Abstract
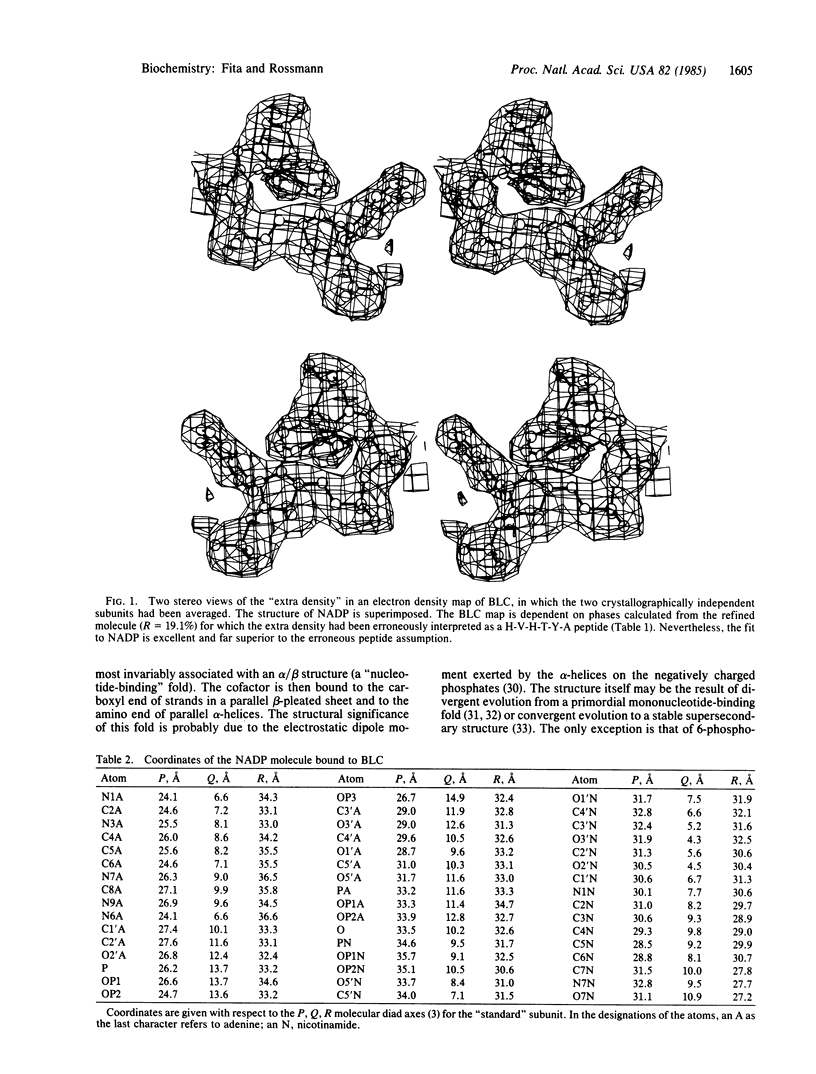
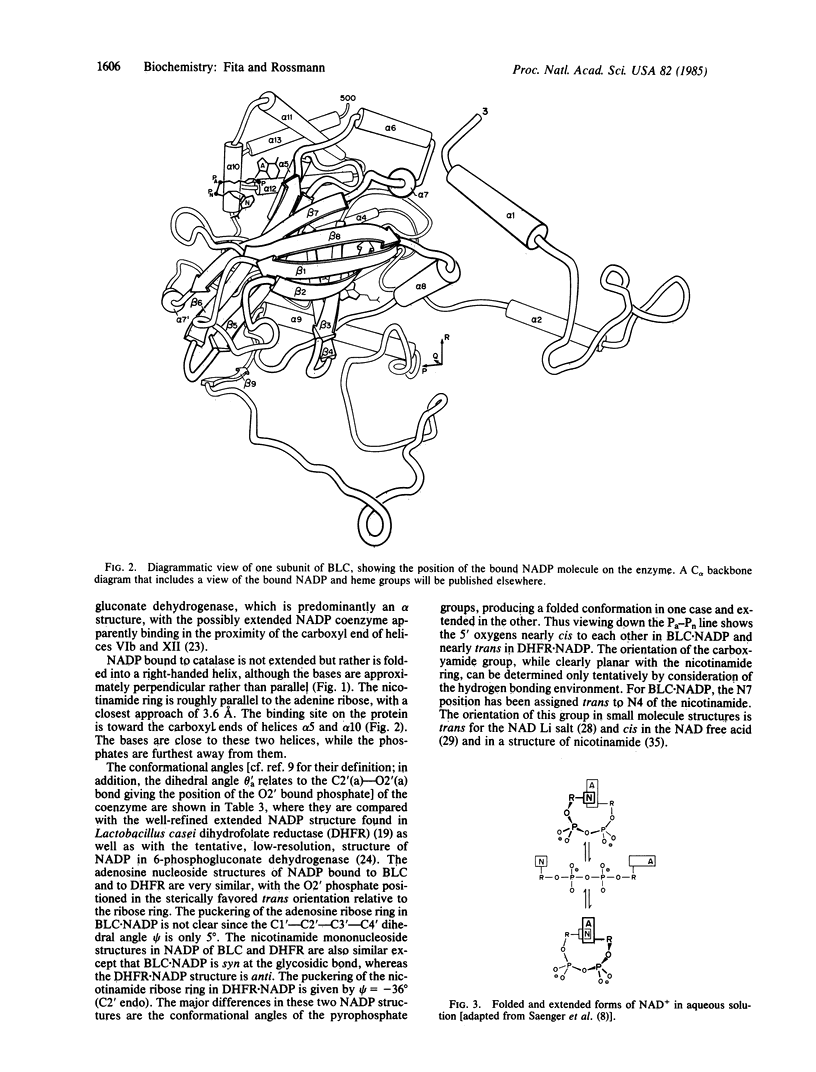
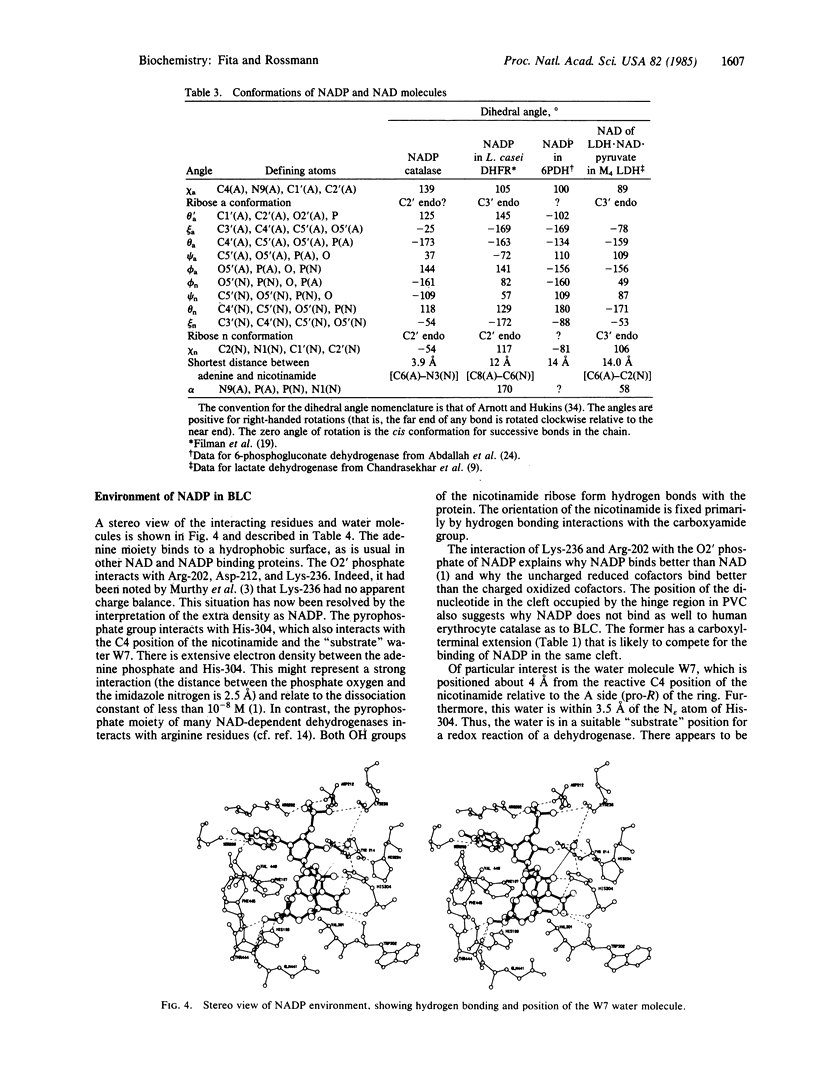
Beef liver and human erythrocyte catalases (EC 1.11.1.6) bind NADP tenaciously [Kirkman, H. N. & Gaetani, G. F. (1984) Proc. Natl. Acad. Sci. USA 81, 4343-4348]. The position of NADP on beef liver catalase corresponds to the carboxyl-terminal polypeptide hinge in Penicillium vitale fungal catalase, which connects the common catalase structure to the additional flavodoxin-like domain. In contrast to nearly all other known structures of protein-bound NADP, NAD, and FAD, the NADP molecule of beef liver catalase is folded into a right-handed helix and bound, in part, in the vicinity of the carboxyl end of two alpha-helices. A water molecule (W7) occupies a pseudosubstrate site close to the C4 position of the nicotinamide and is hydrogen bonded to His-304. Although the NADP and heme groups approach each other to within 13.7 A, there is no direct interaction. The function of the NADP remains a mystery.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abdallah M. A., Adams M. J., Archibald I. G., Biellmann J. F., Helliwell J. R., Jenkins S. E. Binding of coenzyme and substrate and coenzyme analogues to 6-phosphogluconate dehydrogenase from sheep liver. An X-ray study at 0.6 nm resolution. Eur J Biochem. 1979 Jul;98(1):121–130. doi: 10.1111/j.1432-1033.1979.tb13168.x. [DOI] [PubMed] [Google Scholar]
- Abdallah M. A., Biellmann J. F., Nordström B., Brändén C. I. The conformation of adenosine diphosphoribose and 8-bromoadenosine diphosphoribose when bound to liver alcohol dehydrogenase. Eur J Biochem. 1975 Jan 15;50(3):475–481. doi: 10.1111/j.1432-1033.1975.tb09885.x. [DOI] [PubMed] [Google Scholar]
- Adams M. J., Archibald I. G., Bugg C. E., Carne A., Gover S., Helliwell J. R., Pickersgill R. W., White S. W. The three dimensional structure of sheep liver 6-phosphogluconate dehydrogenase at 2.6 A resolution. EMBO J. 1983;2(6):1009–1014. doi: 10.1002/j.1460-2075.1983.tb01535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnott S., Hukins D. W. Conservation of conformation in mono and poly-nucleotides. Nature. 1969 Nov 29;224(5222):886–888. doi: 10.1038/224886a0. [DOI] [PubMed] [Google Scholar]
- Biesecker G., Harris J. I., Thierry J. C., Walker J. E., Wonacott A. J. Sequence and structure of D-glyceraldehyde 3-phosphate dehydrogenase from Bacillus stearothermophilus. Nature. 1977 Mar 24;266(5600):328–333. doi: 10.1038/266328a0. [DOI] [PubMed] [Google Scholar]
- Blumenstein M., Raftery M. A. Natural abundance 13C nuclear magnetic resonance spectra of nicotinamide adenine dinucleotide and related nucleotides. Biochemistry. 1973 Sep 11;12(19):3585–3590. doi: 10.1021/bi00743a001. [DOI] [PubMed] [Google Scholar]
- Catterall W. A., Hollis D. P., Walter C. F. Nuclear magnetic resonance study of the conformation of nicotinamide--adenine dinucleotide and reduced nicotinamide--adenine dinucleotide in solution. Biochemistry. 1969 Oct;8(10):4032–4036. doi: 10.1021/bi00838a021. [DOI] [PubMed] [Google Scholar]
- Cedergren-Zeppezauer E., Samama J. P., Eklund H. Crystal structure determinations of coenzyme analogue and substrate complexes of liver alcohol dehydrogenase: binding of 1,4,5,6-tetrahydronicotinamide adenine dinucleotide and trans-4-(N,N-dimethylamino)cinnamaldehyde to the enzyme. Biochemistry. 1982 Sep 28;21(20):4895–4908. doi: 10.1021/bi00263a011. [DOI] [PubMed] [Google Scholar]
- Chandrasekhar K., McPherson A., Jr, Adams M. J., Rossmann M. G. Conformation of coenzyme fragments when bound to lactate dehydrogenase. J Mol Biol. 1973 Jun 5;76(4):503–518. doi: 10.1016/0022-2836(73)90488-9. [DOI] [PubMed] [Google Scholar]
- Filman D. J., Bolin J. T., Matthews D. A., Kraut J. Crystal structures of Escherichia coli and Lactobacillus casei dihydrofolate reductase refined at 1.7 A resolution. II. Environment of bound NADPH and implications for catalysis. J Biol Chem. 1982 Nov 25;257(22):13663–13672. [PubMed] [Google Scholar]
- Grau U. M., Trommer W. E., Rossmann M. G. Structure of the active ternary complex of pig heart lactate dehydrogenase with S-lac-NAD at 2.7 A resolution. J Mol Biol. 1981 Sep 15;151(2):289–307. doi: 10.1016/0022-2836(81)90516-7. [DOI] [PubMed] [Google Scholar]
- Gronenborn A. M., Clore G. M., Jeffery J. An unusual conformation of NAD+ bound to sorbitol dehydrogenase? A time-dependent transferred nuclear Overhauser effect study. J Mol Biol. 1984 Feb 5;172(4):559–572. doi: 10.1016/s0022-2836(84)80023-6. [DOI] [PubMed] [Google Scholar]
- Hofsteenge J., Vereijken J. M., Weijer W. J., Beintema J. J., Wierenga R. K., Drenth J. Primary and tertiary structure studies of p-hydroxybenzoate hydroxylase from Pseudomonas fluorescens. Isolation and alignment of the CNBr peptides; interactions of the protein with flavin adenine dinucleotide. Eur J Biochem. 1980 Dec;113(1):141–150. [PubMed] [Google Scholar]
- Hol W. G., van Duijnen P. T., Berendsen H. J. The alpha-helix dipole and the properties of proteins. Nature. 1978 Jun 8;273(5662):443–446. doi: 10.1038/273443a0. [DOI] [PubMed] [Google Scholar]
- Holden H. M., Banaszak L. J. L-3-hydroxyacyl coenzyme A dehydrogenase. The location of NAD binding sites and the bilobal subunit structure. J Biol Chem. 1983 Feb 25;258(4):2383–2389. [PubMed] [Google Scholar]
- Kirkman H. N., Gaetani G. F. Catalase: a tetrameric enzyme with four tightly bound molecules of NADPH. Proc Natl Acad Sci U S A. 1984 Jul;81(14):4343–4347. doi: 10.1073/pnas.81.14.4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moras D., Olsen K. W., Sabesan M. N., Buehner M., Ford G. C., Rossmann M. G. Studies of asymmetry in the three-dimensional structure of lobster D-glyceraldehyde-3-phosphate dehydrogenase. J Biol Chem. 1975 Dec 10;250(23):9137–9162. doi: 10.2210/pdb1gpd/pdb. [DOI] [PubMed] [Google Scholar]
- Murthy M. R., Reid T. J., 3rd, Sicignano A., Tanaka N., Rossmann M. G. Structure of beef liver catalase. J Mol Biol. 1981 Oct 25;152(2):465–499. doi: 10.1016/0022-2836(81)90254-0. [DOI] [PubMed] [Google Scholar]
- Ohlsson I., Nordström B., Brändén C. I. Structural and functional similarities within the coenzyme binding domains of dehydrogenases. J Mol Biol. 1974 Oct 25;89(2):339–354. doi: 10.1016/0022-2836(74)90523-3. [DOI] [PubMed] [Google Scholar]
- Parthasarathy R., Fridey S. M. Conformational variability of NAD+ in the free and bound states: a nicotinamide sandwich in NAD+ crystals. Science. 1984 Nov 23;226(4677):969–971. doi: 10.1126/science.6239374. [DOI] [PubMed] [Google Scholar]
- Rao S. T., Rossmann M. G. Comparison of super-secondary structures in proteins. J Mol Biol. 1973 May 15;76(2):241–256. doi: 10.1016/0022-2836(73)90388-4. [DOI] [PubMed] [Google Scholar]
- Reid T. J., 3rd, Murthy M. R., Sicignano A., Tanaka N., Musick W. D., Rossmann M. G. Structure and heme environment of beef liver catalase at 2.5 A resolution. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4767–4771. doi: 10.1073/pnas.78.8.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann M. G., Moras D., Olsen K. W. Chemical and biological evolution of nucleotide-binding protein. Nature. 1974 Jul 19;250(463):194–199. doi: 10.1038/250194a0. [DOI] [PubMed] [Google Scholar]
- Schroeder W. A., Shelton J. R., Shelton J. B., Robberson B., Apell G., Fang R. S., Bonaventura J. The complete amino acid sequence of bovine liver catalase and the partial sequence of bovine erythrocyte catalase. Arch Biochem Biophys. 1982 Mar;214(1):397–421. doi: 10.1016/0003-9861(82)90044-3. [DOI] [PubMed] [Google Scholar]
- Schulz G. E., Schirmer R. H., Pai E. F. FAD-binding site of glutathione reductase. J Mol Biol. 1982 Sep 15;160(2):287–308. doi: 10.1016/0022-2836(82)90177-2. [DOI] [PubMed] [Google Scholar]
- Stura E. A., Zanotti G., Babu Y. S., Sansom M. S., Stuart D. I., Wilson K. S., Johnson L. N., Van de Werve G. Comparison of AMP and NADH binding to glycogen phosphorylase b. J Mol Biol. 1983 Oct 25;170(2):529–565. doi: 10.1016/s0022-2836(83)80160-0. [DOI] [PubMed] [Google Scholar]
- Thieme R., Pai E. F., Schirmer R. H., Schulz G. E. Three-dimensional structure of glutathione reductase at 2 A resolution. J Mol Biol. 1981 Nov 15;152(4):763–782. doi: 10.1016/0022-2836(81)90126-1. [DOI] [PubMed] [Google Scholar]
- Vainshtein B. K., Melik-Adamyan W. R., Barynin V. V., Vagin A. A., Grebenko A. I. Three-dimensional structure of the enzyme catalase. Nature. 1981 Oct 1;293(5831):411–412. doi: 10.1038/293411a0. [DOI] [PubMed] [Google Scholar]
- Webb L. E., Hill E. J., Banaszak L. J. Conformation of nicotinamide adenine dinucleotide bound to cytoplasmic malate dehydrogenase. Biochemistry. 1973 Dec 4;12(25):5101–5109. doi: 10.1021/bi00749a013. [DOI] [PubMed] [Google Scholar]
- Wierenga R. K., Drenth J., Schulz G. E. Comparison of the three-dimensional protein and nucleotide structure of the FAD-binding domain of p-hydroxybenzoate hydroxylase with the FAD- as well as NADPH-binding domains of glutathione reductase. J Mol Biol. 1983 Jul 5;167(3):725–739. doi: 10.1016/s0022-2836(83)80106-5. [DOI] [PubMed] [Google Scholar]
- Wierenga R. K., de Jong R. J., Kalk K. H., Hol W. G., Drenth J. Crystal structure of p-hydroxybenzoate hydroxylase. J Mol Biol. 1979 Jun 15;131(1):55–73. doi: 10.1016/0022-2836(79)90301-2. [DOI] [PubMed] [Google Scholar]



