Abstract
Quantification of nanoparticle uptake into cells is necessary for numerous applications in cellular imaging and therapy. Herein, synchrotron X-ray fluorescence (SXRF) microscopy, a promising tool to quantify elements in plant and animal cells, was employed to quantify and characterize the distribution of titanium dioxide (TiO2) nanosphere uptake in a population of single cells. These results were compared with average nanoparticle concentrations per cell obtained by widely used inductively coupled plasma mass spectrometry (ICP-MS). The results show that nanoparticle concentrations per cell quantified by SXRF were of one to two orders of magnitude greater compared with ICP-MS. The SXRF results also indicate a Gaussian distribution of the nanoparticle concentration per cell. The results suggest that issues relevant to the field of single-cell analysis, the limitation of methods to determine physical parameters from large population averages leading to potentially misleading information and the lack of any information about the cellular heterogeneity are equally relevant for quantification of nanoparticles in cell populations.
Keywords: nanoparticles, single cell, concentration, quantification, distribution, X-ray fluorescence
1. Introduction
Recent progress in nanoparticle technology allows the development of approaches to probe and/or manipulate the functions of cells in all their complexity down to the molecular level [1]. These approaches are essential for practical applications to improve health care, through advancements in molecular imaging and personalized molecular medicine [1]. At an early stage in the nanoparticle development, in vitro toxicity and efficacy studies of nanoparticles are performed on model cell lines. These studies provide average effects of the nanoparticle on these cells, and typically allow identification of therapeutic dosages. At those therapeutic dosages, the nanoparticles, if used as a probe or sensor, are assumed to be bioinert; provide information about the changes in the cellular anatomy or physiology without disturbing the cellular homoeostasis. A nanoparticle used to manipulate the function of cells (e.g. as a therapeutic drug) is expected to be present in sufficient concentrations to disturb the cellular homoeostasis. Additionally, in both the above scenarios, the nanoparticle should neither be present at low inefficacious doses nor at high toxic concentrations. Thus, direct quantification of nanoparticles per cell for the above studies is necessary and is routinely performed by random sampling of known number of cells and calculating the average nanoparticle concentration per cell [2]. In general, studies that use these average methods assume their accuracy to be acceptable. Additionally, these average methods do not provide any information of the distributions of nanoparticle in single-cell populations.
Direct quantification of nanoparticles in a single cell and the heterogeneity of the concentration distribution need to be investigated to help in understanding the variability in cell-to-cell nanoparticle concentration, and how these variations affect nanoparticle–cell interactions. These insights in turn could provide guiding principles to improve and allow nanoparticle-based personalized diagnostics and therapeutics. Microscopy and/or spectroscopy techniques and stereology have been applied to track and follow the interactions of nanoparticles within single cells [3,4]. Recently, X-ray fluorescence (XRF) microscopy at synchrotron radiation light sources has enabled the characterization of nanoparticle–cell interactions at the sub-cellular level [5,6]. Synchrotron X-ray fluorescence (SXRF) microscopy is a versatile analytical tool widely used in biomedical research and employs hard X-rays to excite and detect characteristic Kα X-ray fluorescence of important elements in medicine [5,6]. The potential of SXRF to quantify a variety of elements in individual algae, fungal and eukaryote cells has also been reported [7–9]. To the best of our knowledge, a comparison of capabilities of SXRF to quantify nanoparticles in individual cells vis-à-vis a current gold standard and the examination of the heterogeneity of the nanoparticle's distribution in single-cell populations has not been investigated. Herein, using titanium dioxide (TiO2) nanospheres as model nanoparticles we have harnessed the capabilities of SXRF to quantify and analyse the distribution of nanoparticle concentration in a large population of individual cells. We compare these results with those obtained from inductively coupled plasma mass spectrometry (ICP-MS)—a current gold standard to determine average concentrations per cell of these nanoparticles.
2. Results and discussion
The SXRF microscope employed in the current study generates simultaneous elemental maps at a spatial resolution of 7–14 µm [10]. TiO2 nanoparticles were chosen for this study because they are one of the widely studied nanoparticles for various biomedical applications, such as gene therapy, biosensing, cancer therapeutics and bioimaging [11]. Commercially, they are manufactured in large amounts, and thus comprehensive studies on the biological and environmental impact of these nanomaterials have been reported [11]. The titanium (Ti) atom present in TiO2 exhibits excellent X-ray fluorescence, is not a constitutive element of cells and has been observed by SXRF microscopy after uptake by various cancer cell lines to determine the stability and targeting of TiO2 nanoparticles conjugated with DNA [6,9,12]. The electronic supplementary material, figure S1a–d, shows the characterization of the anatase TiO2 (99.5%) nanospheres and cells. The electronic supplementary material, S1a) shows a representative TEM image of TiO2 nanospheres, which are smooth, spherical particles with diameters of ≈20–30 nm. Raman spectra of the TiO2 nanospheres (electronic supplementary material, figure S1b) showed peaks for anatase TiO2 at 136, 166 and 628 cm–1 corresponding to the Eg Raman mode, 382 cm–1 signifying the B1g mode, and 514 cm–1, which corresponds to A1g and B1g Raman modes. A small peak at 454 cm–1, which is the Eg Raman mode peak for rutile TiO2 was also observed [13]. Electronic supplementary material, S1c displays solubility of 1 mg ml−1 concentration of TiO2 nanospheres in water over 24 h without aggregation. This formulation was used to treat the cells. The electronic supplementary material, S1d, shows a differential interference contrast (DIC) microscopy image of SK-BR-3 cells fixed onto Ultralene film for SXRF microscopy. The cells treated with TiO2 nanospheres were not functionalized or treated with additional transfecting agents.
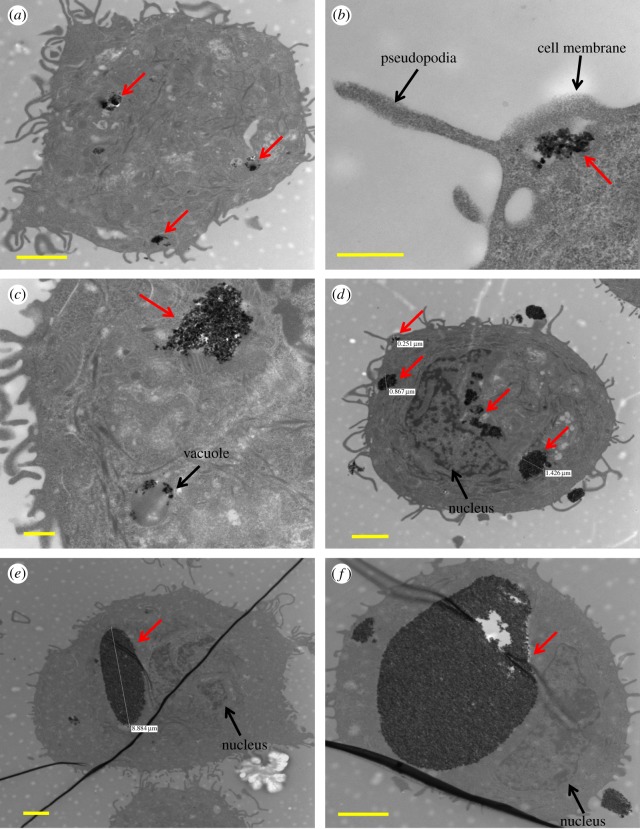
The uptake of the TiO2 nanoparticles by SK-BR-3 cells was confirmed by TEM as displayed in figure 1. TiO2 nanoparticles (red arrows) were within the cytoplasm (figure 1a–f) and vesicular structures (figure 1a,e,f), but not in the nucleus. Cell pseudopodia engulfing the nanoparticles were also observed (figure 1b,d). Analysis of TEM images of many individual cells including those in figure 1a–f indicated that most of the nanoparticles were internalized by cells with a small fraction present on the cell membranes. Qualitatively, cells either showed no uptake of nanoparticles, or small (figure 1a,b), medium (figure 1c,d) and large (figure 1e,f) nanoparticle concentrations and aggregate sizes.
Figure 1.
TiO2 nanoparticles (red arrows) within the cytoplasm (a–f). Nanoparticle aggregates within vesicular structures (a,e,f). Cell pseudopodia engulfing the nanoparticles (b,d). No nanoparticles were seen in the nucleus. (a,d,e,f) Scale bars, 2 µm; (b,c) scale bars, 500 nm.
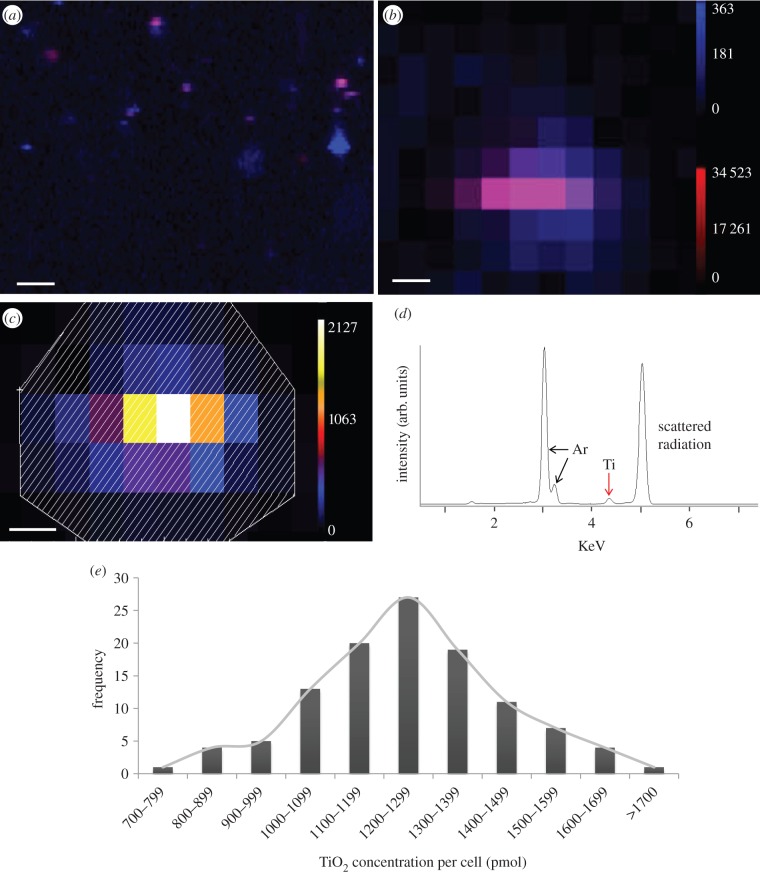
SXRF was used to determine the concentration of nanoparticles in single cells. Initially, SXRF ‘flyscans’ (figure 2a), a rapid-scanning technique using a dwell time of 100 ms per pixel, were used to create an elemental map of an area of about 1–3 mm2. The flyscans of approximately 30% cells (out of approx. 1700 cells) showed only Ca (i.e. no Ti uptake). Step scans, shown in figure 2b, were performed with a longer dwell time (5 s per pixel) on 112 individual cells (complete cell selection criteria can be found in experimental section 7 in the electronic supplemental information). Individual scans were processed for spectral summing (figure 2c) by creating an outline of the cell using interactive data language (IDL)-based beamline software which outputs a summed energy-dispersive spectra (figure 2d) for the highlighted area and contains the total counts for Ti Kα in the cell. Using the information of Ti counts per cell, a known and measured National Institute of Standards and Technology (NIST) standard 1833 for Ti, the concentration of TiO2 per cell, was quantified. The abundance of nanoparticles internalized per cell was heterogeneous with a range between 37 and 84 ng cell−1 or 773–1751 pmol per cell (atomic weight of Ti = 47.9). Stoichiometrically, one mole of Ti gives one mole of TiO2. Thus, the concentration of TiO2 also ranges between 773 and 1751 pmol per cell (figure 2e). The nanoparticle per cell concentration plot exhibits a Gaussian distribution.
Figure 2.
(a) Representative SXRF ‘flyscan’ map showing areas of calcium (blue) and titanium (red), scale bar, 100 µm. (b) SXRF scan of an individual cell showing calcium (blue) and titanium (red) counts, scale bar, 10 µm. (c) SXRF scan of an individual cell showing titanium, scale bar, 10 µm. The shaded area is the region of interest for the spectral sum of titanium counts. (d) Spectral sum of titanium counts found in a single cell. (e) Bar graph showing heterogeneity of concentrations of TiO2 nanospheres internalized by SK-BR-3 cells.
The SXRF results were compared to ICP-MS; a standard method of quantifying average concentration per cell [14]. For this analysis, 30 000 cells (n = 5) were treated in the same manner as described for the SXRF experiments. After 24 h, 15 000 cells were counted by flow cytometry. These cells were digested and prepared for ICP-MS (see experimental section 8 in the electronic supplementary material for details). For the five samples, the total Ti abundance in 15 000 cells, determined by ICP-MS, was between 16 and 24 µg and Ti abundance per cell was calculated to be 1.1 to 1.6 ng cell−1. The average concentration of Ti and hence TiO2 in moles was 23–33 pmol per cell (table 1). This average value does not provide any additional information about the distribution of the nanoparticle per cell concentration in the 15 000 cells. The difference in the nanoparticle per cell concentrations obtained by SXRF and ICP-MS methods was significant and taken together indicate that TiO2 nanospheres were present in single cells at concentrations of one to two orders of magnitude greater than the average concentration per cell values. The lower average nanoparticle concentrations per cell by ICP-MS could be due to systematic errors induced during sample preparation [15], which involves multiple steps (see the experimental section in the electronic supplementary material for these steps). During the sample preparation, cells are digested, involving transfer of the solution a number of times, as well as filtration and dilution to be within the linear range of detection for the instrument. Even though these and other sample preparation steps are undertaken with utmost caution, each of these steps is dependent on the previous one and has potential to introduce compounding error propagation owing to sample loss. Although systematic errors do not affect the precision of the results, they can significantly affect accuracy [16]. Another reason for observed discrepancy in nanoparticle concentration per cell by ICP-MS measurements could be its dependence on the nanoparticle's cell labelling efficiency. Increase in the number of cells with nanoparticle concentrations below the detection limit of ICP-MS for Ti due to poor labelling efficiency could skew the average concentration of TiO2 per cell to lower values (see the electronic supplementary material for representative examples). It should also be stressed that methods such as ICP, per se, are precise (table 1). Additionally, similar limitations, such as underestimation of nanoparticle per cell concentrations and inability to characterize the concentration distribution in cell population, would apply to other methods (e.g. optical fluorescence methods [17]) that provide average concentration information. While SXRF does not allow detection as low as ICP-MS, its detection limit is on par with ICP-optical emission spectrometry, which is in the microgram per millilitre range. Importantly, the nanoparticle concentration per cell values does not change depending on the efficiency of uptake by cells and furthermore, provides potentially valuable information on the distribution characteristics of nanoparticle concentration in a population of cells; hitherto not possible by ICP-based methods. The sample preparation for SXRF avoids the propagation of systematic errors. However, systematic errors could be generated by the technique including variable spatial regions of interest around the cells to determine the elemental abundance and scans of clusters of cells rather than individual cells. However, these user-dependent errors can be avoided by implementing proper cell selection and scanning area criteria.
Table 1.
ICP-MS results for SK-BR-3 cells treated with 100 µg ml−1 TiO2 nanospheres.
| sample number | total titanium in sample (µg) | TiO2 per cell (ng) | Ti per cell (pmol) |
|---|---|---|---|
| 1 | 16 | 1.1 | 23 |
| 2 | 17 | 1.1 | 23 |
| 3 | 24 | 1.6 | 33 |
| 4 | 20 | 1.3 | 28 |
| 5 | 17 | 1.2 | 24 |
Recent advancements in single-cell analysis clearly show heterogeneity in cell populations previously assumed to be identical [18]. There is an emerging consensus that experimental methods that provide information about average population-level cellular characteristics are insufficient, and sometimes potentially misleading [18]. Novel techniques have been explored to address the challenges associated with single-cell analysis [19]. The above results indicate that the concentration of nanoparticles in single cells could be larger than the values estimated by traditional methods, and suggest that similar challenges also exist while quantifying nanoparticles internalized by cells. A critical component of all single-cell analysis, imaging or therapy involving nanoparticles would include accurate determination of the nanoparticle uptake concentration per cell and the distribution of this concentration in cell populations; this information will affect the safety and efficacy of the particular application [17]. Our study indicates that SXRF may be a suitable technique to determine nanoparticle concentration distribution in single-cell populations. It can easily be expanded for use with nanoparticles synthesized using other low or high Z elements [5] and provide fundamental insights into the concentration distribution modes (e.g. monomodal, bimodal), and heterogeneity and the dependence of these parameters on the structure (e.g. spherical, rod-like) and composition (e.g. metallic, ceramic). Furthermore, the ability of SXRF to map changes in elements integral to cellular homoeostasis [20], even at the sub-cellular level [5], could provide integrated understanding of nanoparticle-single-cell population interactions.
Acknowledgements
Portions of this work were performed at Beamline X27A, National Synchrotron Light Source (NSLS), Brookhaven National Laboratory. The authors thank Susan Van Horn (Central Microscopy, Stony Brook University) for her help in Transmission Electron Microscopy. The authors declare no competing financial interests.
Funding statement
X27A is supported in part by the U.S. Department of Energy (DOE)—Geosciences (DE-FG02–92ER14244 to The University of Chicago, CARS). Use of the NSLS was supported by the DOE, Office of Science, Office of Basic Energy Sciences, under Contract no. DE-AC02–98CH10886. This work was supported by the National Institutes of Health (grant no. 1DP2OD007394-01).
References
- 1.Andersson H, Berg AVD. 2004. Microtechnologies and nanotechnologies for single-cell analysis. Curr. Opin. Biotechnol. 15, 44–49. ( 10.1016/j.copbio.2004.01.004) [DOI] [PubMed] [Google Scholar]
- 2.Ibuki Y, Toyooka T. 2012. Nanoparticle uptake measured by flow cytometry. In Nanotoxicity: methods and protocols (ed. Reineke J.), pp. 157–166. Totowa, NJ: Humana Press. [DOI] [PubMed] [Google Scholar]
- 3.Jin H, Heller DA, Strano MS. 2008. Single-particle tracking of endocytosis and exocytosis of single-walled carbon nanotubes in nih-3t3 cells. Nano Lett. 8, 1577–1585. ( 10.1021/nl072969s) [DOI] [PubMed] [Google Scholar]
- 4.Peckys DB, Jonge ND. 2011. Visualizing gold nanoparticle uptake in live cells with liquid scanning transmission electron microscopy. Nano Lett. 11, 1733–1738. ( 10.1021/nl200285r) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bussy C, et al. 2008. Carbon nanotubes in macrophages: imaging and chemical analysis by X-ray fluorescence microscopy. Nano Lett. 8, 2659–2663. ( 10.1021/nl800914m) [DOI] [PubMed] [Google Scholar]
- 6.Paunesku T, et al. 2003. Biology of tio2-oligonucleotide nanocomposites. Nat. Mater. 2, 343–346. ( 10.1038/nmat875) [DOI] [PubMed] [Google Scholar]
- 7.Nunez-Milland DR, Baines SB, Vogt S, Twining BS. 2010. Quantification of phosphorus in single cells using synchrotron x-ray fluorescence. J. Synchrotron Radiat. 17, 560–566. ( 10.1107/S0909049510014020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Twining BS, Baines SB, Fisher NS, Maser J, Vogt S, Jacobsen C, Tovar-Sanchez A, Sañudo-Wilhelmy SA. 2003. Quantifying trace elements in individual aquatic protist cells with a synchrotron x-ray fluorescence microprobe. Anal. Chem. 75, 3806–3816. ( 10.1021/ac034227z) [DOI] [PubMed] [Google Scholar]
- 9.Thurn KT, et al. 2009. Labeling tio2 nanoparticles with dyes for optical fluorescence microscopy and determination of tio2–DNA nanoconjugate stability. Small 5, 1318–1325. ( 10.1002/smll.200801458) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tappero R, et al. 2007. Hyperaccumulator alyssum murale relies on a different metal storage mechanism for cobalt than for nickel. New Phytol. 175, 641–654. ( 10.1111/j.1469-8137.2007.02134.x) [DOI] [PubMed] [Google Scholar]
- 11.Shi H, Magaye R, Castranova V, Zhao J. 2013. Titanium dioxide nanoparticles: a review of current toxicological data. Part. Fibre Toxicol. 10, 15 ( 10.1186/1743-8977-10-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paunesku T, et al. 2007. Intracellular distribution of TiO2 –DNA oligonucleotide nanoconjugates directed to nucleolus and mitochondria indicates sequence specificity. Nano Lett. 7, 596–601. ( 10.1021/nl0624723) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee B-Y, Behler K, Kurtoglu ME, Wynosky-Dolfi MA, Rest RF, Gogotsi Y. 2010. Titanium dioxide-coated nanofibers for advanced filters. J. Nanopart. Res. 12, 2511–2519. ( 10.1007/s11051-009-9820-x) [DOI] [Google Scholar]
- 14.Avti PK, Caparelli ED, Sitharaman B. 2013. Cytotoxicity, cytocompatibility, cell-labeling efficiency, and in vitro cellular magnetic resonance imaging of gadolinium-catalyzed single-walled carbon nanotubes. J. Biomed. Mater. Res. A. ( 10.1002/jbm.a.34643) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.García Alonso JI, Gutiérrez Camblor M, Montes Bayón M, Marchante-Gayón JM, Sanz-Medel A. 1997. Different quantification approaches for the analysis of biological and environmental samples using inductively coupled plasma mass spectrometry. J. Mass Spectrom. 32, 556–564. () [DOI] [PubMed] [Google Scholar]
- 16.Elburg MA, Vroon P, Schersten A. 2005. An empirical method for determining the error introduced by blank corrections on mc-icp-ms measurements. J. Anal. At. Spectrom. 20, 1389–1391. ( 10.1039/b507203d) [DOI] [Google Scholar]
- 17.Elsaesser A, Taylor A, Yanés GSD, Mckerr G, Kim E-M, O'hare E, Howard CV. 2010. Quantification of nanoparticle uptake by cells using microscopical and analytical techniques. Nanomedicine 5, 1447–1457. ( 10.1039/B507203D) [DOI] [PubMed] [Google Scholar]
- 18.Templer RH, Ces O. 2008. New frontiers in single-cell analysis. J. R. Soc. Interface 5, S111–S112. ( 10.1098/rsif.2008.0279.focus) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fritzsch FSO, Dusny C, Frick O, Schmid A. 2012. Single-cell analysis in biotechnology, systems biology, and biocatalysis. Annu. Rev. Chem. Biomol. Eng. 3, 129–155. ( 10.1146/annurev-chembioeng-062011-081056) [DOI] [PubMed] [Google Scholar]
- 20.Fahrni CJ. 2007. Biological applications of x-ray fluorescence microscopy: exploring the subcellular topography and speciation of transition metals. Curr. Opin. Chem. Biol. 11, 121–127. ( 10.1016/j.cbpa.2007.02.039) [DOI] [PubMed] [Google Scholar]