene space revealed genomic signatures of major alterations in pathogenic strategy, metabolism, and development that occurred during the evolution of obligate biotrophy from a facultative, hemi-biotrophic ancestor. Interestingly, some features of Hpa gene space (large numbers of secreted effectors, reduction in degradative enzymes, loss of N & S assimilation) are mirrored in genomes of biotrophic fungi (23, 24, 29, 30). These similarities indicate that convergent adaptations occurred during the independent evolution of biotrophy in fungal and oomycete lineages.
Supplementary Material
Fig. S1. Genome size estimation from Illumina and Sanger read coverage (A) Frequency of nucleotide positions in the Sanger assembly with given Illumina read coverage. (B) Frequency of Sanger reads with given Sanger read coverage. In both (A) and (B), to obtain the mean coverage of the single copy sequences, a Gaussian curve was fitted to the main peak (indicated by shaded points and horizontal bar) by fitting a quadratic function to the natural-log-transformed frequency data (inset). From each fitted quadratic function ax2 + bx + c, the mean of each Gaussian distribution was obtained as −b/2a.
Fig. S2. Identification of single copy core eukaryotic orthologous genes (CEGs) by the CEGMA pipeline ~95% of the CEGs were identified in the Hpa Emoy2 Velvet and v8.3 assemblies. This is comparable to the number of CEGs identified in P. infestans (95%), P. ramorum (96%) and P. sojae (98%). There were 4 CEGs partially identified in the Velvet, v8.3 and P. infestans assemblies, and one partially identified in P. ramorum.
The CEGs are split into 4 groups with Group 1 being the least conserved between organisms, and Group 4 being the most conserved between organisms.
Fig. S3. Sequences of H. arabidopsidis extracellular glucanases identified by mass spectrometry of proteins extracted from intracellular spaces of Arabidopsis leaves infected with H. arabidopsidis (A) HaEGL12-1 (VMD ID #808599); (B) HaEGL12-2 (VMD ID #814377). All identified peptides are highlighted in red. The annotated MS/MS spectrum is that of peptide LLDDQYLVK. Red and blue highlighted amino acids represent identified B- and Y-ions respectively.
Fig. S4. Sequence of HaPect1 protein identified by mass spectrometry of proteins extracted from intracellular spaces of Arabidopsis leaves infected with H. arabidopsidis All identified peptides are highlighted in red. The annotated MS/MS spectrum is that of peptide NQVTIAQATAQK. Red and blue highlighted amino acids represent identified B- and Y-ions respectively.
Fig. S5. Abundance of transcripts from HaEGL12 and HaPect genes during infection of Arabidopsis by H. arabidopsidis Expression was normalized against Hpa Actin2 and expression at t= 0 dpi (days post infection) is set at 1.0. (A) HpEGL12-1, -2, and -3. (B) HpPect1, -2, and -3.
Fig. S6. Minimum Evolution phylogenetic reconstruction of GH12 endoglucanase genes from H. arabidopsidis (Ha), P. sojae (Ps), and P. ramorum (Pr) Values on branches are support in Minimum Evolution, Maximum likelihood and Bayesian analysis, respectively; x denotes conflicting topology.
Fig. S7. Minimum Evolution phylogenetic reconstruction of pectin methyl esterase genes from H. arabidopsidis (Ha), P. sojae (Ps), and P. ramorum (Pr) Values on branches are support in Minimum Evolution, Maximum likelihood and Bayesian analysis, respectively, x denotes conflicting topology.
Fig. S8. Codon substitution pattern heterogeneity for orthologs of H. arabidopsidis pectin methyl esterase genes (two orthologous gene groups). Substitution pattern heterogeneity was computed using MEGA4.0 (codon translation, complete deletion). Significance threshold was set to p = 0.05
Fig. S9. Metabolic pathways (KEGG Atlas Mapping) in H. arabidopsidis (Hpa), P. infestans, P. sojae and P. ramorum Gene products (circles) are joined by enzyme-catalyzed pathways (lines). Components highlighted in blue are present in Hpa and at least one Phytophthora species. Components in red are absent in Hpa but present in at least 2 out of three Phytophthora species. Components in green are present in Hpa but are absent in the Phytophthora species. Greyed circles/lines indicate components absent in all four species.
Fig. S10. Genes for nitrate and sulfate metabolism are present in Phytophthora but absent in H. arabidopsidis. (A) Pathways for assimilation of N and S. Enzyme names are italicized. All enzymes shown are present in one or two copies in Phytophthora sojae, P. ramorum, and P. infestans. White crosses designate genes that are absent in Hpa. Gene IDs for all species are provided in Table S3.
(B) Nitrate and nitrite reductase genes are adjacent in Phytophthora genomes but missing from the syntenic location in the Hpa genome. Regions shown are from P. sojae version 1.1, P. infestans version 1.0 and Hpa version 6. Hpa region spans spanning scaffold_20:236914-269510 (v6) and SuperContig73:223284-255880 (v8.3.2) Colored boxes show order of gene models. Non-coding DNA is not represented. Dark brown, orthologs; gray, non-conserved genes; purple, cluster of nitrate assimilation genes specific to Phytophthora genomes. Black lines join syntenic genes with the same orientation; red lines join genes with reversed orientations.
Table S1. Repeat elements in the genomes of H. arabidopsidis and P. sojae
Table S2. Copy numbers of annotated H. arabidopsidis genes for phospholipid signalling enzymes, secondary metabolite biosynthesis, and ABC transporters, compared to Phytophthora genomes.
Table S3. Genes IDs for nitrogen and sulphur assimilation enzymes in Phytophthora species and H. arabidopsidis
Acknowledgments
We thank Eric Holub for providing the Emoy2 isolate, David Greenshields and Nathan Bruce for technical assistance, Robert Hubley for creating repeat modeller libraries, and participants in the 2007 Annotation Jamboree and in the 2008 and 2009 Oomycete Bioinformatics Training Workshops for sequence annotations. This research was supported by grants EF-0412213, IOS-0744875, IOS-0924861 and MCB-0639226 from the US NSF and 2004-35600-15055 and 2007-35319-18100 from the USDA NIFA to BMT and JMM; BBSRC BB/C509123/1, BB/E024815/1 and EPSRC/BBSRC Systems Biology DTC student EP/F500025/1 to JB; Gatsby GAT2545 and BBSRC BB/F0161901, BB/E024882/1 and BBSRC CASE studentship T12144 to JDGJ. Genome browsers are maintained at the Virginia Bioinformatics Institute (vmd.vbi.vt.edu) and the Sainsbury Laboratories (gbrowse2.tsl.ac.uk/cgi-bin/gb2/gbrowse/hpa_emoy2_publication/). Other support is detailed in the SOM.
References and Notes
- 1.Holub EB. European Journal of Plant Pathology. 2008;122:91. [Google Scholar]
- 2.Coates ME, Beynon JL. Annu Rev Phytopathol. 2010;48:329. doi: 10.1146/annurev-phyto-080508-094422. [DOI] [PubMed] [Google Scholar]
- 3.Clark J, Spencer-Phillips P. Encyclopedia of Microbiology. vol. 2. Academic Press, Inc.; 2000. pp. 117–129. [Google Scholar]
- 4.Giresse X, Ahmed S, Richard-Cervera S, Delmotte F. J. Phytopathol. 2009;158:321. [Google Scholar]
- 5.Goker M, Voglmayr H, Riethmuller A, Oberwinkler F. Fungal Genet Biol. 2007;44:105. doi: 10.1016/j.fgb.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 6.Panstruga R, Dodds PN. Science. 2009;324:748. doi: 10.1126/science.1171652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thines M. PLoS One. 2009;4:e4790. doi: 10.1371/journal.pone.0004790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haas BJ, et al. Nature. 2009;461:393. doi: 10.1038/nature08358. [DOI] [PubMed] [Google Scholar]
- 9.Tyler BM, et al. Science. 2006;313:1261. doi: 10.1126/science.1128796. [DOI] [PubMed] [Google Scholar]
- 10.Callout.
- 11.Bittner-Eddy P, et al. Molecular Plant Pathology. 2003;4:501. doi: 10.1046/j.1364-3703.2003.00194.x. [DOI] [PubMed] [Google Scholar]
- 12.Gijzen M, Nurnberger T. Phytochemistry. 2006;67:1800. doi: 10.1016/j.phytochem.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 13.Kamoun S. Annu Rev Phytopathol. 2006;44:41. doi: 10.1146/annurev.phyto.44.070505.143436. [DOI] [PubMed] [Google Scholar]
- 14.Gaulin E, et al. Plant Cell. 2006;18:1766. doi: 10.1105/tpc.105.038687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang RH, Tripathy S, Govers F, Tyler BM. Proc Natl Acad Sci USA. 2008;105:4874. doi: 10.1073/pnas.0709303105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whisson SC, et al. Nature. 2007;450:115. doi: 10.1038/nature06203. [DOI] [PubMed] [Google Scholar]
- 17.Dou D, et al. Plant Cell. 2008;20:1930. doi: 10.1105/tpc.107.056093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dou D, et al. Plant Cell. 2008;20:1118. doi: 10.1105/tpc.107.057067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sohn KH, Lei R, Nemri A, Jones JD. Plant Cell. 2007;19:4077. doi: 10.1105/tpc.107.054262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allen RL, et al. Science. 2004;306:1957. doi: 10.1126/science.1104022. [DOI] [PubMed] [Google Scholar]
- 21.Rehmany AP, et al. Plant Cell. 2005;17:1839. doi: 10.1105/tpc.105.031807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Win J, et al. Plant Cell. 2007 doi: 10.1105/tpc.107.051037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin F. personal communication.
- 24.Spanu P. personal communication.
- 25.Hardham AR, Hyde GJ. Advances In Botanical Research. 1997;24:353. [Google Scholar]
- 26.Milbrath DG. Journal of Agricultural Sciences. 1923;23:989. [Google Scholar]
- 27.Skalicky V. Preslia. 1966;38:117. [Google Scholar]
- 28.Pazour GJ, Agrin N, Leszyk J, Witman GB. The Journal of Cell Biology. 2005;170:103. doi: 10.1083/jcb.200504008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kamper J, et al. Nature. 2006;444:97. doi: 10.1038/nature05248. [DOI] [PubMed] [Google Scholar]
- 30.Martin F, et al. Nature. 2008;452:88. doi: 10.1038/nature06556. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Genome size estimation from Illumina and Sanger read coverage (A) Frequency of nucleotide positions in the Sanger assembly with given Illumina read coverage. (B) Frequency of Sanger reads with given Sanger read coverage. In both (A) and (B), to obtain the mean coverage of the single copy sequences, a Gaussian curve was fitted to the main peak (indicated by shaded points and horizontal bar) by fitting a quadratic function to the natural-log-transformed frequency data (inset). From each fitted quadratic function ax2 + bx + c, the mean of each Gaussian distribution was obtained as −b/2a.
Fig. S2. Identification of single copy core eukaryotic orthologous genes (CEGs) by the CEGMA pipeline ~95% of the CEGs were identified in the Hpa Emoy2 Velvet and v8.3 assemblies. This is comparable to the number of CEGs identified in P. infestans (95%), P. ramorum (96%) and P. sojae (98%). There were 4 CEGs partially identified in the Velvet, v8.3 and P. infestans assemblies, and one partially identified in P. ramorum.
The CEGs are split into 4 groups with Group 1 being the least conserved between organisms, and Group 4 being the most conserved between organisms.
Fig. S3. Sequences of H. arabidopsidis extracellular glucanases identified by mass spectrometry of proteins extracted from intracellular spaces of Arabidopsis leaves infected with H. arabidopsidis (A) HaEGL12-1 (VMD ID #808599); (B) HaEGL12-2 (VMD ID #814377). All identified peptides are highlighted in red. The annotated MS/MS spectrum is that of peptide LLDDQYLVK. Red and blue highlighted amino acids represent identified B- and Y-ions respectively.
Fig. S4. Sequence of HaPect1 protein identified by mass spectrometry of proteins extracted from intracellular spaces of Arabidopsis leaves infected with H. arabidopsidis All identified peptides are highlighted in red. The annotated MS/MS spectrum is that of peptide NQVTIAQATAQK. Red and blue highlighted amino acids represent identified B- and Y-ions respectively.
Fig. S5. Abundance of transcripts from HaEGL12 and HaPect genes during infection of Arabidopsis by H. arabidopsidis Expression was normalized against Hpa Actin2 and expression at t= 0 dpi (days post infection) is set at 1.0. (A) HpEGL12-1, -2, and -3. (B) HpPect1, -2, and -3.
Fig. S6. Minimum Evolution phylogenetic reconstruction of GH12 endoglucanase genes from H. arabidopsidis (Ha), P. sojae (Ps), and P. ramorum (Pr) Values on branches are support in Minimum Evolution, Maximum likelihood and Bayesian analysis, respectively; x denotes conflicting topology.
Fig. S7. Minimum Evolution phylogenetic reconstruction of pectin methyl esterase genes from H. arabidopsidis (Ha), P. sojae (Ps), and P. ramorum (Pr) Values on branches are support in Minimum Evolution, Maximum likelihood and Bayesian analysis, respectively, x denotes conflicting topology.
Fig. S8. Codon substitution pattern heterogeneity for orthologs of H. arabidopsidis pectin methyl esterase genes (two orthologous gene groups). Substitution pattern heterogeneity was computed using MEGA4.0 (codon translation, complete deletion). Significance threshold was set to p = 0.05
Fig. S9. Metabolic pathways (KEGG Atlas Mapping) in H. arabidopsidis (Hpa), P. infestans, P. sojae and P. ramorum Gene products (circles) are joined by enzyme-catalyzed pathways (lines). Components highlighted in blue are present in Hpa and at least one Phytophthora species. Components in red are absent in Hpa but present in at least 2 out of three Phytophthora species. Components in green are present in Hpa but are absent in the Phytophthora species. Greyed circles/lines indicate components absent in all four species.
Fig. S10. Genes for nitrate and sulfate metabolism are present in Phytophthora but absent in H. arabidopsidis. (A) Pathways for assimilation of N and S. Enzyme names are italicized. All enzymes shown are present in one or two copies in Phytophthora sojae, P. ramorum, and P. infestans. White crosses designate genes that are absent in Hpa. Gene IDs for all species are provided in Table S3.
(B) Nitrate and nitrite reductase genes are adjacent in Phytophthora genomes but missing from the syntenic location in the Hpa genome. Regions shown are from P. sojae version 1.1, P. infestans version 1.0 and Hpa version 6. Hpa region spans spanning scaffold_20:236914-269510 (v6) and SuperContig73:223284-255880 (v8.3.2) Colored boxes show order of gene models. Non-coding DNA is not represented. Dark brown, orthologs; gray, non-conserved genes; purple, cluster of nitrate assimilation genes specific to Phytophthora genomes. Black lines join syntenic genes with the same orientation; red lines join genes with reversed orientations.
Table S1. Repeat elements in the genomes of H. arabidopsidis and P. sojae
Table S2. Copy numbers of annotated H. arabidopsidis genes for phospholipid signalling enzymes, secondary metabolite biosynthesis, and ABC transporters, compared to Phytophthora genomes.
Table S3. Genes IDs for nitrogen and sulphur assimilation enzymes in Phytophthora species and H. arabidopsidis