Abstract
AMSH, a deubiquitinating enzyme (DUB) with exquisite specificity for Lys63-linked polyubiquitin chains, is an endosome-associated DUB that regulates sorting of activated cell-surface signaling receptors to lysosome, a process mediated by the members of the endosomal sorting complexes required for transport (ESCRT) machinery. Whole-exome sequencing of DNA samples from children with microcephaly capillary malformation (MIC-CAP) syndrome identified recessive mutations encoded in the AMSH gene causatively linked to the disease. Herein, we report a number of important observations that significantly advance our understanding of AMSH within the context of the ESCRT machinery. First, we performed mutational and kinetic analysis of the putative residues involved in diubiquitin recognition and catalysis with a view to better understanding the catalytic mechanism of AMSH. Our mutational and kinetic analysis reveals that recognition of the proximal ubiquitin is imperative for the linkage specificity and catalytic efficiency of the enzyme. The MIC-CAP disease mutation, Thr313Ile, shows a substantial loss of catalytic activity without any significant change in thermodynamic stability of the protein, indicating that its perturbed catalytic activity is the basis of the disease. The catalytic activity of AMSH is stimulated upon binding to the ESCRT-0 member STAM, however, the precise mechanism and its significance are not known. Based on a number of biochemical and biophysical analysis, we are able to propose a model for activation according to which activation of AMSH is enabled by facile, simultaneous binding to two ubiquitin groups in a polyubiquitin substrate, one by the catalytic domain of the DUB (binding to the distal ubiquitin) and the other (the proximal ubiquitin) by the ubiquitin interacting motif (UIM) from STAM. Such a mode of binding would stabilize the ubiquitin chain in a productive orientation, resulting in an enhancement of the activity of the enzyme. These data together provide a mechanism for understanding the recruitment and activation of AMSH at ESCRT-0, providing biochemical and biophysical evidence in support of a role for AMSH when it is recruited to the initial ESCRT complex: it functions to facilitate transfer of ubiquitinated receptors (cargo) from one ESCRT member to the next by disassembling the polyubiquitin chain while leaving some ubiquitin groups still attached to the cargo.
AMSH (associated molecule with a Src homology 3 domain of signal transducing adaptor molecule, STAM) is a member of the JAMM (JAB1/MPN/MOV34) family of deubiquitinating enzymes (DUBs) (1), which regulates ubiquitin signaling by catalyzing the hydrolysis of isopeptide (or peptide) bonds between ubiquitin and target proteins or within polymeric chains of ubiquitin. The JAMM family, being one of the five families of mammalian DUBs, are metalloproteases, whereas the others, UCHs, USPs, OTUs, and MJDs) are cysteine proteases (2–4). Members of the JAMM family show substantial variation in their overall amino-acid sequence, but share, as the name suggests, a conserved JAMM motif as the catalytic domain. Mechanistically, they share distinct similarities to the extensively studied metalloprotease, thermolysin. Like thermolysin, theses enzymes have a Zn2+ in their active site, which is involved in their mechanism of catalysis. The Zn2+ ion is coordinated within the active site usually by two histidines, an acidic residue (aspartic acid or glutamic acid), and a water molecule that eventually is used as the nucleophile for attacking the scissile peptide bond. This catalytic water is held in place by Zn2+ and another acidic residue (glutamic acid in AMSH), which provides a hydrogen bond stabilizing the water. Sequence analysis of the members of the JAMM family reveals that only 7 of 14 proteins have the conserved zinc binding capabilities (5), while only 6 of those 7 exhibit isopeptidase activity toward ubiquitin and ubiquitin-like proteins, AMSH, AMSH-LP (AMSH like protein), BRCC36, RPN11 (POH1), MYSM1, and CSN5 (5–7).
AMSH is one of the two DUBs, the other being UBPY (also known as ubiquitin specific protease 8, USP8) (8) known to be important regulators of the ESCRT (endosomal sorting complexes required for transport) complexes (9). The ESCRT machinery consist of four protein-protein complexes (ESCRT-0, -I, -II, -III) and the AAA ATPase Vps4 (10, 11) that serve several important functions within the cell: endosomal sorting, trafficking, viral budding, cytokinesis, transcriptional regulation, and autophagy (12). The ESCRTs were initially discovered in yeast in the context of their role in endosomal sorting and trafficking of cell-surface receptors to lysosome for degradation, as a means for down-regulating their signals (11). These receptors are first ubiquitinated, and then, shuttled through the ESCRT machinery, until their internalization within endosomes, which then can fuse with the lysosome delivering the receptors for proteolysis (11).
Of the four complexes, only ESCRT-0 and ESCRT-III can specifically recognize DUBs (9). The ESCRT-0 recognition is carried out through the binding of the SH3 binding motif (SBM) of DUBs to the SH3 domain of the ESCRT-0 member STAM (13), while the ESCRT-III recognition is via the MIT (microtubule interacting and transport) domain of DUBs binding to the C-terminal MIT interacting motifs (MIMs of charged multivesicular body proteins (CHMPs) (9). Previous studies have shown that once AMSH binds to STAM, its activity is enhanced (13, 14), however, the precise mechanism is not known (13). The x-ray crystal structure of the MIT domain of AMSH bound to the C-terminal MIM fragment of the ESCRT-III member CHMP3 has been determined (15). The structure reveals that the AMSH-CHMP3 complex is stabilized mainly by polar interactions, manifesting into tight binding between the two proteins, with a dissociation constant (KD) of 60 nM, the highest reported KD for an ESCRT-III MIM -MIT interaction (15).
A clear role for AMSH has not been identified yet, however, whole-exome sequencing analysis has shown that recessive mutations in AMSH leads to microcephaly-capillary malformation (MIC-CAP) syndrome (16). MIC-CAP is discovered at or shortly after birth in which children diagnosed with the disease have severe microcephaly with progressive cortical atrophy, intractable epilepsy, profound developmental delay and multiple small capillary malformations on the skin (16–19). The microcephaly phenotype is attributed to the accumulation of ubiquitinated proteins, suggesting a lost of enzymatic function as was seen in knock-out mice studies (20). Out of the ten patients that were screened, six had missense mutations, two had nonsense mutations, two translational frameshift mutations, and three intronic mutations (16). Interestingly, five out of the six missense mutations were found within the MIT domain of AMSH, and the sixth, Thr313Ile, found within the JAMM domain (16).
Although the exact in vivo function of AMSH is not well understood, its exquisite specificity for Lys63-linked polyubiquitin chains, the same type of chain used for endosomal-lysosomal targeting, is well characterized (21). The structural basis for this recognition was elucidated with the homologous protein, AMSH-LP, bound to Lys63-linked dimer of ubiquitin (22). This structure reveals that the specificity arises from a tri-peptide sequence within the proximal ubiquitin (Gln62, Lys63, and Glu64) interacting directly with a threonine, two phenylalanines, and a serine residue (22) on the proximal binding site of the enzyme (in a diubiquitin motif, the proximal ubiquitin is defined as the one that contributes a Lys residue to be linked to Gly76 of the other ubiquitin, defined as the distal ubiquitin). AMSH and AMSH-LP share 54% identity and 73% sequence similarity, however, AMSH-LP does not have a functional SBM (9, 23) nor MIT domain (24), therefore, not having the ability to bind to the members of the ESCRT machinery. We recently determined the x-ray crystal structure of the catalytic domain of AMSH, and found that the catalytic domains of AMSH and AMSH-LP are structurally very similar, however, much to our surprise, AMSH is thermodynamically less stable than AMSH-LP, which was attributed to structural plasticity of the former (25). This idea of structural plasticity is supported by the second x-ray crystal structure of the catalytic domain of AMSH bearing the active-site glutamate to alanine mutation, which was expected to cause the release of the active-site Zn2+. The structure, however, shows that the tetrahedral coordination around the active-site Zn2+ in this mutant is still maintained by a nearby aspartate residue moving in to provide the fourth ligand for the metal in place of the lost water (25). Though AMSH and AMSH-LP are able to localize to the endosomes in a similar manner through their ability to bind to clathrin (13, 26), the differences between them arising out of AMSH-LP’s inability to bind to any of the ESCRT complexes suggests that AMSH and AMSH-LP are not functionally redundant. Moreover, the active-site cleft of AMSH features three substitutions relative to AMSH-LP, substitutions of residues that are predicted to be used for ubiquitin binding. One of them, Thr313, which in AMSH-LP corresponds to Met325, is also the site of a MIC-CAP mutation.
In the present study, we aim to further establish a role for AMSH as an important regulator of the ESCRT machinery. We have carried out extensive mutational and kinetic analysis of residues within the catalytic domain of AMSH to understand their role in ubiquitin binding to better understand the enzyme’s catalytic mechanism. Also, we have studied the effect of the Thr313Ile mutation found in children with the MIC-CAP syndrome on the activity and thermodynamic stability of AMSH with hopes of understanding the molecular basis of the disease. Our results confirmed that residues within the proximal ubiquitin recognition site of AMSH are the basis of specificity for Lys63-linked polyubiquitin chain. Furthermore, we found that of the three residue differences between AMSH and AMSH-LP in the distal ubiquitin-binding site, Thr313 and Glu316 had the most significant effects on AMSH activity. Finally, using the minimal domains required for AMSH activation, we have shed light on the mechanism of AMSH recruitment and activation at ESCRT-0; we found that AMSH is activated due to an intact SBM-SH3 interaction and an intact UIM from STAM. Based on our results, we have proposed a mechanism and role for AMSH in the context of ESCRT-0.
Materials and Methods
Cloning, Expression, and Purification
The DNA encoding the catalytic domain of AMSH was amplified by PCR using a plasmid that contained full-length DNA as the template (pGEX-6p1-AMSH, a kind gift from Sylvie Urbé, University of Liverpool, UK) and was subcloned into pGEX-6p1 (GE Biosciences) by using standard cloning protocols. The resulting N-terminally fused glutathione S-transferase (GST)-tagged protein was expressed in Escherichia coli Rosetta cells (Novagen) and purified with a glutathione-Sepharose column (GE Biosciences) following manufacturer’s instructions. After removal of the tag by PreScission protease (GE Biosciences), the protein was further purified by size-exclusion chromatography (SEC) using Superdex S75 column (GE Biosciences).
The series of individual point mutations were introduced into the AMSH catalytic domain gene by site-directed mutagenesis using QuikChange Site-Directed Mutagenesis Kit (Stratagene) following manufacturer’s protocol. DNA sequencing confirmed the presence of the mutations. The resulting proteins were purified using standard GST-affinity chromatography followed by SEC (Superdex S75 column).
The DNA encoding the SH3 and UIM-SH3 domains was amplified by PCR using a plasmid that contained the full-length DNA as the template (pGW1Myc2c-STAM2, a kind gift from Craig Blackstone, National Institutes of Neurological Disorders and Stroke (NINDS) at the National Institutes of Health (NIH)) and was subcloned into pGEX-6p1 (GE Biosciences) using standard cloning protocols. The resulting N-terminally fused GST-tagged protein was expressed in E. coli Rosetta cells (Novagen) and purified with a glutathione-Sepharose column (GE Biosciences) following manufacturer’s instructions. After removal of the tag by PreScission protease (GE Biosciences), the protein was further purified by size-exclusion chromatography (SEC) using Superdex S75 column (GE Biosciences).
The A176G mutation was introduced into the UIM-SH3 gene by site-directed mutagenesis using QuikChange Site-Directed Mutagenesis Kit (Stratagene) following manufacturer’s protocol. DNA sequencing confirmed the presence of the mutations. The resulting proteins were purified using standard GST-affinity chromatography followed by SEC (Superdex S75 column).
Human ubiquitin was subcloned into pGEX-6p1 and purified using GST affinity chromatography, and the GST tag was removed by PreScission Protease. The protein was further purified using SEC (Superdex S75 column). Lys63-diubiquitin was enzymatically synthesized from ubiquitin using ATP, human E1, and the E2 complex (Ubc13 and Uev1a) following previously reported procedures (22). The reaction was incubated at 37°C for 2 h and then quenched by dilution with buffer A (50mM sodium acetate, pH 4.5). The quenched reaction mixture was subjected to ion-exchange chromatography on a Mono-S column (GE Biosciences) to obtain Lys63-diubiquitin.
Determination of kinetic parameters and DUB assay
The kinetic parameters were determined by incubating the enzymes (25 nM T313A, 100 nM C282A, N312A, E317A, and F320A, 2 µM E316A and F395A, 3 µM T341A and S346A, and 10 µM F343A and S345A) with four concentrations of diubiquitin, ranging from 20 to 177 µM, in reaction buffer (50 mM TRIS-HCl (pH 7.0), 20 mM KCl, 5 mM MgCl2 and 1 mM DTT). The reaction was carried out at 20°C for 10–75 min depending on activity for initial velocity measurements. Reaction tubes were quenched by the addition of 5X SDS-PAGE sample buffer followed by boiling. The reaction mixtures were visualized by SDS-PAGE gels and scanned. Bands corresponding to monoubiquitin were integrated using Image J software (http://rsb.info.nih.gov/ij/). Ubiquitin standards ranging from 4 to 12.3 µg were used to draw calibration plots, which were used to quantify the amount of ubiquitin produced. Kinetic parameters were calculated by fitting the data in SigmaPlot (Systat Software, San Jose, CA).
The in vitro DUB assay was carried out by incubating AMSH (residues 219–424) to a final enzyme concentration of 100 nM with 1µM of the SH3 domain of STAM2 or UIM-SH3 gene of STAM2, and 20 µM Lys63-diubiquitin in a total reaction volume of 20 µL. All reactions were carried out in reaction buffer (50 mM TRIS-HCl (pH 7.0), 25 mM KCl, 5 mM MgCl2, and 1 mM DTT) for 5 h at 20°C. The reaction was quenched by the addition of 5X SDSPAGE sample buffer followed by boiling and then analyzed by SDS-PAGE.
Analytical Ultracentrifugation
Sedimentation velocity experiments were conducted at 50,000 rpm using the Beckman Coulter XLA and XLI (Beckman Coulter, Fullerton, CA, USA). The samples were monitored by both interference and absorbance optics at 254 and 280 nm. The proteins were dialyzed in 50 mM TRIS-HCl pH 7.6, 50 mM NaCl. Three concentration series for AMSH (residues 219–424) were conducted to evaluate the formation of higher-order species at 24, 48, and 96 µM. The AMSH-SH3 complex was characterized using a constant concentration of 23.5 µM of AMSH and three concentrations of SH3 at 24, 47, and 70 µM. The AMSH-UIM-SH3 complex was characterized using a constant concentration of 23.5 µM of AMSH and three concentrations of 48, 144, and 288µM. The AMSH-ubiquitin complex was characterized using a constant concentration of 23.5 µM of AMSH and two concentrations of ubiquitin at 23 and 92 µM. The AMSH-diubiquitin complex was characterized using a constant concentration of 23.5 µM of AMSH and three diubiquitin concentrations at 24, 48, and 96 µM. The solvent density (1.00170 g.ml-1), viscosity (0.01022 poise), and the partial specific volumes that were used for the analyses, 0.73387 ml.g−1 (AMSH219), 0.71870 ml.g−1 (AMSH-SH3), 0.71701 ml.g−1 (AMSHUIM- SH3), 0.72934 ml.g−1 (AMSH-diubiquitin), and 0.72479 ml.g−1 (AMSH-ubiquitin), were calculated by SEDNTERP v. 20120828 BETA (http://bitcwiki.sr.unh.edu/index.php/Main_Page) (27). The sedimentation coefficients and apparent molecular weights were calculated from size distribution analyses [c(s)] using SEDFIT v. 14.3e (28, 29). The figures were prepared using GUSSI v. 1.0.7b beta with the sedimentation coefficients standardized to s20,w and the data was normalized to the peak area of the complexes.
Sedimentation equilibrium experiments were conducted at 20°C using a 6-channel centerpiece in an AN-60 Ti rotor spun at speeds of 13,200, 29,900, and 42,000 rpm for the AMSH-ubiquitin complex and 11,600, 21,000, and 36,000 rpm for the AMSH-diubiquitin complex. The molar ratios of the AMSH-ubiquitin complex were: 1:2, 1:4, and 1:8, and the molar ratios of the AMSH-diubiquitin complex were: 1:2, 1:4, and 1:5, to determine the molecular weight of the complexes. Absorbance scans at 280 nm were taken every 2 hours for 60 hours. The samples were tested for equilibrium using Sedfit (28, 29). Calculations of the molecular weights were done by SEDPHAT v. 10.58d (30–35) using the Species Analysis and Species Analysis with Mass Conservation Constraints. Errors were calculated using 1- dimensional error surface projections. Final figures were generated in GUSSI.
Isothermal Titration Calorimetry
To determine the KDs of AMSH binding to ubiquitin, ITC experiments were conducted using the MicroCal ITC200 (GE Healthcare Life Sciences). The proteins were dialyzed in the same buffer as was used for AUC. For the AMSH-diubiquitin experiment, 10 µM AMSH was in the cell and 500 µM of diubiquitin was in the syringe. The AMSH-ubiquitin titration had 100 µM AMSH in the cell, and 1 mM ubiquitin in the syringe. The F320A mutant of AMSH with ubiquitin had 100 µM of the enzyme in the cell, and 1 mM of ubiquitin in the syringe. These experiments were done at 20°C, 18 total injections of 1.4 µL per injection, with 180 seconds in between injections to allow for a return to baseline before the subsequent injection. The data was then baseline corrected by NITPIC (36) and loaded into SEDPHAT (30–35) for global analysis and fitting using a 1:1 model. Figures were prepared using GUSSI. To determine the KD for the AMSH-SH3 interaction, 50 µM of AMSH was in the cell and 750 µM of the SH3 domain was in the syringe. KD of AMSH-UIM-SH3 interaction was determined using 100 µM AMSH in the cell and 1 mM UIM-SH3 in the syringe. AMSHK238T-UIM-SH3 experiment was conducted using 50 µM of the enzyme in the cell and 1 mM of UIM-SH3 in the syringe. Characterization of ubiquitin binding to UIM-SH3 was done using 50 µM UIM-SH3 was in the cell and 3.1 mM ubiquitin in the syringe. This data was fit to a two-site model. Confirmation of SH3-ubiquitin binding was done with 100 µM of SH3 in the cell and 3.0 mM ubiquitin in the syringe. The UIM-SH3-Lys63-diubiquitin experiment had 50 µM of UIM-SH3 in the cell and 750 µM diubiquitin in the syringe.
Guanidine Melt using circular dichroism spectroscopy
The stability of the folded state of AMSH244 and AMSH-LP towards GdHCl was determined using 8M stock concentrations of GdHCl (Sigma). Varying concentrations of GdHCl were added to the protein (0.2 mg.mL−1) diluted in 100 mM phosphate buffer pH 7.4 and allowed to sit at room temperature overnight to allow for complete equilibration. Changes in the folded state of the proteins were monitored using circular dichroism by following changes in ellipticity at 220 nm. CD spectra were recorded in a Jasco J-810 Spectropolarimeter in the far UV region (195–260 nm) in a 0.1 cm path length cuvette. Each spectrum was averaged over 4 scans (50 nm.min−1 scan speed, with a 8 second time constant) and corrected by subtraction of a spectrum of the buffer alone. Mean residue molar ellipticity values were calculated using the following equation:
Where θ is the ellipticity in degrees, l is the optical path in cm, C is the concentration in mg/ml, M is the molecular mass and n is in the number of residues in the protein.
The mean residue molar ellipticity [θ] is given in deg.cm2.dmol−1. Unfolding curves were analyzed using a two-state unfolding model, using linear extrapolation to obtain the ΔG value in the absence of GdHCl (37).
RESULTS
Mutational and kinetic analysis of the catalytic domain of AMSH
Previous modeling studies investigating the differences between the ability of AMSH and AMSH-LP to bind and cleave Lys63-linked diubiquitin revealed some interesting results. The active site and proximal ubiquitin binding residues were identical between the two proteins; however, in the distal ubiquitin-binding site, AMSH has three residue differences compared to AMSH-LP. Based on this analysis, we sought to carry out an extensive mutational and kinetic analysis of the putative residues in AMSH that might be involved in diubiquitin cleavage.
Active site
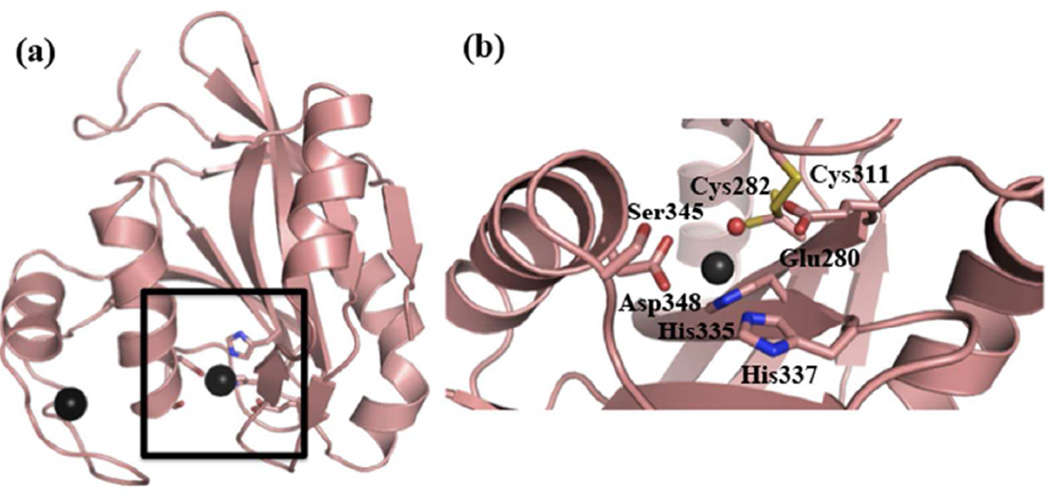
The active site of AMSH consists of a Zn2+ ion, coordinated directly by three residues (Asp348, His 335, and His337) and a water molecule that is hydrogen bonded to Glu280, and a putative oxy-anion stabilizing residue (Ser345) (Figure 1). In order to probe the roles of Asp348 and Glu280, we generated two aspartate mutants (D348A and D348N) and a glutamate mutant (E280A) and, as expected, found that there was no detectable activity in these mutants (Table 1), most likely due to the loss of Zn2+ for the aspartate mutations and the loss of the water molecule in the glutamate mutation.
Figure 1.
The DUB domain of AMSH. (a) Ribbon diagram of the crystal structure of the catalytic domain of AMSH (PDB ID: 3RZU). The active site is highlighted by the black square. (b) An expanded view of the active-site residues of AMSH. The black spheres represent Zn2+, and the red sphere the active-site water molecule.
Table 1.
Kinetic Parameters of AMSH Mutants
| Site | Protein | kcat×10−3 (s−1) | KM (µM) |
|---|---|---|---|
| Wild type | 1400 ± 100 | 32 ±5 | |
| Active site | |||
| Glu280Ala | NAa | NAa | |
| Cys282Ala | 230 ±140 | 45 ±7 | |
| Ser345Ala | 1.4 ±0.1 | 38 ±15 | |
| Asp348Ala | NAa | NAa | |
| Asp348Asn | NAa | NAa | |
| Proximal | |||
| Thr341Ala | 24 ±1 | 15 ±3 | |
| Phe343Ala | 5 ±3 | 21 ±4 | |
| Ser346Ala | 13 ±3 | 23 ±8 | |
| Phe395Ala | 22 ±8 | 18 ±9 | |
| Distal | |||
| Asn312Ala | 430 ±60 | 19 ±2 | |
| Thr313Ala | 2600 ±600 | 82 ±5 | |
| Glu316Ala | 19 ±4 | 31 ±9 | |
| Glu317Ala | 750 ±200 | 19 ±7 | |
| Phe320Ala | 370 ±20 | 98 ±15 | |
| MIC-CAP | |||
| Thr313Ile | 225 ±39 | 21 ±5 |
No observed activity up to 10 µM of protein per 20 µL reaction over 4 hours at 20°C.
Next, we probed the function of the putative oxy-anion hole-stabilizing residue, Ser345. Mutating serine to alanine resulted in a significantly impaired enzyme with a 1000-fold reduction in kcat (Table 1). As described with other families of hydrolases, the oxy-anion hole-stabilizing residue plays a critical role in donating a hydrogen bond to the negatively charged tetrahedral intermediate formed after the initial nucleophilic attack. Substantial reduction of kcat alone upon mutation to alanine, with KM remaining nearly the same, is consistent with Ser345 playing the role as the oxy-anion stabilizing residue in AMSH.
In our previous structural analysis, we noted a potential disulfide bridge between Cys282 and Cys311, 7.4Å away from the active-site Zn2+ (25). Previous studies have shown that N-ethylmaleimide (NEM) inhibits AMSH activity (IC50 of 16.2± 3.2 µM) (21, 38), presumably by modifying one of these two cysteines, perhaps Cys282 because it is proximal to the active-site cleft. Its modification might introduce some steric hindrance for substrate binding thus explaining the inhibitory effect. We sought to determine if Cys282 has any role in the enzyme’s catalytic activity. Previously, when Cys282 was mutated to alanine, we noticed a loss of activity (25), however, a more detailed kinetic analysis of this mutant revealed a more significant reduction in activity, a 6-fold loss in kcat (Table 1), which would suggest that Cys282 does indeed have a role in catalysis. Cys282 is seen making a van der Waals contact with Leu73 of the distal ubiquitin, which may explain the reduction in activity observed here.
Proximal ubiquitin site
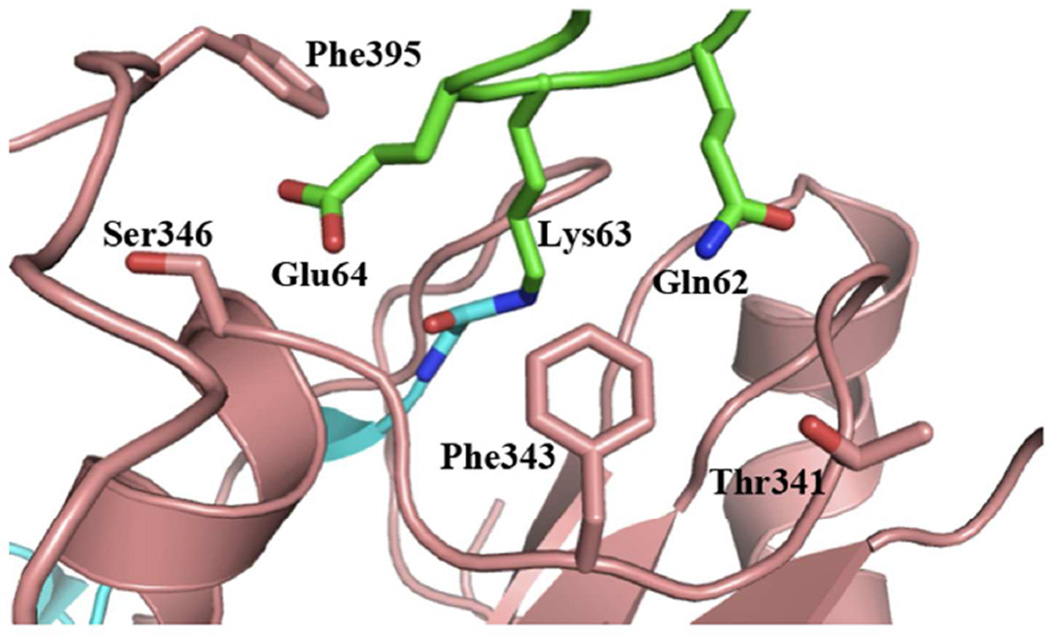
Modeling of the catalytic domain of AMSH onto the structure of AMSH-LP bound to Lys63-linked diubiquitin revealed four residues within AMSH (Thr341, Phe343, Ser346, and Phe395) that could determine its specificity for Lys63-linked polyubiquitin chains by recognizing the tri-peptide sequence motif Gln62-Lys63-Glu64 within the proximal ubiquitin, which encompasses the acceptor Lys63 and its two immediate flanking residues. Individual point mutants (to alanine) were generated and kinetic analysis was performed to probe the functional significance of these residues. Overall, the four resides within AMSH showed a drastic reduction in kcat with similar KM values compared to the wild-type enzyme, confirming their utmost importance to the enzyme’s catalytic mechanism (Figure 2 and Table 1), especially during the rate-determining step of isopeptide bond hydrolysis.
Figure 2.
Residues involved in proximal ubiquitin recognition within the catalytic domain of AMSH. AMSH residues are shown as pink sticks, proximal ubiquitin residues as green sticks, and the distal ubiquitin residues as cyan sticks.
Distal ubiquitin site
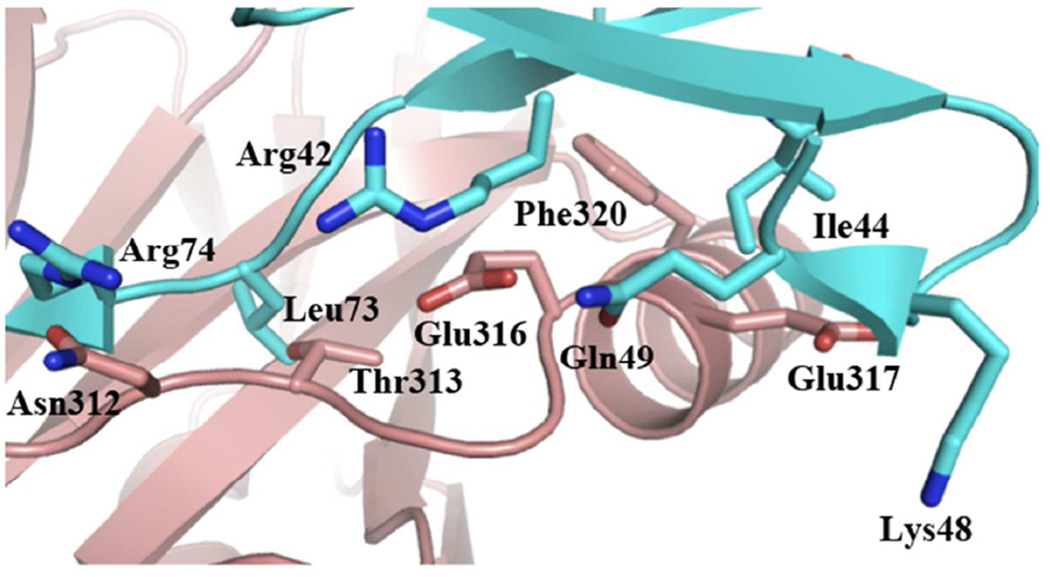
The distal site is where AMSH significantly differs from AMSH-LP in diubiquitin recognition. Three substitutions are found going from AMSH-LP to AMSH: an aspartate to asparagine, a methionine to a threonine, and a valine to glutamate. Two other important residues within the distal site are completely conserved between AMSH and AMSH-LP, a phenylalanine (Phe320, AMSH numbering) and a glutamate (Glu317) (Figure 3). The conserved Phe320 when mutated to alanine exhibited a 4-fold reduction in kcat and 3-fold increase in KM, whereas, the Glu317Ala mutant exhibits somewhat similar activity to the wild type, with only a modest 2-fold reduction in kcat (Table 1).
Figure 3.
Residues involved in distal ubiquitin recognition within the catalytic domain of AMSH. AMSH residues are shown as pink sticks, proximal ubiquitin residues as green sticks, and the distal ubiquitin residues as cyan sticks.
Moving forward, individual point mutations of the three substitutions between AMSH and AMSH-LP revealed some interesting results. Mutating Asn312 to alanine yielded only an approximate 3-fold reduction in kcat (Table 1). Surprisingly, a qualitative diubiquitin cleavage assay revealed that Thr313Ala was apparently more active than the wild type (unpublished data), however, detailed kinetic analysis showed simply an approximate 2-fold increase in kcat, with a ~3-fold loss in KM (Table 1).
Mutating the Glu316 to alanine proved to cause the most significant change in enzymatic activity amongst the distal site residues. Glu316Ala showed a substantial 74-fold reduction in kcat (Table 1). This kcat effect differs strikingly from the distal site residues of AMSH-LP whose mutation to Ala showed a loss primarily in KM (22). The effect of the glutamate mutation in AMSH mirrors that of residues from the proximal site. Inspection of the AMSH-diubiquitin model reveals that Glu316 is within hydrogen-bonding distance from two distal ubiquitin residues: Arg42 is within 3.1Å (εN of Arg42 and εO of Glu316), and Gln49 is within 2.8Å, if the side chain is flipped by 180° (Figure 3). A significant loss in kcat but not in KM is consistent with these hydrogen-bonding interactions contributing to the stabilization of the transition state, perhaps by playing a role in orienting the scissile peptide bond for nucleophilic attack.
Kinetic and thermodynamic characterization of the effect of the MIC-CAP-associated mutation, Thr313Ile
To better understand the molecular basis of the MIC-CAP syndrome, the Thr313Ile (T313I) mutant was generated and analyzed both for its catalytic activity towards Lys63- diubiquitin and its thermodynamic stability. In the absence of a structure, a modeled AMSH-diubiquitin structure suggests that the side-chain hydroxyl from threonine is hydrogen bonded to the backbone NH group of Leu73 in the distal ubiquitin (Fig. S1). The T313I mutant was found to suffer a 6-fold reduction in kcat, with a comparable KM (Table 1). In terms of its thermodynamic stability, T313I was somewhat less stable than the wild type with a ΔGH20 of 2.9 kcal.mol−1 compared to 3.6 kcal.mol−1 for the wild type (Fig. S2 and Supplementary Table 1). This result indicates that the reduced catalytic activity of the mutant could lead to a loss of function of AMSH translating into the disease state.
Biophysical characterization of ubiquitin binding to the catalytic domain of AMSH
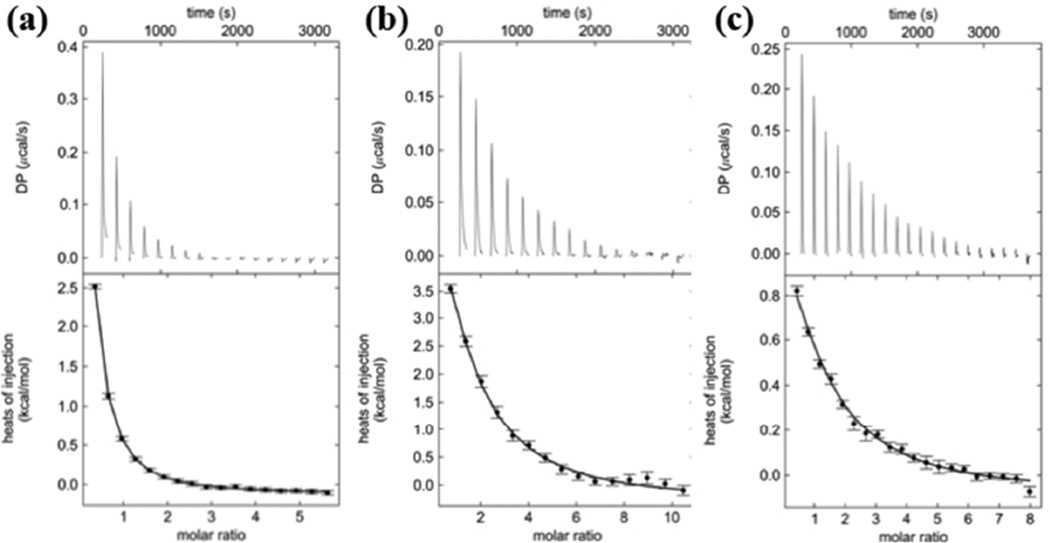
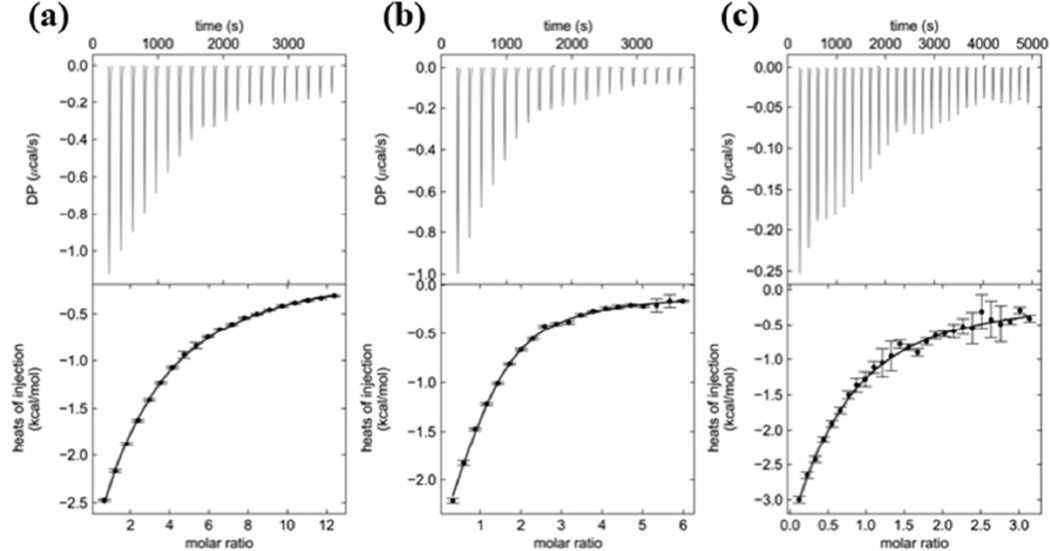
Mutational and kinetic analyses prompted us to seek a better understanding of AMSH-ubiquitin complex formation in solution. Using isothermal titration calorimetry (ITC), we analyzed the binding of Lys63-linked diubiquitin and the catalytic domain of AMSH (AMSH 219–424E280A, an inactive mutant to ensure diubiquitin is not hydrolyzed), and obtained an equilibrium dissociation constant (KD) of 19 ± 4 µM (Figure 4b and Table 2). As a control, ubiquitin and the catalytic domain of AMSH was analyzed and it was determined that it binds AMSH with similar affinity of 19 ± 3 µM (Figure 4a and Table 2). Both sedimentation velocity and sedimentation equilibrium experiments using analytical ultracentrifugation (AUC) confirmed the ITC results (Fig. S3, Fig. S4, and Supplementary Table 2). Almost identical binding affinities between diubiquitin and ubiquitin to the catalytic domain of AMSH suggest that there is only one binding site for ubiquitin. To probe which ubiquitin binding site is used, another ITC experiment was done with AMSHPhe320Ala (Phe320 at the distal site is mutated to Ala) and ubiquitin. We observed a ~4-fold decrease in affinity (KD of 81 ± 15 µM) (Figure 4c and Table 2), consistent with what was observed from kinetics, suggesting that the distal ubiquitin makes the most significant contribution to diubiquitin binding, and the single binding site observed in our ITC experiments with ubiquitin corresponds to binding at the distal site. These data suggest that AMSH alone cannot discriminate between its polyubiquitin substrate and its ubiquitin product.
Figure 4.
Isothermal titration calorimetry (ITC) thermograms of ubiquitin binding to the catalytic domain of AMSH. (a) ITC thermogram of ubiquitin binding to the catalytic domain of AMSH revealing a KD of 19 ± 3 µM. (b) ITC thermogram of Lys63-linked diubiquitin binding to the catalytic domain of AMSH revealing a KD of 19 ± 4 µM. (c) ITC thermogram of a Phe320Ala mutant of the catalytic domain of AMSH binding to ubiquitin revealing a KD of 81 ± 15 µM.
Table 2.
Thermodynamic Parameters Deduced from ITC Data
| Protein | Titrant | KD (µM) | ΔH (kcal.mol−1) | ΔS (cal.mol−1.K−1) | N |
|---|---|---|---|---|---|
| AMSH | Ub | 19 ± 3 | 19.3 ± 7.8 | 86.4 | 1 |
| AMSH | DiUb | 19 ± 4 | 13.7 ± 1.8 | 67.7 | 1 |
| AMSHF320A | Ub | 81 ± 15 | 2.6 ± 0.4 | 27.5 | 1 |
| AMSH | SH3 | 1.4 ± 0.04 | −15.1 ± 0.1 | −23.7 | 1 |
| AMSH | UIM-SH3 | 1.9 ± 0.1 | −15.8 ± 0.1 | −26.9 | 1 |
| AMSHK238A | SH3 | NAa | NAa | NAa | NAa |
| AMSHK238T | SH3 | NAa | NAa | NAa | NAa |
| SH3 | Ub | 62 ± 7 | −4.1 ± 0.2 | 5.3 | 1 |
| UIM-SH3 | Ub | 273 ± 16 | −18.2 ± 1.0 | −44.7 | 2 |
| UIM-SH3 | DiUb | 54 ± 21 | −13.0 ± 6.6 | −23.9 | 1 |
No observed binding at 50 µM of enzyme and 1 mM of UIM-SH3
The intact minimal STAM construct UIM-SH3 is necessary for AMSH activation
The I44 patch, a hydrophobic surface centered on the Ile44 residue in ubiquitin, is ubiquitously used by proteins that specifically bind to ubiquitin, including DUBs. Inspection of our structural model representing AMSH-diubiquitin complex reveals that the I44 patch of the distal ubiquitin is satisfied, with the Ile44 residue engaged in van der Waals interaction with Phe320; however, Ile44 of the proximal ubiquitin is unoccupied (Fig. S5). Looking at the domain structure of the ESCRT-0 member, STAM, one finds a UIM (ubiquitin-interacting motif) N-terminally adjacent to its SH3 domain (Fig. S6). We sought to understand if the UIM, separated from the SH3 domain by a short linker, could act as an adaptor to AMSH by interacting with the proximal ubiquitin while AMSH engages the distal one. To probe this, we used a combination of biophysical techniques and biochemical assays to assess three individual events: (1) AMSH recruitment to STAM via the SH3 domain and a longer STAM segment in which the UIM is fused to the SH3 domain (UIM-SH3), (2) ubiquitin binding to UIM-SH3, and finally, (3) the ternary complex of the catalytic domain of AMSH, UIM-SH3, and Lys63-linked diubiquitin.
AMSH binds to the SH3 domain of STAM2
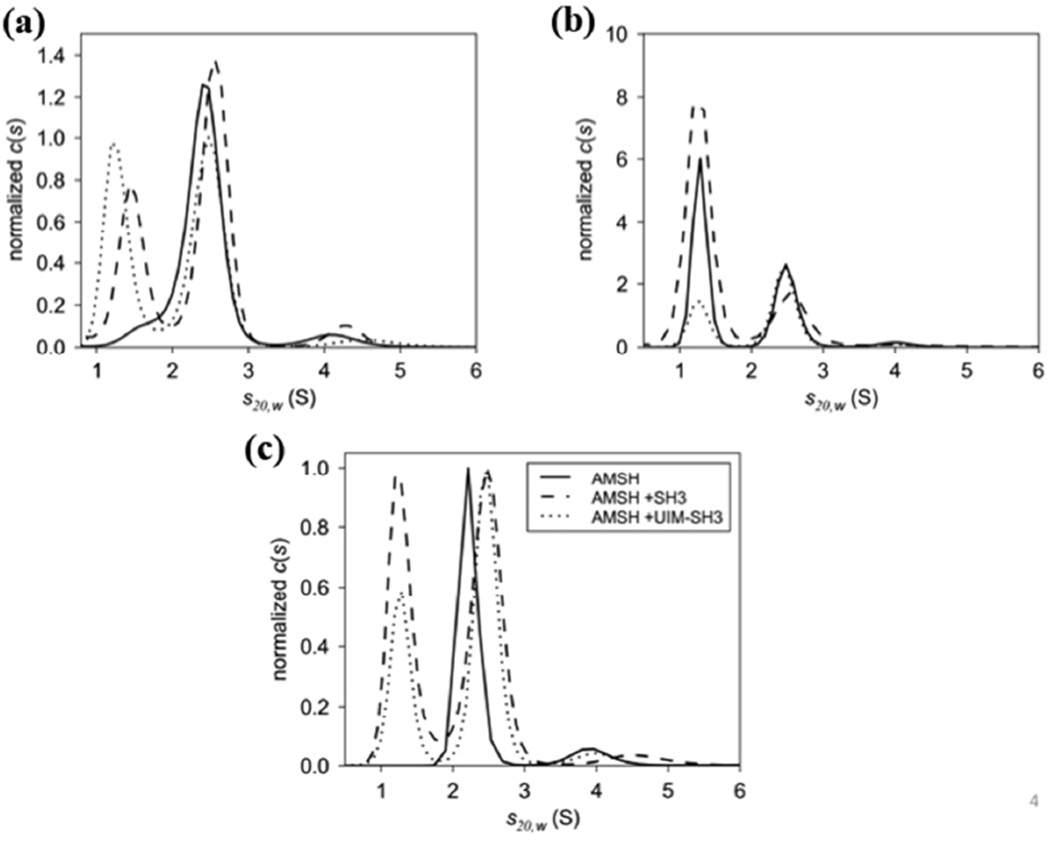
To confirm that we have the minimal domains required for the AMSH-STAM interaction we carried out ITC and AUC experiments. We determined that the SH3 domain of STAM binds the catalytic domain of AMSH (AMSH219E280A) with a KD of 1.4 ± 0.04 µM (Table 2 and Fig. S7). Using the longer UIM-SH3 construct, we obtained an identical KD of 1.9 ± 0.1 µM (Table 2 and Fig. S7), both of which are consistent with a previous ITC study, which showed that a peptide representing the SBM of AMSH binds the SH3 domain with 7 µM affinity (14). Using an orthogonal and complementary technique, we confirmed complex formation by sedimentation velocity experiments using AUC and determined that the catalytic domain of AMSH forms a 1:1 complex. The respective s20,w values of 2.5S and 2.6S for the SH3 domain and UIM-SH3 (Figure 5a and b) suggest that the UIM has no role in AMSH recruitment to STAM, as expected.
Figure 5.
c(s) distributions of the catalytic domain of AMSH binding to (a) the SH3 domain of STAM and (b) UIM-SH3. Three concentration series were used to assess the formation of the AMSH:SH3 and AMSH:UIM-SH3 complexes revealing 1:1 complexes at 2.5S and 2.6S, respectively. Excess SH3 and UIM-SH3 are present at 1.3S. The data for both c(s) distributions were normalized to the peak area of the complexes. (C) Overlay of AMSH, AMSH:SH3, and AMSH:UIM-SH3 revealing changes in s-value of the AMSH:SH3 and AMSH:UIM-SH3 complexes compared to AMSH alone at 2.2S. The c(s) distributions were normalized to the peak area of the complexes.
Both SH3 domain and UIM of STAM bind ubiquitin independently
Secondly, we characterized ubiquitin binding to the UIM of STAM. Since UIMs are only 30-residue domains, much too small for bacterial expression, we used UIM-SH3 to investigate UIM-ubiquitin binding by ITC. Somewhat surprisingly, we found that both the UIM and SH3 domains bind ubiquitin independently. This observation was based on two pieces of evidence. (1) The UIM-SH3 construct binds ubiquitin with a KD of 273 ± 16 µM, in agreement with previous biosensor measurements of a STAM-derived UIM peptide binding to ubiquitin that provided a KD of 182 µM (39). (2) Interestingly, our measurement of SH3-ubiquitin binding by ITC resulted in a KD of 62 ± 7 µM (Figure 6b and Table 2). It has been shown previously that a subset of SH3 domains bind ubiquitin (40, 41), and a recent study using NMR titration experiments showed that the SH3 domain of STAM does in fact bind ubiquitin, and that this interaction can be competed off by USP8 binding to the SH3 domain of STAM (42). Taken together, these data seem to indicate that both the SH3 domain and the UIM bind ubiquitin independently, with the former having higher affinity than the latter, which would explain the overall KD of 273 ± 15 µM obtained as the binding affinity of the UIM-SH3 construct for ubiquitin. It is possible that the two binding events corresponding to the two binding sites on UIM-SH3 have similar enthalpy of binding, and with binding affinities not drastically different between them, the ITC experiment is unable to resolve them distinctly.
Figure 6.
ITC thermograms of ubiquitin binding to UIM-SH3 of STAM. (a) Thermogram of ubiquitin binding to UIM-SH3 revealing a KD of 273 ± 16 µM. (b) Thermogram of ubiquitin binding to the SH3 domain of STAM revealing a KD of 62 ± 7 µM. (c) Thermogram of Lys63- linked diubiquitin binding to UIM-SH3 revealing a KD of 54 ± 21 µM.
Alternatively, it is possible that the UIM and SH3 domain fold onto each other generating a weaker interface for ubiquitin than either of them alone. This seems unlikely since UIM-SH3 binds to Lys63-linked diubiquitin with a KD of 54 ± 20 µM (Figure 6c and Table 2), an affinity higher than that of UIM-SH3 for ubiquitin. These results are consistent with the principle of avid binding of polyubiquitin chains at ESCRT-0, thus indicating that both the UIM and SH3 domain in UIM-SH3 are accessible for ubiquitin binding.
UIM and SH3 domains are necessary for stimulating the activity of AMSH
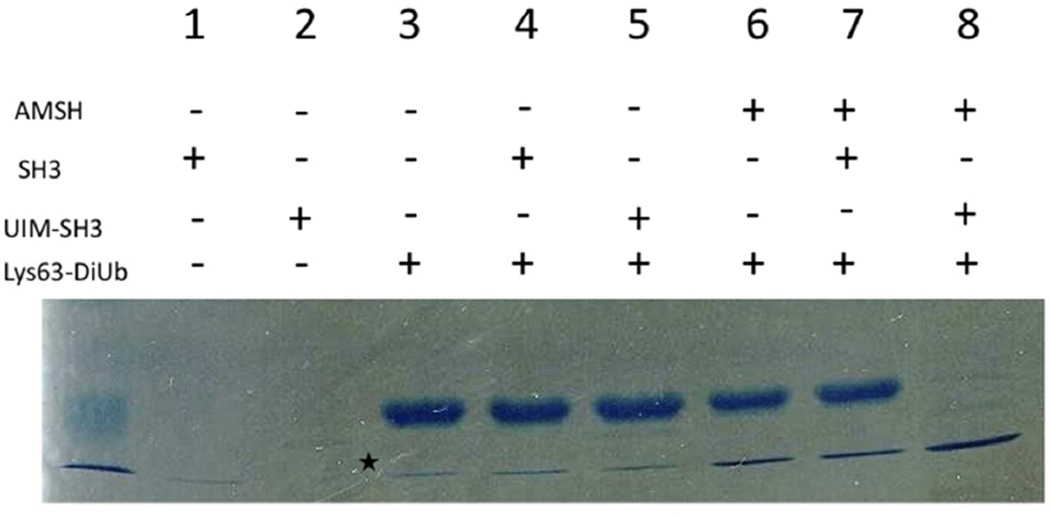
Using the catalytic domain of AMSH, UIM-SH3 and Lys63-linked diubiquitin, we attempted to recapitulate AMSH recruitment to ESCRT-0 in vitro. We carried out a Lys63- diubiquitin DUB cleavage assay with the AMSH:UIM-SH3 complex. The initial experiment comparing the enzyme’s activity alone and in the presence of the SH3 domain and then, UIMSH3, revealed a remarkable difference in DUB activity of AMSH. In the presence of UIM-SH3, it turned over nearly all of the Lys63-diubiquitin to ubiquitin, whereas, AMSH alone or in the presence of simply the SH3 domain had a significant amount of diubiquitin remaining, suggesting a stimulatory role for UIM-SH3 (Figure 7).
Figure 7.
DUB activity assay by monitoring diubiquitin cleavage. SDS-PAGE gel comparing the activity of the catalytic domain of AMSH alone and in the presence of the SH3 domain and UIM-SH3 of STAM. Only the lane with UIM-SH3 reveals activation. The asterisk indicates ubiquitin contamination in the diubiquitin purification.
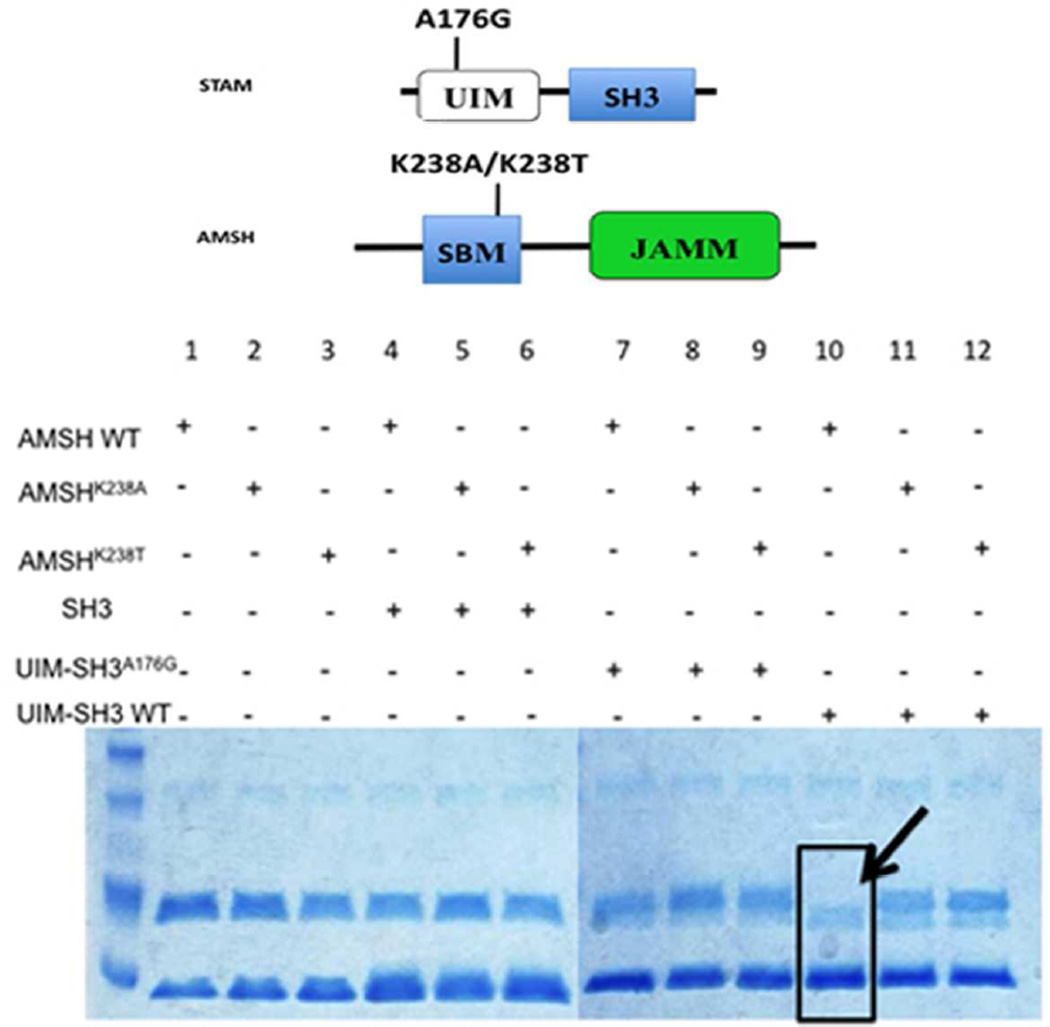
Diving deeper into the mechanism of activation, a similar in vitro assay was performed, this time using two SBM, and one UIM mutant versions of the UIM-SH3 construct (Figure 8). Two individual point mutations within the SBM of AMSH were introduced (Lys238Ala and Lys238Thr) to obliterate the SH3-SBM interaction. Lys238 is a completely conserved residue in the canonical SBM motif known to bind SH3 domains, mutating this to threonine made AMSH look like AMSH-LP in terms of its SBM. AMSH-LP has the conserved set of residues within its SBM, except the critical Lys replaced by Thr. We confirmed that there was no binding between the AMSH SBM mutants and the SH3 domain using ITC (Fig. S8). Secondly, we introduced a mutation within the UIM of UIM-SH3 (Ala176Gly) to interrupt ubiquitin binding (Figure 8). The Ala to Gly mutation has been shown previously to cause significant reduction in ubiquitin binding (39). The diubiquitin cleavage reactions were performed at 37°C for 15 minutes using 1 µM enzyme, 20 µM Lys63-linked diubiquitin as the substrate, and 5µM STAM binding partner (SH3, UIMA176GSH3, or UIM-SH3). SDS-PAGE analysis revealed that only in the presence of the wild-type enzyme and UIM-SH3 is diubiquitin completely hydrolyzed to ubiquitin, hence, an intact SBM-SH3 interaction and a functional UIM are necessary for AMSH activation (Figure 8).
Figure 8.
Catalytic activation of AMSH in presence of UIM-SH3. (a) Domain diagram of the minimal AMSH and STAM proteins, indicating the locations of the introduced mutations. (b) SDS-PAGE investigating the effects of mutants on the catalytic activation of AMSH. Only in the presence of the wild-type enzyme and wild-type UIM-SH3 is the activity of AMSH enhanced, indicated by complete disappearance of the diubiquitin substrate (black arrow). All lanes have Lys63-linked diubiquitin.
Furthermore, we wanted to understand this activation phenomenon in more detail, in terms of kcat and KM. To this end, we carried out another kinetic assay in which the catalytic domain of AMSH was pre-incubated in the presence of 20-fold excess UIM-SH3 to ensure that equilibrium favors the formation of the AMSH-UIM-SH3 complex. We saw 6-fold activation in AMSH in the presence of UIM-SH3, contributed by a somewhat greater change in kcat than in KM (Supplementary Table 3). The kcat effect is not entirely surprising because the UIM interacts with the proximal ubiquitin, and as we have shown in our mutational and kinetic analysis, the proximal site plays a significant role in properly aligning the isopeptide bond within the active site of AMSH, as determined by the significant loss in kcat upon mutating the residues involved in binding.
DISCUSSION
AMSH is one of the two DUBs recruited to the human ESCRT machinery to regulate the endosomal-lysosomal degradation pathway (9). It is a JAMM family DUB,(1) having exquisite specificity for recognizing and cleaving Lys63-linked polyubiquitin chains (9), which serve as signals for ESCRT-mediated sorting to lysosome. However, a clearly defined role for AMSH has yet to be elucidated. A homologous protein, AMSH-LP, sharing 54% identity and 73% sequence similarity (9, 23), of the same family of DUBs, has the same ubiquitin linkage specificity, and from our previous work, is structurally almost identical in its catalytic domain to AMSH (25). Even though the catalytic domains of AMSH and AMSH-LP are structurally nearly identical, we showed that AMSH is significantly less stable than AMSH-LP, and consequently perhaps conformationally more plastic, and the enzymes differ in their ubiquitin recognition (25). As a result of these differences, we sought to further investigate AMSH kinetically and biophysically to advance our understanding of the enzyme with a view to understanding its role in the context of the ESCRT machinery.
Our kinetic analysis using site-directed mutagenesis of conserved residues in the proximal ubiquitin binding site has shown that the specificity of AMSH for Lys63-linked polyubiquitin chains arises from its recognition of the proximal ubiquitin, similar to the case of AMSH-LP (22). Lys63-linked chain specificity for AMSH plays a significant role in understanding its function as an ESCRT-DUB. Since Lys63-linked polyubiquitin chains are targeting signals for ESCRT-mediated degradation, the specific DUB activity of AMSH may play a central role in the persistent functionality of the ESCRT machinery. Mutation of the proximal ubiquitin binding residues causes drastic reduction in kcat, suggesting that recognition of the tri-peptide sequence motif Gln62-Lys63-Glu64 within the proximal ubiquitin plays a significant role in the ability of AMSH to cleave Lys63-linked polyubiquitin chains. Recognition of Lys63 isopeptide bond and its two flanking residues in the proximal ubiquitin would mean that AMSH could only efficiently hydrolyze bonds between successive ubiquitins in a polymeric chain, and not the last ubiquitin directly attached to a protein receptor (the cargo). This impediment toward completely deubiquitinating a ubiquitinated receptor could have multiple functional implications (discussed below). At the outset, it calls into question the functional role of AMSH when it is recruited to ESCRT-III, where complete deconjugation of a ubiquitinated cargo is the absolute desire, since ubiquitin will otherwise end up in intra-luminal vesicles (ILVs) attached to the cargo and will be subsequently degraded in the lysosome. It seems unlikely that AMSH can have a significant catalytic role with respect to hydrolyzing the last ubiquitin attached to the cargo, yet, AMSH binds to ESCRT-III component CHMP3 with relatively high affinity.
Our data on mutational analysis of the distal ubiquitin binding residues offer some interesting insights. Of the three residues different between AMSH and AMSH-LP, two of them, Glu316 and Thr313, contribute significantly to catalysis playing different roles than the corresponding residues in AMSH-LP. The AMSH residue Glu316 contributes to stabilization of the transition state as indicated by the largely kcat effect when it is mutated to Ala, in contrast to mostly a KM effect observed when the corresponding residues Val in AMSH-LP mutated to Ala (22). Thr313 in our AMSH-diubiquitin model is seen making a hydrogen bonding contact with the backbone NH group of Leu73 of the distal ubiquitin using its side-chain hydroxyl group and its methyl group is engaged in van der Waals contact with the aliphatic side groups of Leu73 of ubiquitin. Its substitution with Ile as seen in children with the MIC-CAP syndrome is expected to preserve the van der Waals contact but loose the hydrogen bond. Our data show that the substitution of the Thr to an Ile has minimal effect on protein folding and stability, but results in a significantly reduced catalytic efficiency. Going from AMSH to AMSH-LP, the Thr residue is replaced by Met, which could contribute only van der Waals interaction with the substrate. Thus, it appears that the hydrogen-bonding interaction of Thr in AMSH has a unique role whose loss leads to a dramatic effect resulting in a loss of function substantial enough to cause the disease. Overall, these results indicate that subtle differences between very similar enzymes can have profound functional effects.
We found the minimal domain of STAM that is required to stimulate the activity of AMSH. Previous work has shown that STAM has a role in AMSH activation towards Lys63 polymeric chains (13, 14); however, these studies were not able to fully elucidate the mechanism of activation. Our study begins to divulge the mechanism underlying activation. This work suggests a simple model invoking simultaneous recognition of two ubiquitin groups in a polyubiquitin chain by AMSH and the UIM of STAM could explain the catalytic activation of the DUB. The UIM of STAM, separated from the SH3 domain by a short linker, could act as an adaptor for AMSH by interacting with the proximal ubiquitin, while AMSH engages the distal one. Such an arrangement would create a more extensive binding interface for diubiquitin in the AMSH:STAM complex than in the enzyme alone, causing catalytic activation. It appears that such activation is necessary since, as our ITC data show, AMSH has no preference for binding to Lys63-linked diubiquitin, therefore by extrapolation to Lys63-linked polyubiquitin chains over the ubiquitin product.
Prior to activation, AMSH is in a more latent state, but when it is recruited to STAM its full activity is unveiled. AMSH is known to have diverse subcellular localization profiles. Perhaps the free form of the enzyme needs to be in a less-active state so as not to hydrolyze the Lys63 chains that are present in the cytosol other than endosomes. Once it is recruited to the endosomes, its true activity comes alive, as seen by the 6-fold enhancement in activity upon binding to the STAM derived UIM-SH3 construct. A significant implication of this mechanism of activation is that the activation will be absent when AMSH is trying to cleave the last ubiquitin attached to the cargo. While efficiently cleaving between two ubiquitin groups in a Lys63-linked polyubiquitin chain, AMSH might show a severe impediment in hydrolyzing the last ubiquitin attached directly to a cargo, on account of two factors: (1) its high specificity for the Lys63-linked chain between two ubiquitins, which in turn would make it a poor enzyme when ubiquitin is attached to a non-ubiquitin protein, the cargo; and, (2) the lack of an activation effect when cleaving ubiquitin attached to a non-ubiquitin moiety.
Finally, bringing all our data together, we can envision a mechanism for recruitment and activation for AMSH that will ultimately define a function for the enzyme. ESCRT-0 has the defined function of ubiquitinated cargo clustering, capable of harboring up to eight ubiquitin moieties at a time (43, 44), which now, with the addition of the SH3 domain could be ten ubiquitins. Our ITC data show that the SH3 domain can actually bind ubiquitin tighter than the UIM. Subsequently, AMSH is recruited to STAM. The AMSH-SH3 binding affinity is stronger than SH3-ubiquitin, making it possible for AMSH to effectively displace ubiquitin from the SH3 domain (the binding interface on SH3 domain for the two proteins show substantial overlap) (42) leading to its recruitment to ESCRT-0. With the UIM from STAM acting as an adaptor to the enzyme, facilitating enzyme activity enhancement, AMSH begins to efficiently disassemble the polyubiquitin chain attached to the cargo. Deubiquitination of the chain will continue until the last ubiquitin directly attached to the cargo. Thus, the recruitment of AMSH at ESCRT-0 will lead to substantial chain trimming but not complete deconjugation of ubiquitin from the cargo. As discussed in the next paragraph, this would promote the cargo’s passage from ESCRT-0 to ESCRT-I and subsequent complexes.
Our proposed mechanism defines AMSH as the DUB that facilitates cargo passage from ESCRT-0 onto the next complex. This idea is supported by previous data that shows that avidly bound ubiquitin chains comprise of a binding affinity of ~20 µM affinity (45), whereas, ESCRT-I subunit, UBAP1, binds ubiquitin anywhere from 70–140 µM (46). The binding affinity of ubiquitin at ESCRT-0 needs to be reduced at least 5–10-fold in order for cargo destined for lysosomal degradation to be transferred to ESCRT-I. When going from Lys63-linked tetraubiquitin to diubiquitin, ESCRT-0 has a ~6-fold reduced affinity, and a remarkable, 46-fold reduction in affinity for ubiquitin (45). Therefore, we presume that because of the specificity and activation of AMSH, this would be the enzyme that would be most suited for promoting cargo passage, all in support of an idea proposed previously (47). By occupying the binding site on ESCRT-0, AMSH will serve to keep USP8 off the initial ESCRT complex. The recruitment of USP8 at ESCRT-0 would be detrimental to cargo’s passage to lysosome because USP8 has no hindrance in complete deconjugation. We suggest that USP8’s role is specifically at the ESCRTIII level where complete deconjugation is desired.
The main conclusion from our study implies that loss of function mutations in AMSH would lead to impairment of ubiquitin-dependent sorting to lysosome via the ESCRT pathway. One possible outcome of this impairment is accumulation of ubiquitinated proteins (cargo). Indeed, patient cell lines with AMSH mutation show accumulation of aggregated ubiquitin-protein conjugates (16). Furthermore, impairment of ESCRT-mediated endocytic sorting to lysosome is expected to cause hyperactivation of signaling across lipid bilayer. As suggested by the authors of the paper describing MIC-CAP mutations, the capillary abnormalities associated with the syndrome could be a consequence of hyperactive RAS-MAPK signaling induced in humans by impaired AMSH function (16). However, because AMSH has several roles within the cell it would be difficult to assign loss of function in one pathway as the exclusive molecular basis of MIC-CAP syndrome.
In summary, using a combination of biochemical and biophysical studies, guided by a structural model, we are able to learn many important aspects of AMSH: (1) The T313I mutation underlying the MIC-CAP syndrome leads to a significant loss of catalytic activity owing to loss of a hydrogen-bonding interaction with ubiquitin. (2) Recognition of proximal ubiquitin contributes significantly to catalysis. (3) Activation of AMSH is enabled by facile, simultaneous binding to two ubiquitin groups in a polyubiquitin substrate, one by the catalytic domain of the DUB (binding to the distal ubiquitin) and the other (the proximal ubiquitin) by the UIM from STAM. (4) Taken together, the above two points strongly indicate that AMSH will suffer a severe loss of catalytic efficiency when cleaving the last ubiquitin attached to cargo compared to a Lys63-linked polyubiquitin chain substrate. These studies provide biochemical and biophysical evidence in support of a hypothesis which postulates that AMSH is recruited to the initial ESCRT complex to facilitate transfer of cargo from one ESCRT member to the next, but not to completely deubiquitinate it (9). Its recruitment therefore would facilitate cargo shuttling rather than release from ESCRT and subsequent recycling back to the plasma membrane.
Supplementary Material
ACKNOWLEDGMENT
We would also like to acknowledge Dr. Myung-il Kim for his help in optimization of the Lys63- linked diubiquitin chain synthesis. Financial support from the American Heart Association Predoctoral fellowship (12PRE12060249) to C.W. Davies is gratefully acknowledged. Finally, financial support from the National Institutes of Health (1R01RR026273) to C. Das is gratefully acknowledged.
ABBREVIATIONS
- AMSH
associated molecule with a Src homology 3 domain of STAM
- AMSH-LP
AMSH-like protein
- AUC
analytical ultracentrifugation
- CD
circular dichroism
- DUBs
deubiquitinating enzymes
- ESCRT
endosomal sorting complexes required for transport
- ITC
isothermal titration calorimetry
- STAM
signal transducing adaptor molecule
- MIC-CAP
microcephaly malformation syndrome
- UBPY/USP8
ubiquitin specific protease 8.
Footnotes
Supporting Information Available
Cartoon representations of the MIC-CAP-associated mutation and a model of AMSH bound to Lys63-linked diubiquitin. A fraction unfolded GdHCl melting curve and stability data table. ITC thermograms of AMSH binding to the SH3 and UIM-SH3, and AMSHK238T with UIM-SH3. Sedimentation velocity and equilibrium data and data table. AMSH and STAM domain diagram. AMSH activation data table. This material is available free of charge via the Internet at http://pubs.acs.org
REFERENCES
- 1.Maytal-Kivity V, Reis N, Hofmann K, Glickman MH. MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC Biochem. 2002;3:28. doi: 10.1186/1471-2091-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Komander D. Mechanism, specificity and structure of the deubiquitinases. Subcell Biochem. 2010;54:69–87. doi: 10.1007/978-1-4419-6676-6_6. [DOI] [PubMed] [Google Scholar]
- 3.Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 4.Wilkinson KD. DUBs at a glance. J Cell Sci. 2009;122:2325–2329. doi: 10.1242/jcs.041046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 6.Zhu P, Zhou W, Wang J, Puc J, Ohgi KA, Erdjument-Bromage H, Tempst P, Glass CK, Rosenfeld MG. A histone H2A deubiquitinase complex coordinating histone acetylation and H1 dissociation in transcriptional regulation. Mol Cell. 2007;27:609–621. doi: 10.1016/j.molcel.2007.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, Livingston DM, Greenberg RA. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;316:1198–1202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Avvakumov GV, Walker JR, Xue S, Finerty PJ, Jr, Mackenzie F, Newman EM, Dhe-Paganon S. Amino-terminal dimerization, NRDP1-rhodanese interaction, and inhibited catalytic domain conformation of the ubiquitin-specific protease 8 (USP8) J Biol Chem. 2006;281:38061–38070. doi: 10.1074/jbc.M606704200. [DOI] [PubMed] [Google Scholar]
- 9.Clague MJ, Urbe S. Endocytosis: the DUB version. Trends Cell Biol. 2006;16:551–559. doi: 10.1016/j.tcb.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 10.Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH. Membrane scission by the ESCRT-III complex. Nature. 2009;458:172–177. doi: 10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saksena S, Sun J, Chu T, Emr SD. ESCRTing proteins in the endocytic pathway. Trends Biochem Sci. 2007;32:561–573. doi: 10.1016/j.tibs.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 12.Roxrud I, Stenmark H, Malerod L. ESCRT & Co. Biol Cell. 2010;102:293–318. doi: 10.1042/BC20090161. [DOI] [PubMed] [Google Scholar]
- 13.McCullough J, Row PE, Lorenzo O, Doherty M, Beynon R, Clague MJ, Urbe S. Activation of the endosome-associated ubiquitin isopeptidase AMSH by STAM, a component of the multivesicular body-sorting machinery. Curr Biol. 2006;16:160–165. doi: 10.1016/j.cub.2005.11.073. [DOI] [PubMed] [Google Scholar]
- 14.Kim MS, Kim JA, Song HK, Jeon H. STAM-AMSH interaction facilitates the deubiquitination activity in the C-terminal AMSH. Biochem Biophys Res Commun. 2006;351:612–618. doi: 10.1016/j.bbrc.2006.10.068. [DOI] [PubMed] [Google Scholar]
- 15.Solomons J, Sabin C, Poudevigne E, Usami Y, Hulsik DL, Macheboeuf P, Hartlieb B, Gottlinger H, Weissenhorn W. Structural basis for ESCRT-III CHMP3 recruitment of AMSH. Structure. 2011;19:1149–1159. doi: 10.1016/j.str.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDonell LM, Mirzaa GM, Alcantara D, Schwartzentruber J, Carter MT, Lee LJ, Clericuzio CL, Graham JM, Jr, Morris-Rosendahl DJ, Polster T, Acsadi G, Townshend S, Williams S, Halbert A, Isidor B, David A, Smyser CD, Paciorkowski AR, Willing M, Woulfe J, Das S, Beaulieu CL, Marcadier J, Geraghty MT, Frey BJ, Majewski J, Bulman DE, Dobyns WB, O'Driscoll M, Boycott KM. Mutations in STAMBP, encoding a deubiquitinating enzyme, cause microcephaly-capillary malformation syndrome. Nat Genet. 2013;45:556–562. doi: 10.1038/ng.2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carter MT, Boycott KM. Microcephaly-capillary malformation syndrome: a story of rapid emergence of a new recognizable entity. Am J Med Genet A. 2011;155A:2078–2079. doi: 10.1002/ajmg.a.34149. [DOI] [PubMed] [Google Scholar]
- 18.Mirzaa GM, Paciorkowski AR, Smyser CD, Willing MC, Lind AC, Dobyns WB. The microcephaly-capillary malformation syndrome. Am J Med Genet A. 2011;155A:2080–2087. doi: 10.1002/ajmg.a.34118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Isidor B, Barbarot S, Beneteau C, Le Caignec C, David A. Multiple capillary skin malformations, epilepsy, microcephaly, mental retardation, hypoplasia of the distal phalanges: report of a new case and further delineation of a new syndrome. Am J Med Genet A. 2011;155A:1458–1460. doi: 10.1002/ajmg.a.34048. [DOI] [PubMed] [Google Scholar]
- 20.Suzuki S, Tamai K, Watanabe M, Kyuuma M, Ono M, Sugamura K, Tanaka N. AMSH is required to degrade ubiquitinated proteins in the central nervous system. Biochem Biophys Res Commun. 2011;408:582–588. doi: 10.1016/j.bbrc.2011.04.065. [DOI] [PubMed] [Google Scholar]
- 21.McCullough J, Clague MJ, Urbe S. AMSH is an endosome-associated ubiquitin isopeptidase. J Cell Biol. 2004;166:487–492. doi: 10.1083/jcb.200401141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sato Y, Yoshikawa A, Yamagata A, Mimura H, Yamashita M, Ookata K, Nureki O, Iwai K, Komada M, Fukai S. Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature. 2008;455:358–362. doi: 10.1038/nature07254. [DOI] [PubMed] [Google Scholar]
- 23.Kikuchi K, Ishii N, Asao H, Sugamura K. Identification of AMSH-LP containing a Jab1/MPN domain metalloenzyme motif. Biochem Biophys Res Commun. 2003;306:637–643. doi: 10.1016/s0006-291x(03)01009-x. [DOI] [PubMed] [Google Scholar]
- 24.Agromayor M, Martin-Serrano J. Interaction of AMSH with ESCRT-III and deubiquitination of endosomal cargo. J Biol Chem. 2006;281:23083–23091. doi: 10.1074/jbc.M513803200. [DOI] [PubMed] [Google Scholar]
- 25.Davies CW, Paul LN, Kim MI, Das C. Structural and thermodynamic comparison of the catalytic domain of AMSH and AMSH-LP: nearly identical fold but different stability. J Mol Biol. 2011;413:416–429. doi: 10.1016/j.jmb.2011.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakamura M, Tanaka N, Kitamura N, Komada M. Clathrin anchors deubiquitinating enzymes, AMSH and AMSH-like protein, on early endosomes. Genes Cells. 2006;11:593–606. doi: 10.1111/j.1365-2443.2006.00963.x. [DOI] [PubMed] [Google Scholar]
- 27.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. Analytical Ultracentrifugation in Biochemistry and Polymer Science. Royal Society of Chemistry. 1992:90–125. [Google Scholar]
- 28.Brown PH, Schuck P. Macromolecular size-and-shape distributions by sedimentation velocity analytical ultracentrifugation. Biophys J. 2006;90:4651–4661. doi: 10.1529/biophysj.106.081372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balbo A, Minor KH, Velikovsky CA, Mariuzza RA, Peterson CB, Schuck P. Studying multiprotein complexes by multisignal sedimentation velocity analytical ultracentrifugation. Proc Natl Acad Sci U S A. 2005;102:81–86. doi: 10.1073/pnas.0408399102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dam J, Schuck P. Sedimentation velocity analysis of heterogeneous proteinprotein interactions: sedimentation coefficient distributions c(s) and asymptotic boundary profiles from Gilbert-Jenkins theory. Biophys J. 2005;89:651–666. doi: 10.1529/biophysj.105.059584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dam J, Velikovsky CA, Mariuzza RA, Urbanke C, Schuck P. Sedimentation velocity analysis of heterogeneous protein-protein interactions: Lamm equation modeling and sedimentation coefficient distributions c(s) Biophys J. 2005;89:619–634. doi: 10.1529/biophysj.105.059568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Houtman JC, Brown PH, Bowden B, Yamaguchi H, Appella E, Samelson LE, Schuck P. Studying multisite binary and ternary protein interactions by global analysis of isothermal titration calorimetry data in SEDPHAT: application to adaptor protein complexes in cell signaling. Protein Sci. 2007;16:30–42. doi: 10.1110/ps.062558507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vistica J, Dam J, Balbo A, Yikilmaz E, Mariuzza RA, Rouault TA, Schuck P. Sedimentation equilibrium analysis of protein interactions with global implicit mass conservation constraints and systematic noise decomposition. Anal Biochem. 2004;326:234–256. doi: 10.1016/j.ab.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 35.Schuck P. On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Anal Biochem. 2003;320:104–124. doi: 10.1016/s0003-2697(03)00289-6. [DOI] [PubMed] [Google Scholar]
- 36.Keller S, Vargas C, Zhao H, Piszczek G, Brautigam CA, Schuck P. High-precision isothermal titration calorimetry with automated peak-shape analysis. Anal Chem. 2012;84:5066–5073. doi: 10.1021/ac3007522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pace CN. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol. 1986;131:266–280. doi: 10.1016/0076-6879(86)31045-0. [DOI] [PubMed] [Google Scholar]
- 38.Arnst JL, Davies CW, Raja SM, Das C, Natarajan A. High-throughput compatible fluorescence resonance energy transfer-based assay to identify small molecule inhibitors of AMSH deubiquitinase activity. Anal Biochem. 2013 doi: 10.1016/j.ab.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fisher RD, Wang B, Alam SL, Higginson DS, Robinson H, Sundquist WI, Hill CP. Structure and ubiquitin binding of the ubiquitin-interacting motif. J Biol Chem. 2003;278:28976–28984. doi: 10.1074/jbc.M302596200. [DOI] [PubMed] [Google Scholar]
- 40.He Y, Hicke L, Radhakrishnan I. Structural basis for ubiquitin recognition by SH3 domains. J Mol Biol. 2007;373:190–196. doi: 10.1016/j.jmb.2007.07.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stamenova SD, French ME, He Y, Francis SA, Kramer ZB, Hicke L. Ubiquitin binds to and regulates a subset of SH3 domains. Mol Cell. 2007;25:273–284. doi: 10.1016/j.molcel.2006.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lange A, Ismail MB, Riviere G, Hologne M, Lacabanne D, Guilliere F, Lancelin JM, Krimm I, Walker O. Competitive binding of UBPY and ubiquitin to the STAM2 SH3 domain revealed by NMR. FEBS Lett. 2012 doi: 10.1016/j.febslet.2012.07.047. [DOI] [PubMed] [Google Scholar]
- 43.Mayers JR, Fyfe I, Schuh AL, Chapman ER, Edwardson JM, Audhya A. ESCRT-0 assembles as a heterotetrameric complex on membranes and binds multiple ubiquitinylated cargoes simultaneously. J Biol Chem. 2010 doi: 10.1074/jbc.M110.185363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wollert T, Hurley JH. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature. 2010;464:864–869. doi: 10.1038/nature08849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ren X, Hurley JH. VHS domains of ESCRT-0 cooperate in high-avidity binding to polyubiquitinated cargo. EMBO J. 2010 doi: 10.1038/emboj.2010.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Agromayor M, Soler N, Caballe A, Kueck T, Freund SM, Allen MD, Bycroft M, Perisic O, Ye Y, McDonald B, Scheel H, Hofmann K, Neil SJ, Martin- Serrano J, Williams RL. The UBAP1 subunit of ESCRT-I interacts with ubiquitin via a SOUBA domain. Structure. 2012;20:414–428. doi: 10.1016/j.str.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hurley JH. Nipped in the bud: how the AMSH MIT domain helps deubiquitinate lysosome-bound cargo. Structure. 2011;19:1033–1035. doi: 10.1016/j.str.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.