Abstract
Ornithine decarboxylase (OrnDCase; L-ornithine carboxy-lyase, EC 4.1.1.17) is the first and rate-limiting enzyme in the biosynthesis of polyamines in mammalian cells. During cell growth the enzyme is regulated by rapid changes in the level of its mRNA and protein. To explore the molecular basis of these changes, we cloned a full-length cDNA copy of the major 2.4-kilobase OrnDCase mRNA from mouse cells and determined its sequence. The cDNA contains 2465 nucleotides derived from OrnDCase mRNA, consisting of a 737-nucleotide-long 5' noncoding segment, a coding segment of 1383 nucleotides terminated by a TAG triplet, and a 342-nucleotide 3' noncoding segment. The encoded protein of 461-amino acid residues has a molecular weight of 51,105 and has a potential site for phosphorylation by casein kinase II. In the unusually long 5' leader sequence, there are four ATG triplets, each of which is followed by an in-phase termination signal; the presence of these upstream ATGs could explain the low in vitro translational activity of OrnDCase mRNA reported earlier. A restriction digest of mouse genomic DNA was probed with a defined OrnDCase coding sequence, revealing a multimembered family of OrnDCase-related genes.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Gubler U., Hoffman B. J. A simple and very efficient method for generating cDNA libraries. Gene. 1983 Nov;25(2-3):263–269. doi: 10.1016/0378-1119(83)90230-5. [DOI] [PubMed] [Google Scholar]
- Hathaway G. M., Traugh J. A. Casein kinases--multipotential protein kinases. Curr Top Cell Regul. 1982;21:101–127. [PubMed] [Google Scholar]
- Hinnebusch A. G. Evidence for translational regulation of the activator of general amino acid control in yeast. Proc Natl Acad Sci U S A. 1984 Oct;81(20):6442–6446. doi: 10.1073/pnas.81.20.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu N., Messing J. The making of strand-specific M13 probes. Gene. 1982 Mar;17(3):271–277. doi: 10.1016/0378-1119(82)90143-3. [DOI] [PubMed] [Google Scholar]
- Hubbard S. C., Ivatt R. J. Synthesis and processing of asparagine-linked oligosaccharides. Annu Rev Biochem. 1981;50:555–583. doi: 10.1146/annurev.bi.50.070181.003011. [DOI] [PubMed] [Google Scholar]
- Isomaa V. V., Pajunen A. E., Bardin C. W., Jänne O. A. Ornithine decarboxylase in mouse kidney. Purification, characterization, and radioimmunological determination of the enzyme protein. J Biol Chem. 1983 Jun 10;258(11):6735–6740. [PubMed] [Google Scholar]
- Kahana C., Nathans D. Isolation of cloned cDNA encoding mammalian ornithine decarboxylase. Proc Natl Acad Sci U S A. 1984 Jun;81(12):3645–3649. doi: 10.1073/pnas.81.12.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontula K. K., Torkkeli T. K., Bardin C. W., Jänne O. A. Androgen induction of ornithine decarboxylase mRNA in mouse kidney as studied by complementary DNA. Proc Natl Acad Sci U S A. 1984 Feb;81(3):731–735. doi: 10.1073/pnas.81.3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984 Jan 25;12(2):857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Selection of initiation sites by eucaryotic ribosomes: effect of inserting AUG triplets upstream from the coding sequence for preproinsulin. Nucleic Acids Res. 1984 May 11;12(9):3873–3893. doi: 10.1093/nar/12.9.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Liu C. C., Simonsen C. C., Levinson A. D. Initiation of translation at internal AUG codons in mammalian cells. Nature. 1984 May 3;309(5963):82–85. doi: 10.1038/309082a0. [DOI] [PubMed] [Google Scholar]
- McConlogue L., Coffino P. A mouse lymphoma cell mutant whose major protein product is ornithine decarboxylase. J Biol Chem. 1983 Oct 25;258(20):12083–12086. [PubMed] [Google Scholar]
- McConlogue L., Gupta M., Wu L., Coffino P. Molecular cloning and expression of the mouse ornithine decarboxylase gene. Proc Natl Acad Sci U S A. 1984 Jan;81(2):540–544. doi: 10.1073/pnas.81.2.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
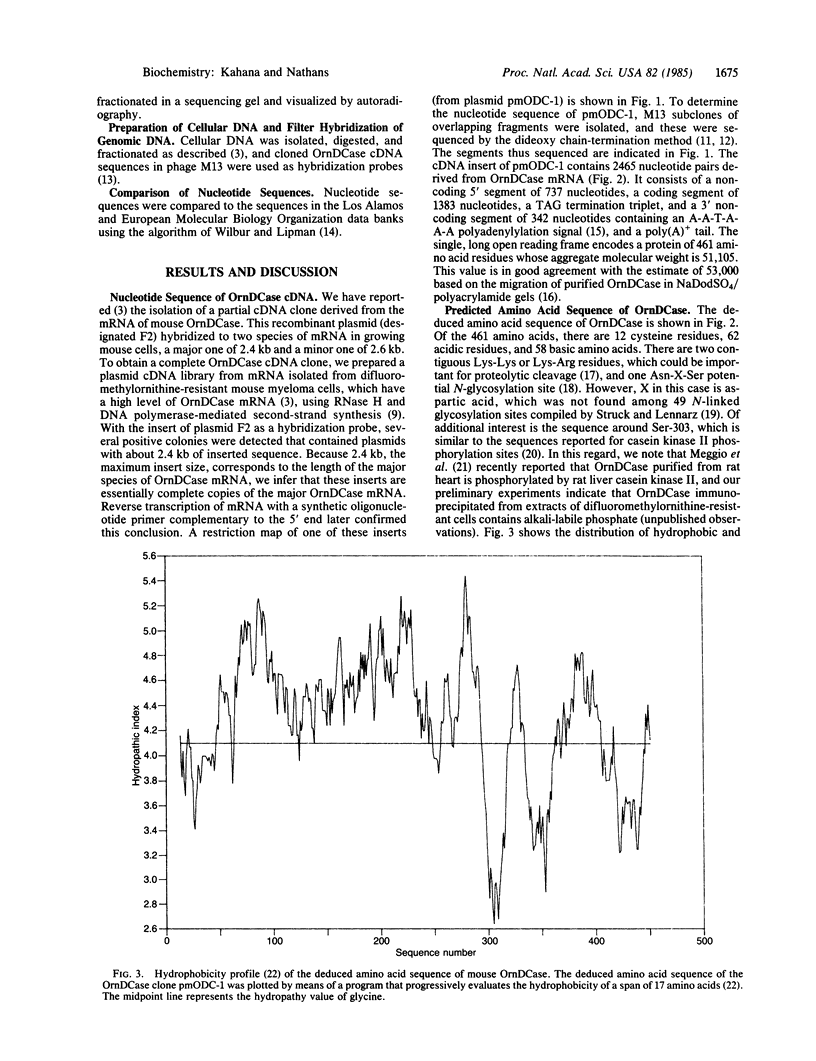
- Meggio F., Flamigni F., Caldarera C. M., Guarnieri C., Pinna L. A. Phosphorylation of rat heart ornithine decarboxylase by type-2 casein kinase. Biochem Biophys Res Commun. 1984 Aug 16;122(3):997–1004. doi: 10.1016/0006-291x(84)91190-2. [DOI] [PubMed] [Google Scholar]
- Okayama H., Berg P. High-efficiency cloning of full-length cDNA. Mol Cell Biol. 1982 Feb;2(2):161–170. doi: 10.1128/mcb.2.2.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poncz M., Solowiejczyk D., Ballantine M., Schwartz E., Surrey S. "Nonrandom" DNA sequence analysis in bacteriophage M13 by the dideoxy chain-termination method. Proc Natl Acad Sci U S A. 1982 Jul;79(14):4298–4302. doi: 10.1073/pnas.79.14.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. Sequence at the 3' end of globin mRNA shows homology with immunoglobulin light chain mRNA. Nature. 1974 Nov 29;252(5482):359–362. doi: 10.1038/252359a0. [DOI] [PubMed] [Google Scholar]
- Reynolds G. A., Basu S. K., Osborne T. F., Chin D. J., Gil G., Brown M. S., Goldstein J. L., Luskey K. L. HMG CoA reductase: a negatively regulated gene with unusual promoter and 5' untranslated regions. Cell. 1984 Aug;38(1):275–285. doi: 10.1016/0092-8674(84)90549-x. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Seely J. E., Pösö H., Pegg A. E. Purification of ornithine decarboxylase from kidneys of androgen-treated mice. Biochemistry. 1982 Jul 6;21(14):3394–3399. doi: 10.1021/bi00257a023. [DOI] [PubMed] [Google Scholar]
- Steiner D. F., Quinn P. S., Chan S. J., Marsh J., Tager H. S. Processing mechanisms in the biosynthesis of proteins. Ann N Y Acad Sci. 1980;343:1–16. doi: 10.1111/j.1749-6632.1980.tb47238.x. [DOI] [PubMed] [Google Scholar]
- Thireos G., Penn M. D., Greer H. 5' untranslated sequences are required for the translational control of a yeast regulatory gene. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5096–5100. doi: 10.1073/pnas.81.16.5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt R., Stanton L. W., Marcu K. B., Gallo R. C., Croce C. M., Rovera G. Nucleotide sequence of cloned cDNA of human c-myc oncogene. Nature. 1983 Jun 23;303(5919):725–728. doi: 10.1038/303725a0. [DOI] [PubMed] [Google Scholar]
- Wilbur W. J., Lipman D. J. Rapid similarity searches of nucleic acid and protein data banks. Proc Natl Acad Sci U S A. 1983 Feb;80(3):726–730. doi: 10.1073/pnas.80.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Straaten F., Müller R., Curran T., Van Beveren C., Verma I. M. Complete nucleotide sequence of a human c-onc gene: deduced amino acid sequence of the human c-fos protein. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3183–3187. doi: 10.1073/pnas.80.11.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]