Abstract
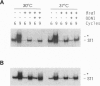
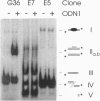
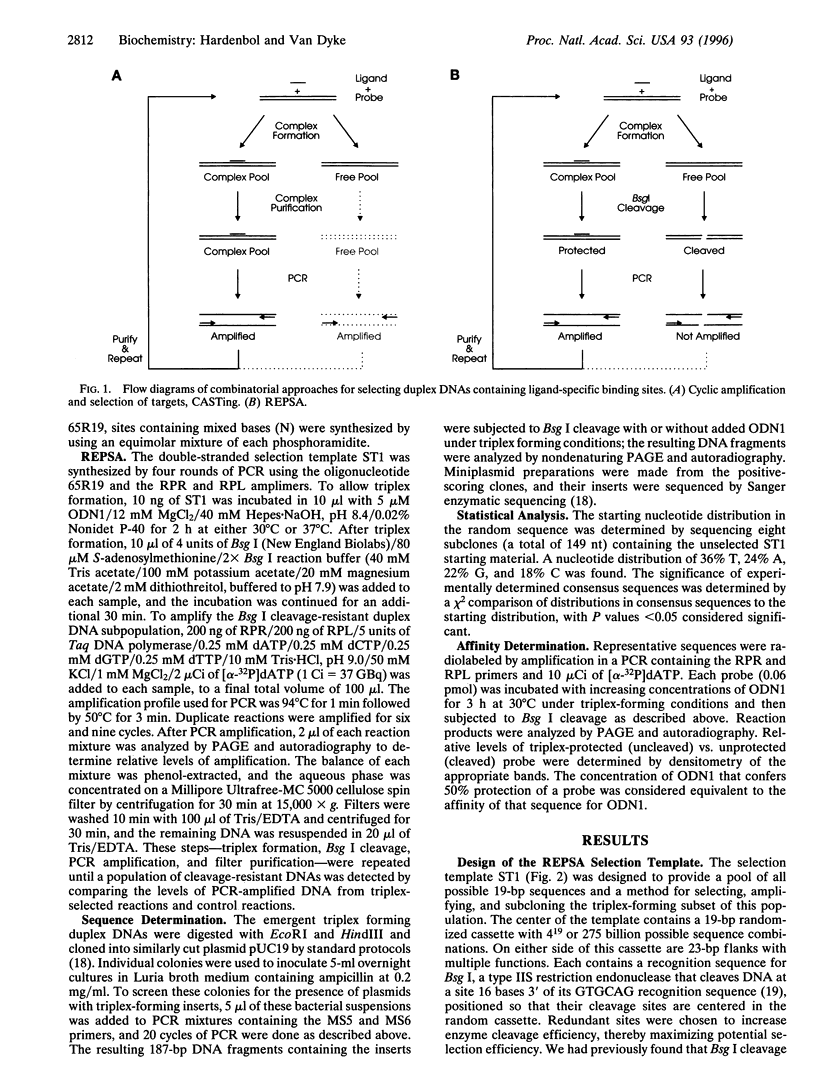
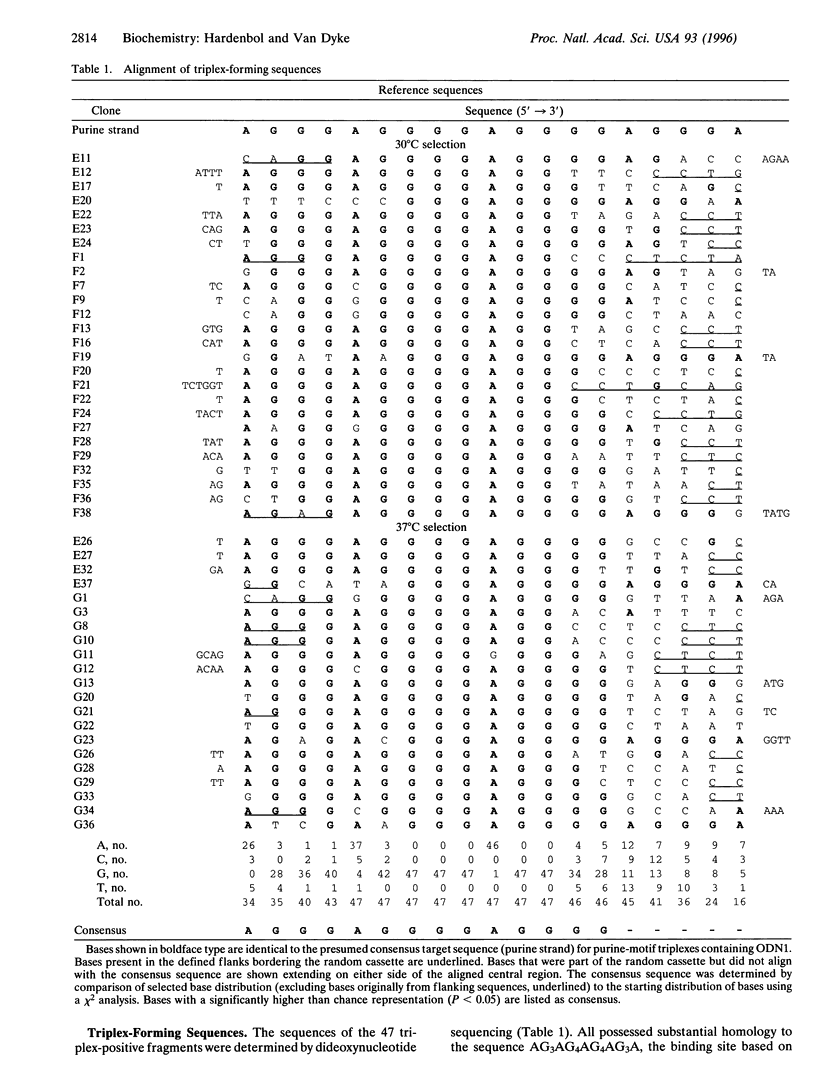
We have devised a combinatorial method, restriction endonuclease protection selection and amplification (REPSA), to identify consensus ligand binding sequences in DNA. In this technique, cleavage by a type IIS restriction endonuclease (an enzyme that cleaves DNA at a site distal from its recognition sequence) is prevented by a bound ligand while unbound DNA is cleaved. Since the selection step of REPSA is performed in solution under mild conditions, this approach is amenable to the investigation of ligand-DNA complexes that are either insufficiently stable or not readily separable by other methods. Here we report the use of REPSA to identify the consensus duplex DNA sequence recognized by a G/T-rich oligodeoxyribonucleotide under conditions favoring purine-motif triple-helix formation. Analysis of 47 sequences indicated that recognition between 13 bases on the oligonucleotide 3' end and the duplex DNA was sufficient for triplex formation and indicated the possible existence of a new base triplet, G.AT. This information should help identify appropriate target sequences for purine-motif triplex formation and demonstrates the power of REPSA for investigating ligand-DNA interactions.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beal P. A., Dervan P. B. Second structural motif for recognition of DNA by oligonucleotide-directed triple-helix formation. Science. 1991 Mar 15;251(4999):1360–1363. doi: 10.1126/science.2003222. [DOI] [PubMed] [Google Scholar]
- Beal P. A., Dervan P. B. The influence of single base triplet changes on the stability of a pur.pur.pyr triple helix determined by affinity cleaving. Nucleic Acids Res. 1992 Jun 11;20(11):2773–2776. doi: 10.1093/nar/20.11.2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickle T. A., Krüger D. H. Biology of DNA restriction. Microbiol Rev. 1993 Jun;57(2):434–450. doi: 10.1128/mr.57.2.434-450.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell T. K., Weintraub H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science. 1990 Nov 23;250(4984):1104–1110. doi: 10.1126/science.2174572. [DOI] [PubMed] [Google Scholar]
- Cheng A. J., Van Dyke M. W. Oligodeoxyribonucleotide length and sequence effects on intermolecular purine-purine-pyrimidine triple-helix formation. Nucleic Acids Res. 1994 Nov 11;22(22):4742–4747. doi: 10.1093/nar/22.22.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durland R. H., Kessler D. J., Gunnell S., Duvic M., Pettitt B. M., Hogan M. E. Binding of triple helix forming oligonucleotides to sites in gene promoters. Biochemistry. 1991 Sep 24;30(38):9246–9255. doi: 10.1021/bi00102a017. [DOI] [PubMed] [Google Scholar]
- Ellington A. D., Szostak J. W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990 Aug 30;346(6287):818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- Ellington A. D., Szostak J. W. Selection in vitro of single-stranded DNA molecules that fold into specific ligand-binding structures. Nature. 1992 Feb 27;355(6363):850–852. doi: 10.1038/355850a0. [DOI] [PubMed] [Google Scholar]
- François J. C., Saison-Behmoaras T., Thuong N. T., Hélène C. Inhibition of restriction endonuclease cleavage via triple helix formation by homopyrimidine oligonucleotides. Biochemistry. 1989 Dec 12;28(25):9617–9619. doi: 10.1021/bi00451a011. [DOI] [PubMed] [Google Scholar]
- Hanvey J. C., Shimizu M., Wells R. D. Site-specific inhibition of EcoRI restriction/modification enzymes by a DNA triple helix. Nucleic Acids Res. 1990 Jan 11;18(1):157–161. doi: 10.1093/nar/18.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Smith C. L., Cantor C. R. Sequence-specific DNA purification by triplex affinity capture. Proc Natl Acad Sci U S A. 1992 Jan 15;89(2):495–498. doi: 10.1073/pnas.89.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenan D. J., Tsai D. E., Keene J. D. Exploring molecular diversity with combinatorial shape libraries. Trends Biochem Sci. 1994 Feb;19(2):57–64. doi: 10.1016/0968-0004(94)90033-7. [DOI] [PubMed] [Google Scholar]
- Maher L. J., 3rd, Wold B., Dervan P. B. Inhibition of DNA binding proteins by oligonucleotide-directed triple helix formation. Science. 1989 Aug 18;245(4919):725–730. doi: 10.1126/science.2549631. [DOI] [PubMed] [Google Scholar]
- Mavrothalassitis G., Beal G., Papas T. S. Defining target sequences of DNA-binding proteins by random selection and PCR: determination of the GCN4 binding sequence repertoire. DNA Cell Biol. 1990 Dec;9(10):783–788. doi: 10.1089/dna.1990.9.783. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K., Mizuuchi M., Gellert M. Cruciform structures in palindromic DNA are favored by DNA supercoiling. J Mol Biol. 1982 Apr 5;156(2):229–243. doi: 10.1016/0022-2836(82)90325-4. [DOI] [PubMed] [Google Scholar]
- Nishikawa N., Oishi M., Kiyama R. Construction of a human genomic library of clones containing poly(dG-dA).poly(dT-dC) tracts by Mg(2+)-dependent triplex affinity capture. DNA polymorphism associated with the tracts. J Biol Chem. 1995 Apr 21;270(16):9258–9264. doi: 10.1074/jbc.270.16.9258. [DOI] [PubMed] [Google Scholar]
- Pei D. H., Ulrich H. D., Schultz P. G. A combinatorial approach toward DNA recognition. Science. 1991 Sep 20;253(5026):1408–1411. doi: 10.1126/science.1716784. [DOI] [PubMed] [Google Scholar]
- Pollock R., Treisman R. A sensitive method for the determination of protein-DNA binding specificities. Nucleic Acids Res. 1990 Nov 11;18(21):6197–6204. doi: 10.1093/nar/18.21.6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak J. W. In vitro genetics. Trends Biochem Sci. 1992 Mar;17(3):89–93. doi: 10.1016/0968-0004(92)90242-2. [DOI] [PubMed] [Google Scholar]
- Szybalski W., Kim S. C., Hasan N., Podhajska A. J. Class-IIS restriction enzymes--a review. Gene. 1991 Apr;100:13–26. doi: 10.1016/0378-1119(91)90345-c. [DOI] [PubMed] [Google Scholar]
- Thiesen H. J., Bach C. Target Detection Assay (TDA): a versatile procedure to determine DNA binding sites as demonstrated on SP1 protein. Nucleic Acids Res. 1990 Jun 11;18(11):3203–3209. doi: 10.1093/nar/18.11.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai D. E., Harper D. S., Keene J. D. U1-snRNP-A protein selects a ten nucleotide consensus sequence from a degenerate RNA pool presented in various structural contexts. Nucleic Acids Res. 1991 Sep 25;19(18):4931–4936. doi: 10.1093/nar/19.18.4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990 Aug 3;249(4968):505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Wright W. E., Funk W. D. CASTing for multicomponent DNA-binding complexes. Trends Biochem Sci. 1993 Mar;18(3):77–80. doi: 10.1016/0968-0004(93)90156-h. [DOI] [PubMed] [Google Scholar]