Abstract
Background
Cervical cancer is the second leading cause of cancer deaths among women worldwide. We sought to describe the most common oncogenic mutations in cervical cancers, and to explore genomic differences between the two most common histological subtypes: adenocarcinoma and squamous cell carcinoma.
Methods
A high-throughput genotyping platform, termed Oncomap, was used to interrogate 80 cervical tumors for 1250 known mutations in 139 cancer genes. Samples were analyzed using a mass spectrometry-based genotyping platform (Sequenom), and validated with an orthogonal chemistry. EGFR mutations were further validated by massively parallel sequencing (Illumina). Human papilloma virus (HPV) genotyping was also performed.
Results
Validated mutations were detected in 60.0% (48/80) of tumors examined. The highest mutation rates were PIK3CA (31.3%), KRAS (8.8%), and EGFR (3.8%). PIK3CA mutation rates were not significantly different in adenocarcinoma and squamous cell carcinomas (25.0% vs. 37.5%, respectively, p=0.33). In contrast, KRAS mutations were identified only in adenocarcinoma (17.5% vs. 0%, p=0.01), and a novel EGFR mutation was detected only in squamous cell carcinomas (0% vs. 7.5%, p=0.24). There were no associations between HPV-16 or HPV-18 and somatic mutations or overall survival. In adjusted analyses, PIK3CA mutations were associated with shorter survival—67.1 vs. 90.3 months (HR=9.1, 95% CI 2.8–29.5, p<0.001).
Conclusions
Cervical cancers harbor high rates of potentially targetable oncogenic mutations. In addition, cervical squamous cell carcinoma and adenocarcinoma have distinct molecular profiles, suggesting that clinical outcomes may be improved with the use of more tailored treatment strategies, including PI3-kinase and MEK inhibitors.
Keywords: cervical cancer, adenocarcinoma, squamous cell carcinoma, somatic mutations, PI3K, PIK3CA, EGFR, KRAS, DNA mutational analysis, human papilloma virus (HPV), neoplasm, mutation
Introduction
Cervical cancer is the second leading cause of cancer deaths among women worldwide resulting in 275,000 deaths annually.1 While screening programs have decreased the incidence of squamous cell cervical cancer, the incidence of adenocarcinoma of the cervix has risen from 5 to 24%.2, 3 Several studies have shown that adenocarcinoma confers a worse prognosis with higher rates of nodal involvement, distant metastases, and decreased survival across stages, compared with squamous cell carcinoma.3,4–8 Despite this, few studies have examined whether squamous cell carcinoma and adenocarcinoma have distinct molecular profiles that might explain the observed clinical differences.
Previous studies in cervical cancer have focused on identifying isolated somatic mutations or evidence of gene amplification, and exploring their clinical relevance. To date, activating mutations in the EGFR gene (i.e. exons 19–21) have not been identified,9, 10 but one study found evidence of EGFR amplification in 10.2% of squamous cell carcinomas which was associated with shorter overall survival.9, 10 Activating mutations and amplification of PIK3CA, the gene that encodes the catalytic subunit of phosphatidylinositol 3-kinase (PI3K), have also been reported in 23–36% of cervical cancer specimens.11–13 In observational studies, PI3K pathway activation has been associated with higher rates of local recurrence after radiotherapy and decreased survival.13, 14 Finally, KRAS mutations have been identified in two independent studies, with rates varying between 6–14%,15, 16 and have been associated with worse outcomes after radiation.17
In this study, we performed a systematic molecular analysis of cervical cancers to determine the rates and spectrum of somatic mutations present. Specifically, high-throughput parallel mutation detection was performed on 80 cervical cancer tumors (40 adenocarcinoma and 40 squamous cell carcinoma) to identify the rates of “targetable” oncogene and tumor suppressor gene mutations in cervical cancer. Given the rising incidence of adenocarcinomas, we also examined the mutational differences between adenocarcinoma and squamous cell carcinoma of the cervix, as well as the HPV status associated with these tumors.
Materials and Methods
Tumor and Patient Data Collection
Pathology records from an existing pathology database were reviewed between 2005 and 2011 in the Division of Women’s and Perinatal Pathology at Brigham and Women’s Hospital, Boston, MA to identify cases of cervical adenocarcinoma or squamous cell carcinoma. Clinical data were extracted from electronic medical records. The Dana-Farber/Harvard Cancer Center (DF/HCC) Institutional Review Board (IRB) granted approval to analyze the formalin fixed paraffin embedded (FFPE) samples and collect clinical data. Because all of the samples were de-identified, the IRB granted a waiver to analyze the samples without patient consent.
DNA extraction and quantification
Cases of cervical cancer were obtained by a trained gynecologic pathologist (BEH, MSH, ARL, CMQ), who reviewed pathology reports and hematoxylin-and-eosin (H&E) stained slides to confirm the diagnosis. Corresponding FFPE tissue blocks were retrieved and reviewed by a trained gynecologic pathologist (BEH or MSH) to confirm sufficient tumor was present. For each case, areas with the highest percentage of tumor (and when available, normal adjacent tissue) were selected. All blocks were cored for DNA extraction. A total of 80 samples were sufficient for coring and DNA extraction.
Genomic DNA was extracted from the cored FFPE patient tissue samples with QIAamp DNA FFPE Tissue Kit (Qiagen) according to the manufacturer’s protocol. Briefly, cores were deparaffinized in xylene and further lysed in denaturing buffer containing proteinase K. The tissue lysate was incubated at 90°C to reverse formalin crosslinking. Using QiaCube, the lysate was applied to the DNA binding column and the column was washed serially, and then eluted in 30 ul of distilled water. Genomic DNA was quantified using Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen) per manufacturer’s protocol. 250 ng of genomic DNA was used for the analysis.
HPV Genotyping
HPV genotyping was performed using the F-HPV typing™ Multiplex Fluorescent-PCR Kit for Human Papilloma Virus (HPV) Genotyping (Genomed AG, Switzerland), as per the manufacturer’s instructions. This assay uses 15 primers that amplify in the E6 and E7 regions of the HPV genome and can specifically recognize HPV types 6, 11, 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68. A total of 200 ng of gDNA was used as input for the PCR reaction. Automated electrophoresis and detection was performed on an ABI 3730XL using GeneMapper version 4.0 software (Applied Biosystems, Foster City, CA) at the Dana-Farber Cancer Institute’s (DFCI) Molecular Biology Core Facilities. The HPV F-PCR products were detected on an electrophoretogram, and each HPV type was identified based on the size and color of the corresponding amplicon. Sixty-five samples had sufficient DNA to perform HPV genotyping.
Mutation Detection
Oncomap version 4, which interrogates 1250 known mutations in 139 validated oncogenes and tumor suppressors, was performed on all samples. All studies were performed in the Center for Cancer Genome Discovery (CCGD) at the DFCI. Whole genome amplification (WGA) was performed using the GenomePlex Complete WGA kit (Sigma) based on chemical fragmentation followed by adapter mediated PCR amplification. For each sample, a PCR reaction to assess DNA quality was performed.
Samples were run on the mass spectrometry-based genotyping platform (Sequenom) and analyzed according to current standardized protocols.18 Sample identity and the possible introduction of artifacts by WGA were evaluated using a 48 Single Nucelotide Polymorphism (SNP)s panel comparing the pre-WGA to the post-WGA DNA. If ≥3 SNP discrepancies were identified between SNPs found in pre- and post-WGA samples, this sample was discarded. Mutations were validated by a different, more sensitive chemistry using a multi-base extension [homogeneous Mass EXTEND® (hME), Sequenom], on native (unamplified) genomic DNA
Since EGFR mutations have not been previously reported in cervical cancer,9, 10 we sequenced two samples with EGFR mutations that were validated by Sequenom (i.e. both iPLEX and hME mass spectrometry chemistries) and four samples harboring EGFR mutations identified exclusively by hME chemistry. Sequencing was performed on an Illumina Hiseq 2000. One sample with a Sequenom validated EGFR mutation had insufficient remaining DNA for sequencing. Briefly, 200 ng of DNA was fragmented to 150 bp and ligated to specific adaptors with sample specific barcode sequences during library preparation (Illumina TruSeq). Libraries were pooled, enriched for exonic sequences and sequenced for 100 bp in paired-end mode. Mutation analysis for single nucleotide variants (SNV) was performed using MuTect.19
Immunohistochemistry
Immunohistochemistry for PTEN was evaluated in FFPE 5 um sections using a validated immunohistochemistry assay for the 51 samples where unstained slides were available. This was performed since inactivation of the tumor suppressor PTEN can result in abnormal PI3K pathway activation, and the hot-spot mutation analysis had limited coverage for PTEN mutations. Antigen retrieval was performed using EDTA at pH 9.00 and a 1:50 dilution of the DAKO PTEN antibody clone 6H2.1 (catalog # M3627). Complete absence of staining in the tumor cells in the presence of internal positive control (stromal cells, lymphocytes) was interpreted as PTEN protein loss. Cases in which all or a subset of the tumor showed immunoreactivity were scored as PTEN protein positive. Negative controls (i.e., incubation in the absence of primary antibody) were also performed.
Statistical methods
Fisher’s exact test was used to test for associations between validated mutations and categorical variables (e.g., stage, grade, lymph node involvement, HPV genotype), and the Wilcoxon rank sum test was used for continuous variables (e.g., age, tumor size). Time to event distributions were estimated using the Kaplan-Meier method, and Cox proportional hazard models were fitted to estimate progression-free and overall distributions; the log-rank test was used to assess differences in these time-to-event distributions. A two-sided p<0.05 was considered statistically significant, without adjustment for multiple comparisons.
Results
Patient and Disease Characteristics
Patients’ sociodemographic and clinical characteristics are shown in Table 1. The cohort consisted of 80 patients with cervical adenocarcinoma (n=40) and squamous cell carcinoma (n=40). Overall, patient age and race at diagnosis were consistent with population-based data from cancers registries in Surveillance Epidemiology and End Results (SEER) areas,20 except that our cohort had a higher proportion of early stage disease compared with data collected in SEER areas (e.g., stage I: 57.6% vs. 47.0%; stage II/III: 33.8% vs. 36%; and stage IV: 8.8% vs. 12.0%).
Table 1.
Patient Demographics and Disease Characteristics
| Characteristic | Total (N=80) |
Squamous cell carcinoma (SCC) (N=40) |
Adenocarcinoma (AC) (N=40) |
|---|---|---|---|
| Age, median (Q1, Q3) | 46.0 (37.0, 54.3) | 51.5 (42.5, 59.3) | 39.5 (35.0, 48.0) |
| Race, N (%)1 | |||
| White | 63 (84.0) | 31 (83.8) | 32 (84.2) |
| Black | 8 (10.7) | 4 (10.8) | 4 (10.5) |
| Other | 4 (5.3) | 2 (5.4) | 2 (5.3) |
| Married, N (%) | 45 (56.3) | 17 (42.5) | 28 (70.0) |
| FIGO Stage, N (%)1 | |||
| IA2 | 11 (13.8) | 2 (5.0) | 9 (22.5) |
| IB1 | 32 (40.0) | 8 (20.0) | 24 (60.0) |
| IB2 | 3 (3.8) | 3 (7.5) | 0 (0.0) |
| IIA | 8 (10.0) | 7 (17.5) | 1 (2.5) |
| IIB | 14 (17.5) | 11 (27.5) | 3 (7.5) |
| IIIB | 5 (6.3) | 4 (10.0) | 1 (2.5) |
| IV | 7 (8.8) | 5 (12.5) | 2 (5.0) |
| Grade, N (%)1 | |||
| Well differentiated | 22 (29.3) | 3 (8.1) | 19 (50.0) |
| Moderately differentiated | 34 (45.3) | 23 (62.2) | 11 (28.9) |
| Poorly differentiated | 19 (25.3) | 11 (29.7) | 8 (21.1) |
| Lymph Node Involvement, N (%) | 27 (33.8) | 20 (50.0) | 7 (17.5) |
| Pelvis | 27 (33.8) | 20 (50.0) | 7 (17.5) |
| Para-aortic | 10 (12.5) | 9 (11.3) | 1 (2.5) |
| Distant | 2 (2.5) | 2 (5.0) | 0 (0.0) |
| Human Papilloma Virus, N (%) | |||
| None | 3 (4.6) | 1 (3.0) | 2 (6.3) |
| Single infection | |||
| 16 | 41 (63.1) | 20 (60.6) | 21 (65.6) |
| 18 | 3 (4.6) | 0 (0.0) | 3 (9.4) |
| 45 | 2 (3.1) | 1 (3.0) | 1 (3.1) |
| Other2 | 4 (6.2) | 4 (12.1) | 0 (0.0) |
| Multiple infections | |||
| 16 &18 | 2 (3.1) | 0 (0.0) | 2 (6.3) |
| 16 & 33 | 3 (4.6) | 2 (6.1) | 1 (3.1) |
| 18 & 33 | 1 (1.5) | 1 (3.0) | 0 (0.0) |
| Other mixed types3 | 6 (9.2) | 4 (12.1) | 2 (6.3) |
| Recurrent Disease, N (%) | 22 (27.5) | 15 (37.5) | 7 (17.5) |
| Deceased, N (%)4 | 23 (28.8) | 19 (47.5) | 4 (10.0) |
Missing data: race n=5 (3 SCC, 2 AC); grade n=5 (3 SCC, 2 AC); HPV genotyping n=15 (7 SCC, 8 AC) due to insufficient DNA. Due to rounding, the sum total of columns may exceed 100.0%
Other single infections include: HPV 33, 39, 58, 68, each present in 1 SCC case.
Other mixed types include: HPV 16&52, 16&58, 16&68, 33&39, each present in 1 SCC case; and 16&45 and 16&59, each present in 1 AC case.
One patient died of another cancer.
In our samples, HPV DNA was detected in 95.4% of tumors; 80.6% of which were single infections. HPV-16 was detected in 70.8% of tumors, consistent with prior population-based studies which have detected HPV-16 in 53.2% –68.2% of cervical cancers.21, 22 HPV-18 was present in 7.7% of samples, and more frequently detected in adenocarcinomas compared with squamous cell carcinomas (15.6% vs. 0.0%, respectively; p=0.02).
The median follow up time was 39 months, during which time 27.5% (22/80) of patients died due to cervical cancer; one patient died from a second cancer. Significant differences between patients diagnosed with adenocarcinoma and squamous cell carcinoma included the fact that patients with adenocarcinoma were younger and more likely to be married (39.5 vs. 51.5 years, p=0.002 and 70.0% vs. 42.5%, p=0.02, respectively), more likely to have lower grade and earlier stage disease at diagnosis (50.0% vs. 8.1% well-differentiated and 82.5% vs. 25.0% Stage IBI or lower disease, respectively; p=0.001), and less likely to have nodal involvement (17.5% vs. 50.0%, respectively; p=0.004).
Spectrum and Frequency of Mutations in Cervical Cancer
Here we report mutations which were validated by two orthogonal chemistries: Sequenom iPLEX and hME chemistries (unless otherwise stated). Validated mutations were detected in 48 of the 80 (60.0%) tumors examined, and 7 (8.8%) harbored concurrent mutations in 2 or more genes. As shown in Table 2, the genes with the highest mutation rates were PIK3CA (31.3%), KRAS (8.8%), and EGFR (3.8%). The PIK3CA mutations were activating mutations predominately located in the exon 9 helical domain hot spot (E545K and E542K), and 3.8% of tumors contained co-mutations within the helical domain (data not shown). Notably, no mutations were identified in the exon 20 catalytic domain (H1047R) of PIK3CA. KRAS mutations were classic G12 and G13 missense mutations within the guanine exchange factor (GEF) domain.
Table 2.
Validated Mutations and PTEN Loss Detected by Histological Subtype
| Gene | Total (N=80) | Squamous cell carcinoma (SCC) (N=40) |
Adenocarcinoma (AC) (N=40) |
P-value |
|---|---|---|---|---|
| Any PIK3CA mutation1 | 25 (31.3) | 15 (37.5) | 10 (25.0) | 0.33 |
| E542K | 10 (12.5) | 6 (15.0) | 4 (10.0) | |
| E545K | 16 (20.0) | 10 (25.0) | 6 (15.0) | |
| E453K | 1 (1.3) | 0 (0.0) | 1 (2.5) | |
| 3R88Q | 1 (1.3) | 0 (0.0) | 1 (2.5) | |
| Any KRAS mutation | 7 (8.8) | 0 (0.0) | 7 (17.5) | 0.01 |
| G12A | 1 (1.3) | 0 (0.0) | 1 (2.5) | |
| G12D | 3 (3.8) | 0 (0.0) | 3 (7.5) | |
| G12V | 2 (2.5) | 0 (0.0) | 2 (5.0) | |
| G13D | 1 (1.3) | 0 (0.0) | 1 (2.5) | |
| EGFR mutation2 | 3 (3.8) | 3 (7.5) | 0 (0.0) | 0.24 |
| Any PTEN loss3 | 4 (7.8) | 3 (13.0) | 1 (3.6) | 0.32 |
3/80 (3.8%) of samples had combined PIK3CA mutations (e.g., E542K and E545K).
Two additional squamous cell carcinomas had EGFR S703F detected with hME chemistry or Illumina sequencing.
51 samples were available for immunohistochemistry to detect PTEN loss (28 AC and 23 SCC).
Five percent of tumors harbored mutations in STK11 (which encodes the LKB protein)23 within the kinase domain, although they were only present in hME and therefore require further validation. STK11 mutations were present in both adenocarcinomas and squamous cell carcinomas (5.0% vs. 5.0%, p=1.0; data not shown). PTEN loss by immunohistochemistry was identified in 5 samples across histological subtypes (), while only 1 sample included a nonsense mutation in the phosphatase domain of PTEN (R130*). There were no BRAF mutations identified in any of the samples.
Differences in Mutations between Adenocarcinoma and Squamous Cell Carcinoma
The distribution of mutations detected in adenocarcinoma and squamous cell carcinoma is shown in Table 2. Both PIK3CA mutations and PTEN loss were found at slightly lower rates in adenocarcinoma compared with squamous cell carcinoma, although these findings were not significantly different (25.0% vs. 37.5%, p=0.33 and 3.6% vs. 13.0%, p=0.32, respectively). In contrast, KRAS mutations were detected in adenocarcinoma only (17.5% vs. 0%, p=0.01), and EGFR mutations were restricted to patients with squamous cell carcinoma (0.0% vs. 7.5%, p=0.24) although the latter result did not meet statistical significance, likely due to the small number of cases with positive findings.
EGFR Mutations Identified in Squamous Cell Carcinomas
All validated EGFR mutations resulted in a missense mutation, creating an EGFR isoform which lacked both transmembrane and kinase domains.24 This mutation was validated using 3 different techniques (mass spectrometry iPLEX, mass spectrometry hME chemistry, and Illumina sequencing) in 3 samples, all of which were squamous cell carcinomas; EGFR S703F was also detected by hME chemistry or Illumina sequencing in 2 additional squamous cell carcinomas (data not shown). This mutation was also detected in DNA from adjacent normal tissue identified by a gynecologic pathologist in the one sample where DNA from normal tissue was available, and is therefore a germline SNP in this specimen.
Clinical Associations
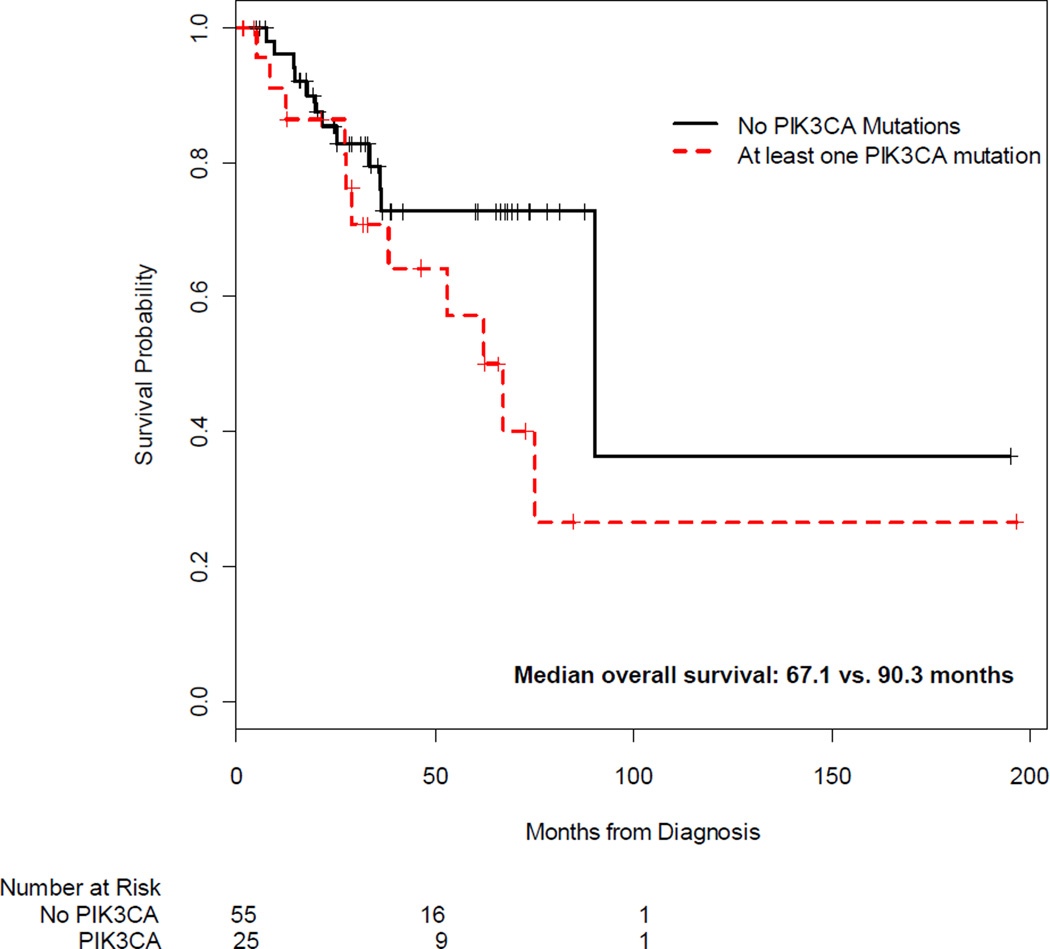
Associations between validated mutations, clinical outcomes, and HPV genotype were explored. As shown in Figure 1, PIK3CA mutations were associated with shorter survival; patients with mutations had a median overall survival of 67.1 months compared with 90.3 months in patients who did not harbor PIK3CA mutations. In multivariable Cox regression models, adjusted for stage (I/II vs. III/IV) and any lymph node involvement, the estimated OS hazard ratio associated with any PIK3CA mutation was 9.1 (95% CI 2.8–28.5, p<0.0001), compared with those without PIK3CA mutations. In contrast, we did not detect an association between KRAS mutations and overall survival (HR=0.31, p=0.26). Although larger studies have previously documented that HPV-18 is an independent risk factor for cancer relapse and death in cervical cancer,25, 26 we did not detect an association between HPV-16 or HPV-18 and overall survival. Similarly, we did not detect any associations between these HPV subtypes and the frequency of PIK3CA or KRAS mutations.
Figure 1.
Overall Survival by PIK3CA Mutation Status
Discussion
Our findings suggest that cervical cancers harbor high rates of potentially targetable oncogenic mutations. In this study, both cervical squamous cell carcinoma and adenocarcinoma had high rates of PIK3CA mutations which were associated with decreased survival. Similarly, PTEN loss was observed in both histologic subtypes, although the latter was found in fewer cases overall. In contrast, KRAS mutations were detected exclusively in adenocarcinoma and were not associated with differences in survival. The EGFR variants were limited to patients with squamous cell carcinoma, supporting the hypothesis of histotype-specific molecular mechanisms involved in the pathogenesis of cervical cancer.
We found PIK3CA mutations in 31.1% of the samples, present in both squamous cell carcinomas and adenocarcinomas. The PIK3CA mutations were located in the exon 9 helical domain (E545K and E542K), two well-described hotspot mutations that result in constitutive activation of cellular signaling.27 Five percent of these samples had co-mutations within the helical domain; we did not observe any mutations in exon 20 (H1047R) which has been associated with an increased response rate to PI3K/AKT/mTOR inhibitors.28 Similarly, we did not observe co-occurrence of mutations in PIK3CA and KRAS or BRAF, as previously described in other tumor types.11, 29 However, PI3K signaling has been identified as important in HPV transformation models,30 and should be further explored cervical cancer development. Our finding that PIK3CA mutations were associated with a survival disadvantage validates results from another recent study which observed decreased survival among patients with PIK3CA mutations in early stage cancers.13 Given the described roles of PI3K in cellular proliferation and survival, PI3KCA mutations may impart a more aggressive and treatment-resistant phenotype as suggested by other reports.13, 31
The high prevalence of PI3KCA mutations suggests that PI3K targeted agents should be explored in cervical cancer, particularly in light of recent findings from a phase I trial population which demonstrated that 40% (2/5) of cervical cancer patients with PIK3CA mutations had a clinical response to PI3K/AKT/mTOR inhibitors.32 The presence of PIK3CA mutations and loss of the tumor suppressor PTEN in both adenocarcinoma and squamous cell carcinoma suggests that PI3K targeted agents might be integrated into the treatment of cervical cancer independent of histological subtypes. Future studies should explore the use of PI3K inhibitors in early stage disease—e.g., as an adjuvant to radiation or combination chemoradiotherapy31, 33—particularly since we did not observe co-mutations between PIK3CA and KRAS.
KRAS mutations were identified exclusively in cervical adenocarcinomas, where they were present in 17.5% of samples. Most mutations were missense mutations of codon G12, well-described activating mutations, which have been associated with a worse prognosis in metastatic colorectal34, 35 and non-small lung cancer36 and resistance to EGFR-targeted therapies in colorectal cancer.37 Although we did not detect an association between KRAS mutations and survival in this study, this may be due to the relative rarity of mutations in our sample. Future studies should examine whether KRAS mutations have prognostic or therapeutic implications in larger samples of cervical adenocarcinomas, especially in light of a previous study which documented worse outcomes in KRAS mutant cervical cancers treated with radiation.17 The development of targeted therapeutics for KRAS mutations is an active area of research with promising preclinical data,38, 39 and some early suggestions of success with MEK inhibitors.40
EGFR mutations were identified in 7.5% of cervical squamous cell carcinomas in this study. Consistent with previous studies, mutations were not identified within the activating loop of the kinase domain.9, 10 Instead we detected a missense mutation in exon 15 of the EGFR gene, which produces an alternate spliced transcript (isoform D) that lacks both the transmembrane and intracellular kinase domains. Its presence in both tumor and adjacent normal tissue suggests that EGFR S703F may be a germline mutation. To date, this isoform is not well studied, but membranous expression of EGFR isoforms has been associated with decreased survival in prior studies of ovarian cancer and lymph-node negative cervical cancer.24, 41 Future studies are needed to validate this finding, and to explore the biologic and clinical importance of this mutation.
While our study is the first to comprehensively examine the molecular mutations present in cervical cancer, and to compare the two most common histological subtypes, we recognize a few limitations in our study. Our cohort was relatively small and predominantly early stage disease; thus the findings may be underpowered to detect associations between HPV subtype and survival described in larger studies.25, 26 We chose to use mass spectrometric genotyping given limited tumor tissue and DNA since it was the most robust, clinically relevant, and cost-effective panel of assays that could interrogate important somatic mutations in cancer. While 82% of the known events in PIK3CA (as calculated by the frequency of variants; mostly in amino acids 542, 545, and 1047) reported in COSMIC can be interrogated by Oncomap, we recognize that there are a finite number of specific point mutations that can be assayed (designated a priori within a subset of cancer genes) which may result in false negative mutation rates in some samples. Other limitations include difficulties in designing genotyping assays that identify small insertions or deletions larger than ~50 bp in size and an inability to detect most tumor suppressor gene mutations (which may occur not just “hotspot” regions, but anywhere within the gene) or additional genomic alterations such as gene amplifications or deletions. This may explain our low rates of LKB1 mutation detection relative to prior reports,23 and the differences in rates of PTEN loss observed by immunostaining and mutational analysis in this study. In addition, molecular alterations associated with HPV infection such as mutation and/or loss of p53, pRB, and Notch tumor suppressor genes as well as their associated gene expression changes could not explored in this study.42 Future studies should validate these findings in larger populations of patients with long term follow-up using whole exome sequencing.
In conclusion, our data reveal distinct genomic alterations in squamous cell carcinoma and adenocarcinoma of the cervix, which should encourage further studies to better understand these mutations and exploit them for clinical use. Our findings suggest that the use of more tailored treatment strategies, such as PI3K and MEK inhibitors, should be explored in cervical cancer. While current management strategies do not take histological classification of tumors into account, our study suggests that efforts to identify and target distinct molecular sub-populations within cervical cancer may provide an important opportunity to improve outcomes in women with both early and late-stage disease.
Acknowledgments
Conflicts of Interest and Source of Funding: This research was supported in part by (MRSG-13-013) from the American Cancer Society and a Conquer Cancer Foundation of ASCO Career Development Award to Dr. Wright and philanthropic funds from the Team Maureen Cervical Cancer Fund and the Friends of the Dana-Farber Cancer Institute. The funding organizations had no role in the design and conduct of the study; collection, analysis, or preparation of the data; or preparation, review, or approval of the manuscript. Any opinions, findings, and conclusions expressed in this material are those of the authors and do not necessarily reflect those of the American Cancer Society, the American Society of Clinical Oncology or the Conquer Cancer Foundation. The following authors reported paid consultancies: Dr. Hahn (Novartis, Blueprint Medicines, Thermo Fisher), Dr. Myers (Sanofi Aventis, EISAI). Dr. Wagle reports a paid consultancy and stock ownership (Foundation Medicine). Dr. Myers is a paid employee of Novartis.
Footnotes
Authorship contributions: Drs. Wright and Dahlberg had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Wright, Matulonis, Hirsch.
Analysis and interpretation of data: Wright, Howitt, Myers, Dahlberg, Palescandolo, Van Hummelen, MacConaill, Wagle, Jones, Quick, Laury, Katz, Hahn, Matulonis, Hirsch.
Drafting of the manuscript: Wright, Myers.
Critical revision of the manuscript for important intellectual content: Wright, Howitt, Myers, Dahlberg, Palescandolo, Van Hummelen, MacConaill, Jones, Wagle, Quick, Laury, Katz, Hahn, Matulonis, Hirsch.
Statistical analysis: Wright, Palescandolo, Van Hummelen, Dahlberg.
Obtained funding: Wright and Matulonis.
Administrative, technical, or material support: Wright, Matulonis, Hahn, Hirsch, Howitt, Laury, Quick, Jones.
Additional contributions: We thank Courtney Doyle, BA; Christina Go, BSc; Christina Roden, BSc; Aaron Thorner, PhD; Zachary Herbert, MSc; Matthew Ducar, BSc; and Ravali Adusimilli, BSc for additional administrative, technical, and analytic support.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Wang SS, Sherman ME, Hildesheim A, Lacey JV, Jr, Devesa S. Cervical adenocarcinoma and squamous cell carcinoma incidence trends among white women and black women in the United States for 1976–2000. Cancer. 2004;100(5):1035–1044. doi: 10.1002/cncr.20064. [DOI] [PubMed] [Google Scholar]
- 3.Galic V, Herzog TJ, Lewin SN, et al. Prognostic significance of adenocarcinoma histology in women with cervical cancer. Gynecol Oncol. 125(2):287–291. doi: 10.1016/j.ygyno.2012.01.012. [DOI] [PubMed] [Google Scholar]
- 4.Oka K, Nakano T, Hoshi T. Analysis of response to radiation therapy of patients with cervical adenocarcinoma compared with squamous cell carcinoma. MIB-1 and PC10 labeling indices. Cancer. 1996;77(11):2280–2285. doi: 10.1002/(SICI)1097-0142(19960601)77:11<2280::AID-CNCR15>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 5.Eifel PJ, Burke TW, Morris M, Smith TL. Adenocarcinoma as an independent risk factor for disease recurrence in patients with stage IB cervical carcinoma. Gynecol Oncol. 1995;59(1):38–44. doi: 10.1006/gyno.1995.1265. [DOI] [PubMed] [Google Scholar]
- 6.Park JY, Kim DY, Kim JH, Kim YM, Kim YT, Nam JH. Outcomes after radical hysterectomy in patients with early-stage adenocarcinoma of uterine cervix. Br J Cancer. 102(12):1692–1698. doi: 10.1038/sj.bjc.6605705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galic V, Herzog TJ, Lewin SN, et al. Prognostic significance of adenocarcinoma histology in women with cervical cancer. Gynecol Oncol. 2012;125(2):287–291. doi: 10.1016/j.ygyno.2012.01.012. [DOI] [PubMed] [Google Scholar]
- 8.Lee YY, Choi CH, Kim TJ, et al. A comparison of pure adenocarcinoma and squamous cell carcinoma of the cervix after radical hysterectomy in stage IB-IIA. Gynecol Oncol. 2011;120(3):439–443. doi: 10.1016/j.ygyno.2010.11.022. [DOI] [PubMed] [Google Scholar]
- 9.Arias-Pulido H, Joste N, Chavez A, et al. Absence of epidermal growth factor receptor mutations in cervical cancer. Int J Gynecol Cancer. 2008;18(4):749–754. doi: 10.1111/j.1525-1438.2007.01111.x. [DOI] [PubMed] [Google Scholar]
- 10.Iida K, Nakayama K, Rahman MT, et al. EGFR gene amplification is related to adverse clinical outcomes in cervical squamous cell carcinoma, making the EGFR pathway a novel therapeutic target. Br J Cancer. 2011;105(3):420–427. doi: 10.1038/bjc.2011.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Janku F, Lee JJ, Tsimberidou AM, et al. PIK3CA mutations frequently coexist with RAS and BRAF mutations in patients with advanced cancers. PLoS One. 2011;6(7):e22769. doi: 10.1371/journal.pone.0022769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bertelsen BI, Steine SJ, Sandvei R, Molven A, Laerum OD. Molecular analysis of the PI3K-AKT pathway in uterine cervical neoplasia: frequent PIK3CA amplification and AKT phosphorylation. Int J Cancer. 2006;118(8):1877–1883. doi: 10.1002/ijc.21461. [DOI] [PubMed] [Google Scholar]
- 13.McIntyre JB, Wu J, Craighead PS, et al. PIK3CA mutational status and overall survival in patients with cervical cancer treated with radical chemoradiotherapy. Gynecol Oncol. doi: 10.1016/j.ygyno.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 14.Kim TJ, Lee JW, Song SY, et al. Increased expression of pAKT is associated with radiation resistance in cervical cancer. Br J Cancer. 2006;94(11):1678–1682. doi: 10.1038/sj.bjc.6603180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pappa KI, Choleza M, Markaki S, et al. Consistent absence of BRAF mutations in cervical and endometrial cancer despite KRAS mutation status. Gynecol Oncol. 2006;100(3):596–600. doi: 10.1016/j.ygyno.2005.09.029. [DOI] [PubMed] [Google Scholar]
- 16.Kang S, Kim HS, Seo SS, Park SY, Sidransky D, Dong SM. Inverse correlation between RASSF1A hypermethylation, KRAS and BRAF mutations in cervical adenocarcinoma. Gynecol Oncol. 2007;105(3):662–666. doi: 10.1016/j.ygyno.2007.01.045. [DOI] [PubMed] [Google Scholar]
- 17.Wegman P, Ahlin C, Sorbe B. Genetic alterations in the K-Ras gene influence the prognosis in patients with cervical cancer treated by radiotherapy. Int J Gynecol Cancer. 21(1):86–91. doi: 10.1097/IGC.0b013e3182049924. [DOI] [PubMed] [Google Scholar]
- 18.MacConaill LE, Campbell CD, Kehoe SM, et al. Profiling critical cancer gene mutations in clinical tumor samples. PLoS One. 2009;4(11):e7887. doi: 10.1371/journal.pone.0007887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31(3):213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howlader NNA, Krapcho M, Neyman N, Aminou R, Altekruse SF, Kosary CL, Ruhl J, Tatalovich Z, Cho H, Mariotto A, Eisner MP, Lewis DR, Chen HS, Feuer EJ, Cronin KA. SEER Cancer Statistics Review, 1975–2009 (Vintage 2009 Populations) Available from URL: http://seer.cancer.gov/csr/1975_2009_pops09/
- 21.Wheeler CM, Hunt WC, Joste NE, Key CR, Quint WG, Castle PE. Human papillomavirus genotype distributions: implications for vaccination and cancer screening in the United States. J Natl Cancer Inst. 2009;101(7):475–487. doi: 10.1093/jnci/djn510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwartz SM, Daling JR, Shera KA, et al. Human papillomavirus and prognosis of invasive cervical cancer: a population-based study. J Clin Oncol. 2001;19(7):1906–1915. doi: 10.1200/JCO.2001.19.7.1906. [DOI] [PubMed] [Google Scholar]
- 23.Wingo SN, Gallardo TD, Akbay EA, et al. Somatic LKB1 mutations promote cervical cancer progression. PLoS One. 2009;4(4):e5137. doi: 10.1371/journal.pone.0005137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Halle C, Lando M, Svendsrud DH, et al. Membranous expression of ectodomain isoforms of the epidermal growth factor receptor predicts outcome after chemoradiotherapy of lymph node-negative cervical cancer. Clin Cancer Res. 17(16):5501–5512. doi: 10.1158/1078-0432.CCR-11-0297. [DOI] [PubMed] [Google Scholar]
- 25.Lai CH, Chang CJ, Huang HJ, et al. Role of human papillomavirus genotype in prognosis of early-stage cervical cancer undergoing primary surgery. J Clin Oncol. 2007;25(24):3628–3634. doi: 10.1200/JCO.2007.11.2995. [DOI] [PubMed] [Google Scholar]
- 26.Kang WD, Kim CH, Cho MK, et al. HPV-18 is a poor prognostic factor, unlike the HPV viral load, in patients with stage IB-IIA cervical cancer undergoing radical hysterectomy. Gynecol Oncol. 2011;121(3):546–550. doi: 10.1016/j.ygyno.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 27.Courtney KD, Corcoran RB, Engelman JA. The PI3K pathway as drug target in human cancer. J Clin Oncol. 2010;28(6):1075–1083. doi: 10.1200/JCO.2009.25.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janku F, Wheler JJ, Naing A, et al. PIK3CA Mutation H1047R Is Associated with Response to PI3K/AKT/mTOR Signaling Pathway Inhibitors in Early-Phase Clinical Trials. Cancer Res. 73(1):276–284. doi: 10.1158/0008-5472.CAN-12-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao X, Morikawa T, Lochhead P, et al. Prognostic role of PIK3CA mutation in colorectal cancer: cohort study and literature review. Clin Cancer Res. 18(8):2257–2268. doi: 10.1158/1078-0432.CCR-11-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henken FE, Banerjee NS, Snijders PJ, et al. PIK3CA-mediated PI3-kinase signalling is essential for HPV-induced transformation in vitro. Mol Cancer. 2011;10:71. doi: 10.1186/1476-4598-10-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwarz JK, Payton JE, Rashmi R, et al. Pathway-specific analysis of gene expression data identifies the PI3K/Akt pathway as a novel therapeutic target in cervical cancer. Clin Cancer Res. 18(5):1464–1471. doi: 10.1158/1078-0432.CCR-11-2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Janku F, Wheler JJ, Westin SN, et al. PI3K/AKT/mTOR inhibitors in patients with breast and gynecologic malignancies harboring PIK3CA mutations. J Clin Oncol. 30(8):777–782. doi: 10.1200/JCO.2011.36.1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xia S, Zhao Y, Yu S, Zhang M. Activated PI3K/Akt/COX-2 pathway induces resistance to radiation in human cervical cancer HeLa cells. Cancer Biother Radiopharm. 25(3):317–323. doi: 10.1089/cbr.2009.0707. [DOI] [PubMed] [Google Scholar]
- 34.Smith JC, Brooks L, Hoff PM, et al. KRAS mutations are associated with inferior clinical outcome in patients with metastatic colorectal cancer, but are not predictive for benefit with cediranib. Eur J Cancer. 2013;49(10):2424–2432. doi: 10.1016/j.ejca.2013.02.023. [DOI] [PubMed] [Google Scholar]
- 35.Bruera G, Cannita K, Di Giacomo D, et al. Worse prognosis of KRAS c.35 G > A mutant metastatic colorectal cancer (MCRC) patients treated with intensive triplet chemotherapy plus bevacizumab (FIr-B/FOx) BMC Med. 2013;11:59. doi: 10.1186/1741-7015-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mascaux C, Iannino N, Martin B, et al. The role of RAS oncogene in survival of patients with lung cancer: a systematic review of the literature with meta-analysis. Br J Cancer. 2005;92(1):131–139. doi: 10.1038/sj.bjc.6602258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Misale S, Yaeger R, Hobor S, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 486(7404):532–536. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Corcoran RB, Cheng KA, Hata AN, et al. Synthetic lethal interaction of combined BCL-XL and MEK inhibition promotes tumor regressions in KRAS mutant cancer models. Cancer Cell. 2013;23(1):121–128. doi: 10.1016/j.ccr.2012.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zimmermann G, Papke B, Ismail S, et al. Small molecule inhibition of the KRAS-PDEdelta interaction impairs oncogenic KRAS signalling. Nature. 2013;497(7451):638–642. doi: 10.1038/nature12205. [DOI] [PubMed] [Google Scholar]
- 40.Janne PA, Shaw AT, Pereira JR, et al. Selumetinib plus docetaxel for KRAS-mutant advanced non-small-cell lung cancer: a randomised, multicentre, placebo-controlled, phase 2 study. Lancet Oncol. 2013;14(1):38–47. doi: 10.1016/S1470-2045(12)70489-8. [DOI] [PubMed] [Google Scholar]
- 41.Noske A, Schwabe M, Weichert W, et al. An intracellular targeted antibody detects EGFR as an independent prognostic factor in ovarian carcinomas. BMC Cancer. 11:294. doi: 10.1186/1471-2407-11-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buitrago-Perez A, Garaulet G, Vazquez-Carballo A, Paramio JM, Garcia-Escudero R. Molecular Signature of HPV-Induced Carcinogenesis: pRb p53 and Gene Expression Profiling. Curr Genomics. 2009;10(1):26–34. doi: 10.2174/138920209787581235. [DOI] [PMC free article] [PubMed] [Google Scholar]