Abstract
Alterations in Ca2+ homeostasis and accumulation of unfolded proteins in the endoplasmic reticulum (ER) lead to an ER stress response. Prolonged ER stress may lead to cell death. Glucose-regulated protein (GRP) 78 (Bip) is an ER lumen protein whose expression is induced during ER stress. GRP78 is involved in polypeptide translocation across the ER membrane, and also acts as an apoptotic regulator by protecting the host cell against ER stress-induced cell death, although the mechanism by which GRP78 exerts its cytoprotective effect is not understood. The present study was carried out to determine whether one of the mechanisms of cell death inhibition by GRP78 involves inhibition of caspase activation. Our studies indicate that treatment of cells with ER stress inducers causes GRP78 to redistribute from the ER lumen with subpopulations existing in the cytosol and as an ER transmembrane protein. GRP78 inhibits cytochrome c-mediated caspase activation in a cell-free system, and expression of GRP78 blocks both caspase activation and caspase-mediated cell death. GRP78 forms a complex with caspase-7 and -12 and prevents release of caspase-12 from the ER. Addition of (d)ATP dissociates this complex and may facilitate movement of caspase-12 into the cytoplasm to set in motion the cytosolic component of the ER stress-induced apoptotic cascade. These results define a novel protective role for GRP78 in preventing ER stress-induced cell death.
Keywords: Endoplasmic reticulum stress, Caspase-12, Caspase-7, Apoptosis, Glucose-regulated protein 78, Thapsigargin, Brefeldin
1. Introduction
Alterations in Ca2+. homeostasis and accumulation of unfolded proteins in the endoplasmic reticulum (ER) elicit cellular stress responses, particularly ER stress signals, to protect cells against changes in Ca2+. levels and toxic buildup of misfolded proteins [1–3]. Prolonged ER stress leads to cell death and is linked to the pathogenesis of some neurodegenerative disorders that feature misfolded proteins including Alzheimer’s disease, Parkinson’s disease and ALS [1,2]. Cells from all organisms increase the expression of a class of ER stress proteins in response to ER stress which serve as molecular chaperones and are involved in protein translocation, protein folding and assembly, and the regulation of protein secretion [3]. Glucose-regulated protein (GRP) 78 is an ER chaperone protein whose expression is induced during oxidative stress, chemical toxicity, treatment with Ca2+. ionophores and inhibitors of glycosylation [3]. This induction, which is part of the unfolded protein response (UPR), is required to alleviate ER stress, maintain ER function, facilitate protein folding and thus protect cells from the aforementioned toxic insults [3,4]. Several reports suggest that GRP78 may protect the host cell against cell death by suppressing oxyradical accumulation and stabilizing mitochondrial function [5–8]. However, the mechanism by which GRP78 inhibits the ER stress-induced cell death program, and by which this is reversed during pro-longed ER stress leading to cell death, is still unknown.
Caspases comprise a family of cysteine-dependent aspartate-directed proteases expressed in a variety of cell types and tissues. Activation of caspases is a central mechanism in the apoptotic cell death process [9], which may occur either through recruitment into activating complexes or by a direct cleavage by another caspase [9–11]. Earlier studies demonstrated the association of caspase-7 and caspase-12 with the ER compartment [12–16]. Caspase-12 is activated by ER stress signals and participates in ER stress-induced apoptosis [14–16]. The present study was performed to determine whether one of the mechanisms of cell death inhibition by GRP78 involves blocking caspase activation. Our findings suggest that GRP78 inhibits apoptotic signals at least in part by blocking caspase activation.
2. Materials and methods
2.1. Plasmids, cells and culture conditions
The PstI/SspI fragment of hamster GRP78 (gift of Dr. Amy S. Lee) containing the complete coding sequence was subcloned into the parent vector, pTR-UF12, at the NsiI/EcoRV restrictions sites. This places GRP78 under the control of a hybrid CMV enhancer/β-actin promoter, followed by an internal ribosomal entry sequence and the cDNA for enhanced green fluorescent protein. The cassette is flanked by the adeno-associated virus 145 bp inverted terminal repeats in a pBS (Stratagene) backbone. The pTR-UF12 plasmid was a gift from Dr. Barry Byrne, University of Florida Gene Therapy Center. Mouse caspase-12 cDNA was amplified as previously described [16]. The human embryonic kidney 293T cell line was used for transient transfection as described earlier [16].
2.2. Immunofluorescence staining
The human embryonic kidney 293T cells grown in poly-d-lysine-coated culture slides (Becton Dickinson Labware, Bedford, MA, USA) were washed with phosphate-buffered saline and fixed with 4% paraformaldehyde in phosphate-buffered saline for 20 min. For the detection of GRP78, cells were stained with rabbit polyclonal anti-GRP78 antibody (1:100 dilution, Santa Cruz Biotechnology) as primary antibody and Cy3-labeled donkey anti-rabbit IgG (1:400 dilution, Jackson Immunoresearch Labs) as the secondary antibody. The rabbit polyclonal anti-GRP78 antibody that recognizes only GRP78 has previously been employed to determine the localization of the surfactant protein B [17]. Nuclei were counterstained with 0.05% DAPI (Sigma) in 1 × TBS and washed for 30 min in three changes of 1 × TBS. Stained cells were examined using a Nikon Eclipse-800 epifluorescence microscope equipped with appropriate filters and the images captured with a Magnafire CCD color camera (Optronics, Goleta, CA, USA) using PCI-Imaging software.
2.3. Cell-free extracts and Western blotting
Recombinant hamster GRP78 in 37 mM Tris–HCl pH 7.5/37 mM NaCl was obtained from Stressgen Biotechnologies. Cell-free cytoplasmic extracts were prepared as previously described [18]. Electrophoresis and Western blot analysis were carried out as described earlier [16]. Blots were probed with a 1:1000 dilution of the anti-GRP78 polyclonal antibody from Stressgen (Victoria, Canada), a 1:1000 dilution of the anti-caspase-3 monoclonal antibody, 1:500 dilution of anti-caspase-7 monoclonal antibody (all from Transduction/Pharmingen Laboratories, San Diego, CA, USA), and a 1:50 dilution of rat monoclonal anti-caspase-12 antibody (gift of Dr. Junying Yuan). The blots were incubated in a horseradish peroxidase-coupled secondary antibody for 1 h followed by enhanced chemiluminescence detection of the proteins with Hyperfilm ECL detection (Amersham, Arlington Heights, IL, USA).
2.4. Co-immunoprecipitation (co-IP) assay
Cell lysis and IP were performed as previously described [16,19,20] to provide a qualitative assessment of caspase interaction before and after ER stress. In a typical IP experiment we loaded 2.5-fold more starting protein in the IP lane compared to the lysate lane. The presence of the interacting protein was detected by Western blot analysis using the respective antibody.
2.5. Cell fractionation and limited tryptic digestion of microsomal proteins
Fractionation was performed as described earlier [16,21]. The purity of each fraction was assessed by the presence of specific marker proteins: poly(ADP-ribose) polymerase (PARP) for nuclei, protein disulfide isomerase (PDI) and SREBP-1 for ER. Equal amounts of protein were analyzed by electrophoresis. Isolated microsomes from normal and treated cells were incubated with 0.05% trypsin-EDTA for 30 min at 35°C. At the end of the reaction, samples were detected by Western blotting by loading equal amounts of total microsomal protein.
2.6. Quantification of apoptosis
Control and treated cells were stained with Hoechst 33258 and quantified as previously described [18,22].
3. Results
3.1. ER stress-induced cell-free extracts fail to activate caspases
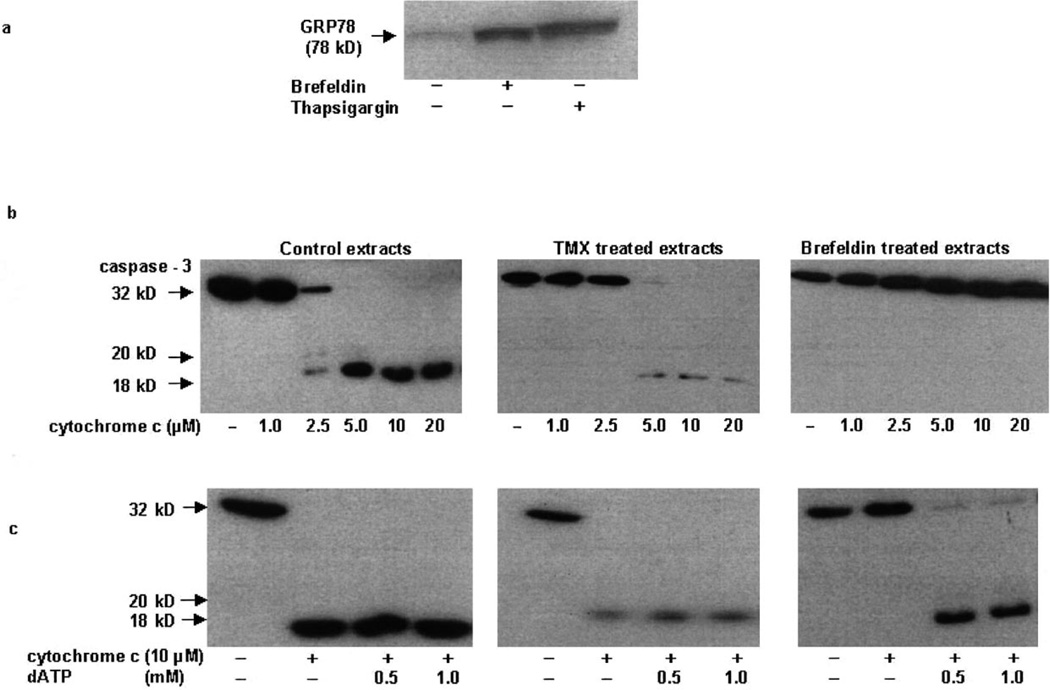
Our initial studies examined the effects of agents that induce ER stress in order to understand the mechanism of cell death in cell-free post-nuclear extracts. Brefeldin-A, a fungal metabolite, and thapsigargin, an inhibitor of the ER Ca-ATPase, induce ER stress and cell death in many cell types [1,3,16]. A high level of GRP78 protein expression is indicative of ER stress [5–8] and accordingly cell-free extracts prepared from cells treated with 2.5 µM brefeldin-A or 2.5 µM thapsigargin displayed increased GRP78 protein expression (Fig. 1a). Addition of cytochrome c alone or together with dATP to post-nuclear extracts initiates proteolysis of caspases and caspase substrates characteristic of apoptosis [18,23]. As shown in Fig. 1b, addition of cytochrome c alone or together with dATP to cell-free extracts from control (vehicle-treated) and tamoxifen (TMX)-treated 293T cells induced caspase-3 processing (Fig. 1b,c). However, cell-free extracts from brefeldin-treated 293T cells, which displayed increased GRP78 protein expression (Fig. 1a), failed to cleave caspase-3 in the presence of various concentrations of cytochrome c (Fig. 1b), and cleavage was seen only when dATP was added together with cytochrome c (Fig. 1c). Similar inhibition of caspase processing was also seen in cell-free extracts made from thapsigargin cells (data not shown).
Fig. 1.
ER stress-induced cell-free extracts fail to activate caspases. a: Cell-free cytosolic extracts (150 µg protein) from untreated, 2.5 µM brefeldin-A- (24 h) and 2.5 µM thapsigargin-treated 293T cells were analyzed by Western blot analysis for GRP78. b: Cell-free cytosolic extracts from untreated, 5 µM TMX- (24 h) and 2.5 µM brefeldin-A- (24 h) treated 293T cells were incubated for 1 h at 37°C with various concentrations of cytochrome c. Samples were analyzed by Western blot analysis for the pro- (32 kDa) and active forms of caspase-3 (20 and 18 kDa). c: Cleavage of caspase-3 was also studied in cell-free extracts that were incubated with a fixed concentration of cytochrome c and varying amounts of dATP. Figures are representative of three independent experiments.
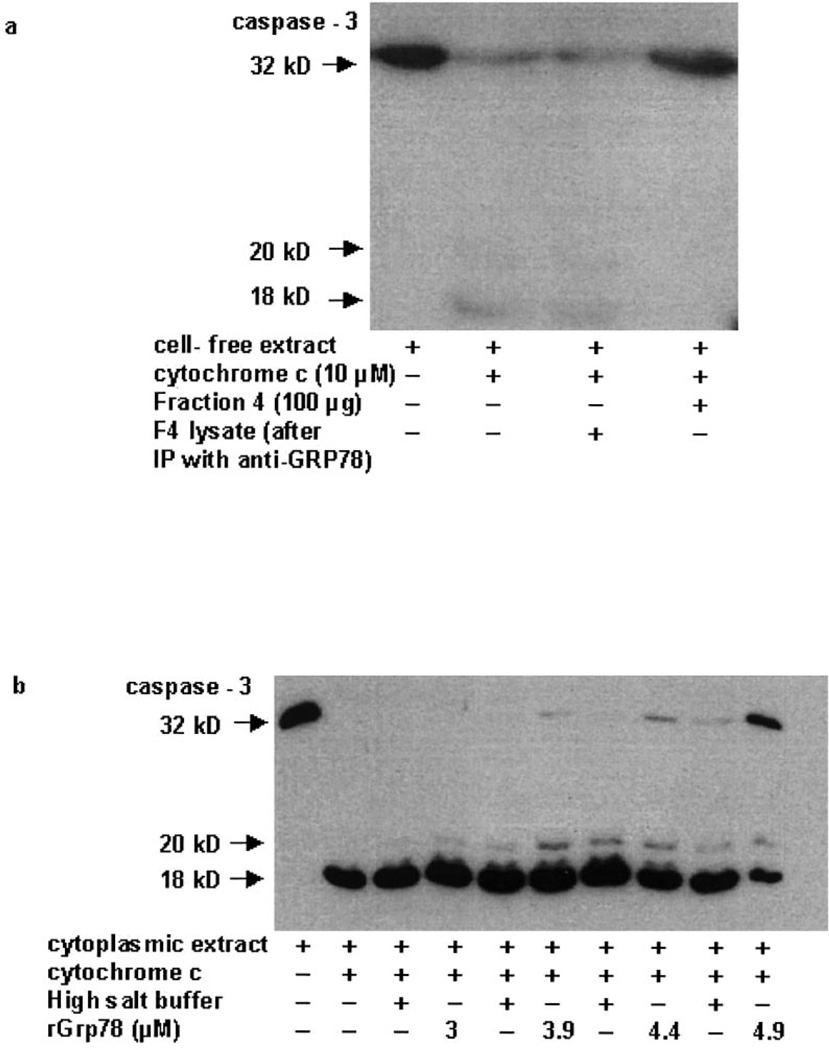
The cell extract from brefeldin-treated 293T cells was partially purified through a Sephadex G-50 column and the fraction that displayed peak GRP78 protein expression was immunoprecipitated with anti-GRP78 antibody. The supernatant obtained was added to a normal cell-free extract in the presence of cytochrome c. A successful immunodepletion of GRP78 from the extracts was achieved with the rabbit polyclonal anti-GRP78 antibody (data not shown). As shown in Fig. 2a (lane 3), the supernatant that had been immunodepleted of GRP78 protein failed to inhibit caspase-3 processing, suggesting that the GRP78 present in the partially purified fraction is required for the inhibition of caspase cleavage. To confirm that GRP78 directly inhibits caspase processing, various concentrations of recombinant purified GRP78 were added to untreated cell-free extracts. While 3 µM GRP78 did not inhibit cytochrome c-mediated caspase-3 cleavage, addition of 4.9 µM GRP78 resulted in inhibition of caspase-3 cleavage as evidenced by an increase in the caspase-3 proform (Fig. 2b). We did not observe complete block of caspase-3 activation by rGRP78, since the high salt buffer in which the recombinant protein was reconstituted (37 mM Tris–HCl pH 7.5/37 mM NaCl) itself showed inhibitory properties at higher concentrations.
Fig. 2.
GRP78 inhibits caspase processing. Cell-free extract from 2.5 µM brefeldin-treated 293T cells was passed through a Sephadex G-50 column and fractions were collected. a: Fraction 4 (100 µg) which showed a high GRP78 expression was immunoprecipitated with anti-GRP78 antibody. The supernatant obtained was added to a normal cell-free extract in the presence of cytochrome c. Samples were analyzed by Western blot analysis for the pro- (32 kDa) and active forms of caspase-3 (20 and 18 kDa). b: Cell-free extracts from 293T cells were incubated with 10 µM cytochrome c in the presence of recombinant GRP78 (3–4.9 µM), or similar amounts of the buffer in which GRP78 was reconstituted. Samples were analyzed by Western blot analysis for the pro- (32 kDa) and active forms of caspase-3 (20 and 18 kDa). Figures are representative of three independent experiments.
3.2. Induction and localization of GRP78 protein in ER stress-induced cells
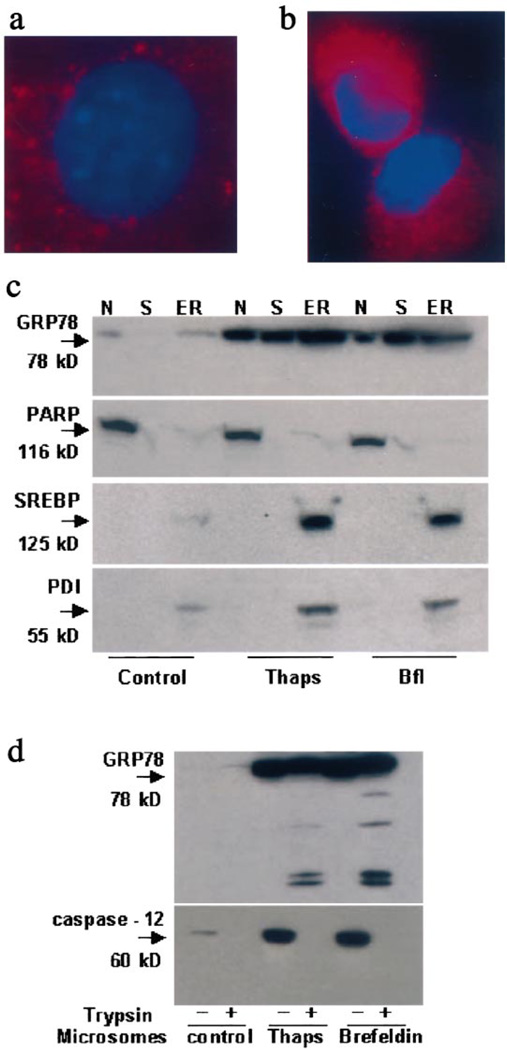
GRP78 often serves as a marker protein for ER, and accordingly immunostaining demonstrated its faint perinuclear expression in untreated cells (Fig. 3a). After treatment with 2.5 µM thapsigargin for 12 h, GRP78 staining became more intense, appeared granular predominantly in the ER (Fig. 3b) and also displayed a diffuse distribution throughout the cytoplasm. Virtually no co-localization of GRP78 and GM130 (a Golgi marker protein) occurred under control or treated conditions (data not shown). To determine the subcellular localization of GRP78 before and after ER stress induction, cell lysates from control, thapsigargin- (24 h) and brefeldin-A-treated (24 h) 293T cells were fractionated into nuclear, microsomal and soluble fractions and analyzed by Western blotting. The quality of the fractionation experiments was controlled by assessing the distribution of standard marker proteins. As shown in Fig. 3c, GRP78 protein expression was low and predominantly localized in the microsomal fraction in control extracts. Some expression was also seen in the nuclear fraction. Following thapsigargin and brefeldin treatment, an induction in GRP78 protein expression was detected in all three fractions analyzed. This suggests that ER stress induction causes a redistribution of GRP78 into the soluble fraction (and possibly into the nuclear fraction, as well).
Fig. 3.
Expression and localization of GRP78 before and after ER stress. Double Immunofluorescent staining for GRP78 before (a) and after ER stress (b) with Cy3 in 293T cells left in control medium for 12 h before fixation in buffered 4% paraformaldehyde. For the detection of GRP78, cells were stained with rabbit polyclonal anti-GRP78 antibody as primary antibody and Cy3-labeled donkey anti-rabbit IgG as the secondary antibody. Nuclei are counterstained using VectaShield® Mounting Medium with DAPI (Vector Laboratories, Burlingame, CA, USA). c: Western blots of nuclear (N), cytoplasmic (S) and microsomal (ER) fractions of 293T human renal epithelial cells before and after treatment with 2.5 µM brefeldin-A or 2.5 µM thapsigargin for 24 h. Cellular fractions were also probed for standard marker proteins namely PARP (nuclei), PDI and SREBP-1 (ER). d: Tryptic cleavage of GRP78 in isolated microsomes. Microsomes isolated from control, thapsigargin- (thaps) and brefeldin-A-treated cell extracts were digested by 0.05% trypsin-EDTA for 30 min at 35°C. At the end of the reaction, samples were detected by Western blotting.
GRP78 within the ER lumen would not be expected to be susceptible to trypsin digestion (in the absence of detergent). In order to elucidate further the localization and topology of GRP78 in the ER compartment after ER stress, we subjected the microsomal fraction from control, brefeldin-A- and thapsigargin-treated 293T cells to limited trypsin digestion. As shown in Fig. 3d, addition of trypsin resulted in partial digestion of GRP78, with several tryptic fragments arising from the digestion. Trypsin treatment was however sufficient to digest caspase-12 completely (Fig. 3d). These data indicate that while GRP78 normally resides primarily within the lumen of the ER, ER stress can cause a subpopulation of GRP78 to exist as a partially protease resistant (presumably transmembrane) protein, and redistribute to the cytoplasmic fraction, as well. Our results do not exclude the possibility that GRP78 appearance in the cytosol and at the ER–cytosolic interface may also reflect upregulation without true redistribution. In either case, the cytoplasmic pool of GRP78 or a subpopulation of GRP78 existing as an ER transmembrane protein could conceivably prevent caspase activation and release.
3.3. Expression of GRP78 blocks caspase cleavage and cell death
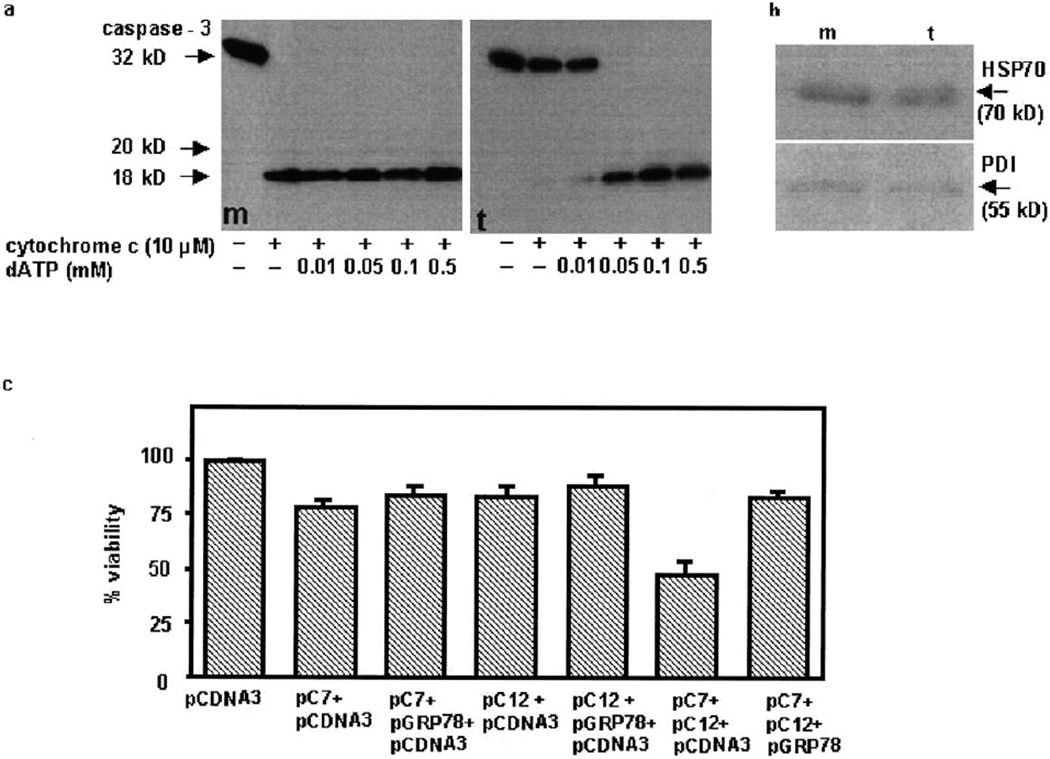
In order to demonstrate further the role of GRP78 in inhibiting caspase processing, 293T cells were transfected with a hamster grp78 expression construct. There was a time-dependent increase in the expression of GRP78 following transfection (data not shown). Since the difference in GRP78 expression between control vector and GRP78 cDNA was greatest at 18–24 h after transfection, further studies were carried out at this time point. Cells were harvested 24 h after pGRP78 transfection and cytoplasmic extracts were prepared. Cleavage of caspase-3, detected by the characteristic procaspase processing from zymogen to active protease (Fig. 4a), occurred in mock-transfected cell extracts (m) incubated with cytochrome c, either alone or together with dATP. In contrast, cleavage of caspase-3 in cell-free extracts from GRP78-transfected cells (t) did not occur with cytochrome c, instead requiring both cytochrome c and dATP. While ATP could also reverse the inhibitory effects of GRP78 (data not shown), dATP was slightly more effective which may be due to the fact that dATP is superior to ATP in activating caspase-9 (in concert with cytochrome c and Apaf-1). It is noteworthy that a recent study demonstrated that GRP94 binds ATP and dATP with nearly equal affinities [24]. In order to rule out the possibility that GRP78 transfection induced the expression of other ER stress proteins that may have anti-apoptotic actions, we determined the expression of PDI (an ER lumen protein) and HSP70 (a cytoplasmic chaperone protein). As shown in Fig. 4b, neither PDI nor HSP70 appeared appreciably different in the GRP78-transfected extracts in comparison to mock-transfected extracts, suggesting that no significant changes in the expression of other stress proteins that contribute to the anti-apoptotic activity had taken place.
Fig. 4.
Expression of GRP78 inhibits caspase activation and cell death. a: Cytochrome c (10 µM) and various concentrations of dATP were added to cell-free cytosolic extracts (150 µg protein) made from 293T cells transiently transfected with GRP78 cDNA (t) or the control vector (m). Extracts were incubated at 37°C for 1 h and then analyzed by Western blot analysis for the pro- and active forms of caspase-3 (p32 to p20 and p18). b: Western blots of mock (m) and GRP78-transfected (t) cell extracts probed with antibodies specific for PDI and HSP70. c: 293T cells were transfected with a total of 3 µg of DNA consisting of either empty vector (pcDNA3) alone, 0.5 µg Flag-tagged pC7, 1 µg pC12, 0.5 µg Flag-tagged pC7+1 µg pC12, or 0.5 µg Flag-tagged pC7+1 µg pC12+1.5 µg pGRP78. Surviving vs. apoptotic cells were quantified after 24 h, as described in Section 2. Data (means ± S.E.M.) are from three independent experiments.
We demonstrated previously that ER stress not only induces the expression of caspase-12 but also causes translocation of cytoplasmic caspase-7 to the ER surface, and prolonged ER stress facilitates movement of active caspase-12 into the cytoplasm [16] and induces cell death. Since GRP78 blocks caspase activation, we reasoned that expression of GRP78 should inhibit caspase-induced cell death. As shown in Fig. 4c, co-transfection of caspase-7 and caspase-12 expression constructs resulted in increased cell death that was blocked by GRP78. The inhibition of caspase-7 and -12-mediated cell death by GRP78 was observed only for about 24 h after transfection and was decreased at longer periods following transfection. One possible explanation may be the susceptibility of GRP78 to caspase digestion (Rao et al., unpublished observations).
3.4. Association of GRP78, caspase-7 and caspase-12
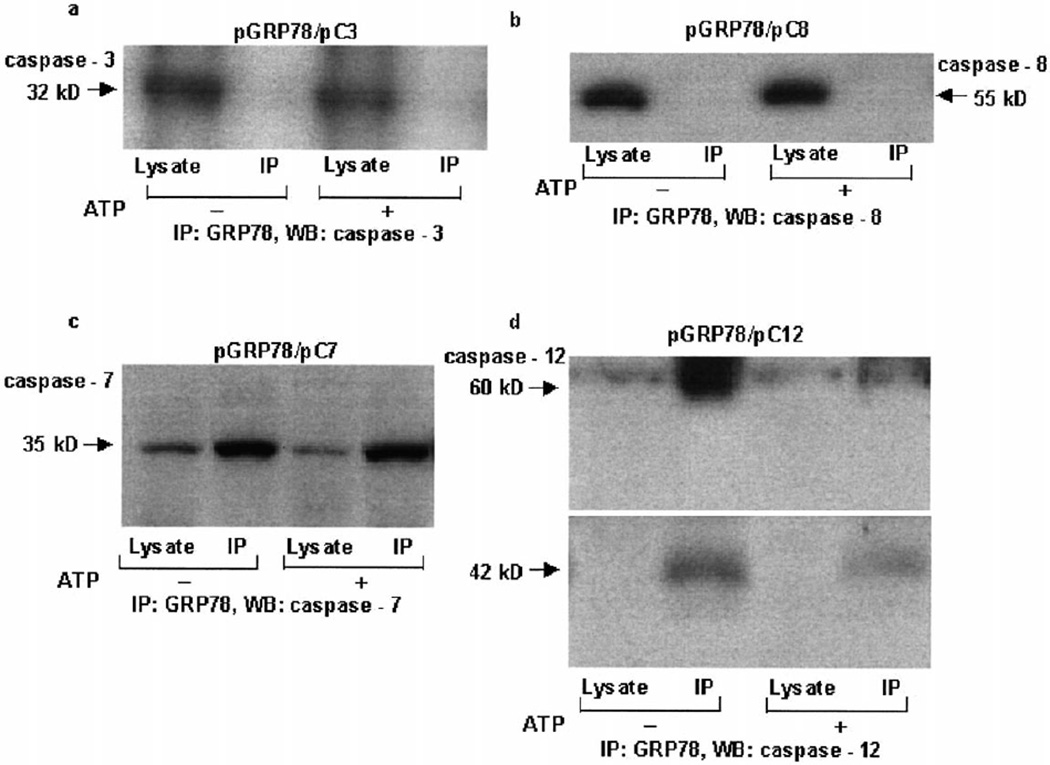
Since GRP78 blocks caspase activation, co-IP experiments were carried out to determine whether GRP78 interacts with components of the apoptosome or with other caspases. IP of GRP78 protein from lysates obtained from transient co-transfections of a hamster GRP78 expression construct with other apoptotic expression constructs did not reveal any association with caspase-3, caspase-8, Apaf-1, or cytochrome c (Fig. 5a,b and data not shown). However, caspase-7 was found to associate with GRP78 (Fig. 5c). Similar association of GRP78 and caspase-7 was also seen in cell extracts from thapsigargin- and brefeldin-treated cells (data not shown). In order to determine whether GRP78 interacted with caspase-12, cell lysates from 293T cells treated with 2.5 µM thapsigargin were immunoprecipitated (IP) with GRP78 polyclonal antibody, and immunoblotted (WB, Western blotting) with anti-caspase-12 monoclonal antibody. As shown in Fig. 5d, GRP78 co-immunoprecipitated with caspase-12. In addition to the caspase-12 zymogen band, activated caspase-12 (42 kDa) also co-immunoprecipitated with GRP78 (Fig. 5d). This raises the possibility that caspase-12 may undergo activation in the multimeric complex [16] by autocatalysis or by the catalytic activity of caspase-7. Addition of ATP or dATP to cell lysates prior to IP caused a reduction in binding of pro- and active forms of caspase-12 to GRP78 (Fig. 5d and data not shown), without affecting the interaction of caspase-7 and GRP78 (Fig. 5c). Thus during ER stress, GRP78 forms a complex with caspase-7 zymogen, caspase-12 zymogen, and caspase-12 and controls activation of the caspase cascade, possibly by preventing their release from this putative inhibitory complex.
Fig. 5.
Interaction of GRP78, caspase-7 and caspase-12. Expression constructs of GRP78 together with caspase-3 (a), caspase-8 (b) or caspase-7 (c) were transiently transfected into 293T cells. After 30 h, cells were lysed and the lysate was immunoprecipitated (IP) with anti-GRP78 rabbit antibody and immunoblotted (WB, Western blotting) with anti-caspase-3 (a), anti-caspase-8 (b) or anti-caspase-7 antibody (c). One set of samples was treated with 1 mM ATP prior to IP. d: Cell lysate from 2.5 µM thapsigargin-treated 293T cells was immunoprecipitated (IP) with anti-GRP78 rabbit antibody and immunoblotted (WB, Western blotting) with rat anti-caspase-12 monoclonal antibody. One set of samples was treated with 1 mM ATP prior to IP. Both caspase-12 and the cross-reacting IgG heavy chain were separated on an 8% Tris–glycine gel. Different exposures were used for the 60 kDa and processed (42 kDa) caspase-12. Figures are representative of two independent experiments.
4. Discussion
While GRP78 exists as an ER lumen protein [3], several reports have suggested that GRP78 and its mutant forms may also be associated with cytosol, membrane and nuclear fractions, as well as the cell surface [25–28]. Calreticulin, an ER lumenal protein, also redistributes from the ER and appears in the cytosol during heat stress [29]. Similarly, a subpopulation of GRP94 associates with the ER membrane during an apoptotic insult, where it becomes a target of calpain activity [21]. Our studies suggest that ER stress leads to the redistribution of GRP78 to the soluble fraction, with a subpopulation of GRP78 becoming partially protease resistant. This effect suggests that, under such conditions, GRP78 may also exist as an ER transmembrane protein. Our data are also compatible with the alternative explanation that GRP78 appearance in cytosol and at the ER–cytosolic interface may reflect upregulation without true redistribution. In either case, however, this would result in the observed complex formation between GRP78, caspase-7, and caspase-12. Either the cytoplasmic pool of GRP78 or a subpopulation of GRP78 existing as an ER transmembrane protein could form a complex with caspases-7 and -12 at the ER surface and prevent their activation and release. Cells are believed to tolerate ER stress insults up to 24 h due to the long half-life of ER stress proteins [30], and in our experiments, control of ER stress-induced cell death by GRP78 could be seen for up to 24 h following treatment. With longer exposure to ER stress agents, apoptosis and associated caspase activity were observed. Prolonged ER stress, together with (d)ATP binding to the complex, may either disrupt the multimeric complex or render it more evanescent, ensuring the liberation of active caspase-12 into the cytosol. Proteolytic cleavage of the cytoprotective GRP78 by caspase-7 or caspase-12 during prolonged ER stress may also occur and could explain the breakdown of this complex and the liberation of active caspase-12. Thus the GRP78–caspase-7–caspase-12 complex represents an inhibitory analogue of the ‘apoptosome’ [31,32]. The apoptosome includes caspase-9 zymogen, Apaf-1, cytochrome c, and (d)ATP, and facilitates activation of caspase-9. In contrast, GRP78 blocks caspases; however, (d)ATP is still pro-apoptotic, since it leads to dissociation of the GRP78/caspase-12 inhibitory complex.
Other ER stress proteins like PDI, ORP-150 and heat shock proteins including HSP27 and HSP70 are also known to have anti-apoptotic activity, but their mechanisms are likely to be different than that of GRP78 [33–37]. The current studies highlight the importance of GRP78 as an anti-apoptotic protein, and shed new light on the mechanisms underlying the relationship between ER stress, the UPR and the cell death program. The results could have broad implications for our understanding of the mechanism by which cells couple ER stress to the cell death program, and the mechanism by which the associated decision between apoptosis inhibition and activation is controlled.
Acknowledgements
We thank Dr. Amy Lee (Department of Biochemistry and Molecular Biology and the USC/Norris Comprehensive Cancer Center, University of Southern California School of Medicine, Los Angeles, CA, USA) for the hamster GRP78 cDNA, Dr. Junying Yuan (Department of Cell Biology, Harvard Medical School, Boston, MA, USA) for the anti-caspase-12 antibody, members of the Bredesen laboratory for helpful comments and discussions, P. Rao for technical help and Molly Susag for administrative assistance. This work was supported by the National Institutes of Health (AG12282, NS33376 and NS35155 to D.E.B., and R01 CA84262 to H.M.E.), Department of Defense (DAMD17-98-8613 to D.E.B.). R.V.R. was supported initially by a National Institutes of Health training Grant (T32AG00252).
Abbreviations
- ER
endoplasmic reticulum
- GRP
glucose-regulated protein
- 293T
human embryonic kidney cells
- PDI
protein disulfide isomerase
- PARP
poly(ADP-ribose) polymerase
References
- 1.Paschen W. Cell Calcium. 2001;29:1–11. doi: 10.1054/ceca.2000.0162. [DOI] [PubMed] [Google Scholar]
- 2.Mattson MP, LaFerla FM, Chan SL, Leissring MA, Shepel PN, Geiger JD. Trends Neurosci. 2000;23:222–229. doi: 10.1016/s0166-2236(00)01548-4. [DOI] [PubMed] [Google Scholar]
- 3.Kaufman RJ. Genes Dev. 1999;13:1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- 4.Sherman MY, Goldberg AL. Neuron. 2001;29:15–32. doi: 10.1016/s0896-6273(01)00177-5. [DOI] [PubMed] [Google Scholar]
- 5.Guo H, Tittle TV, Allen H, Maziarz RT. Exp. Cell Res. 1998;245:57–68. doi: 10.1006/excr.1998.4235. [DOI] [PubMed] [Google Scholar]
- 6.Liu H, Miller E, van de Water B, Stevens JL. J. Biol. Chem. 1998;273:12858–12862. doi: 10.1074/jbc.273.21.12858. [DOI] [PubMed] [Google Scholar]
- 7.Lee J, Bruce-Keller AJ, Kruman Y, Chan SL, Mattson MP. J. Neurosci. Res. 1999;57:48–61. doi: 10.1002/(SICI)1097-4547(19990701)57:1<48::AID-JNR6>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 8.Yu Z, Luo H, Fu W, Mattson MP. Exp. Neurol. 1999;155:302–314. doi: 10.1006/exnr.1998.7002. [DOI] [PubMed] [Google Scholar]
- 9.Alnemri ES, Livingston DJ, Nicholson DW, Salvesen G, Thornberry NA, Wong WW, Yuan J. Cell. 1996;87:171. doi: 10.1016/s0092-8674(00)81334-3. [DOI] [PubMed] [Google Scholar]
- 10.Cryns V, Yuan J. Genes Dev. 1998;12:1551–1570. doi: 10.1101/gad.12.11.1551. [DOI] [PubMed] [Google Scholar]
- 11.Zhivotovsky B, Samali A, Gahm A, Orrenius S. Cell Death Differ. 1999;6:644–651. doi: 10.1038/sj.cdd.4400536. [DOI] [PubMed] [Google Scholar]
- 12.Pai JT, Brown MS, Goldstein JL. Proc. Natl. Acad. Sci. USA. 1996;93:5437–5442. doi: 10.1073/pnas.93.11.5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chandler JM, Cohen GM, MacFarlane M. J. Biol. Chem. 1998;273:10815–10818. doi: 10.1074/jbc.273.18.10815. [DOI] [PubMed] [Google Scholar]
- 14.Nakagawa T, Yuan J. J. Cell. Biol. 2000;150:887–894. doi: 10.1083/jcb.150.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakagawa T, Zhu H, Morishima N, Li E, Xu J, Yankner BA, Yuan J. Nature. 2000;403:98–103. doi: 10.1038/47513. [DOI] [PubMed] [Google Scholar]
- 16.Rao RV, Hermel E, Castro-Obregon S, del Rio G, Ellerby LM, Ellerby HM, Bredesen DE. J. Biol. Chem. 2001;276:33869–33874. doi: 10.1074/jbc.M102225200. [DOI] [PubMed] [Google Scholar]
- 17.Korimilli A, Gonzales LW, Guttentag SH. J. Biol. Chem. 2000;275:8672–8679. doi: 10.1074/jbc.275.12.8672. [DOI] [PubMed] [Google Scholar]
- 18.Ellerby HM, et al. J. Neurosci. 1997;17:6165–6178. [PMC free article] [PubMed] [Google Scholar]
- 19.Ye X, et al. J. Biol. Chem. 1999;274:30202–30208. doi: 10.1074/jbc.274.42.30202. [DOI] [PubMed] [Google Scholar]
- 20.Sperandio S, de Belle I, Bredesen DE. Proc. Natl. Acad. Sci. USA. 2000;97:14376–14381. doi: 10.1073/pnas.97.26.14376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reddy RK, Lu J, Lee AS. J. Biol. Chem. 1999;274:28476–28483. doi: 10.1074/jbc.274.40.28476. [DOI] [PubMed] [Google Scholar]
- 22.Ellerby HM, et al. Nat. Med. 1999;5:1032–1038. doi: 10.1038/12469. [DOI] [PubMed] [Google Scholar]
- 23.Slee EA, et al. J. Cell. Biol. 1999;144:281–292. doi: 10.1083/jcb.144.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosser MF, Nicchitta CV. J. Biol. Chem. 2000;275:22798–22805. doi: 10.1074/jbc.M001477200. [DOI] [PubMed] [Google Scholar]
- 25.Hendershot LM, Wei JY, Gaut JR, Lawson B, Freiden PJ, Murti KG. Mol. Biol. Cell. 1995;6:283–296. doi: 10.1091/mbc.6.3.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morris JA, Dorner AJ, Edwards CA, Hendershot LM, Kaufman RJ. J. Biol. Chem. 1997;272:4327–4334. doi: 10.1074/jbc.272.7.4327. [DOI] [PubMed] [Google Scholar]
- 27.Xiao G, Chung TF, Pyun HY, Fine RE, Johnson RJ. Brain Res. Mol. Brain Res. 1999;72:121–128. doi: 10.1016/s0169-328x(99)00188-6. [DOI] [PubMed] [Google Scholar]
- 28.Triantafilou M, Fradelizi D, Triantafilou K. Hum. Immunol. 2001;62:764–770. doi: 10.1016/s0198-8859(01)00269-5. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki T, Yan Q, Lennarz WJ. J. Biol. Chem. 1998;273:10083–10086. doi: 10.1074/jbc.273.17.10083. [DOI] [PubMed] [Google Scholar]
- 30.Liu H, Bowes RC, 3rd, vandeWater B, Sillence C, Nagelkerke JF, Stevens JL. J. Biol. Chem. 1997;272:21751–21759. doi: 10.1074/jbc.272.35.21751. [DOI] [PubMed] [Google Scholar]
- 31.Li P, Nijhawan D, Budihardjo I, Srinivasula SM, Ahmad M, Alnemri ES, Wang X. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 32.Zou H, Li Y, Liu X, Wang X. J. Biol. Chem. 1999;274:11549–11556. doi: 10.1074/jbc.274.17.11549. [DOI] [PubMed] [Google Scholar]
- 33.Tanaka S, Uehara T, Nomura Y. J. Biol. Chem. 2000;275:10388–10393. doi: 10.1074/jbc.275.14.10388. [DOI] [PubMed] [Google Scholar]
- 34.Ozawa K, et al. J. Biol. Chem. 1999;274:6397–6404. doi: 10.1074/jbc.274.10.6397. [DOI] [PubMed] [Google Scholar]
- 35.Beere HM, et al. Nat. Cell Biol. 2000;2:469–475. doi: 10.1038/35019501. [DOI] [PubMed] [Google Scholar]
- 36.Saleh A, Srinivasula SM, Balkir L, Robbins PD, Alnemri ES. Nat. Cell Biol. 2000;2:476–483. doi: 10.1038/35019510. [DOI] [PubMed] [Google Scholar]
- 37.Bruey JM, et al. Nat. Cell Biol. 2000;2:645–652. doi: 10.1038/35023595. [DOI] [PubMed] [Google Scholar]