Abstract
Colorectal cancers (CRCs) harboring KRAS or BRAF mutations are refractory to current targeted therapies. Using data from a high-throughput drug screen, we have developed a novel therapeutic strategy that combines targeting of the apoptotic machinery using the BCL-2 family inhibitor ABT-263 (navitoclax) in combination with a TORC1/2 inhibitor, AZD8055. This combination leads to efficient apoptosis specifically in KRAS mutant (MT) and BRAF MT but not wild-type (WT) CRC cells. This specific susceptibility results from TORC1/2 inhibition leading to suppression of MCL-1 expression in mutant, but not WT CRCs, leading to abrogation of BIM/MCL-1 complexes. This combination strategy leads to tumor regressions in both KRAS MT colorectal cancer xenograft and genetically-engineered mouse models of CRC, but not in the corresponding KRAS WT CRC models. These data suggest that the combination of BCL-2/XL inhibitors with TORC1/2 inhibitors constitutes a promising targeted therapy strategy to treat these recalcitrant cancers.
Keywords: KRAS, BRAF, colorectal cancer, targeted therapy, ABT-263
Introduction
KRAS mutations are observed in ~ 30–45% of CRCs (1–3). These mutations lead to potent activation of the MEK-ERK signaling pathway (4). Although therapies targeting EGFR have some efficacy in CRCs without KRAS mutations (1, 5–8), these therapies most likely fail because the MEK-ERK pathway is sustained by mutant KRAS in the presence of EGFR inhibitory antibodies. Direct inhibitors of mutant KRAS protein are not yet available; therefore, efforts are often focused on targets in signaling pathways whose inhibition alone or in combination may be effective for this subset of cancers (9–16). Indeed, multiple approaches, including the combination of PI3K and MEK pathway inhibitors, are being examined in clinical trials. Mutant BRAF, which is directly downstream of KRAS, also leads to hyperactivation of the MEK-ERK pathway. BRAF mutations occur in roughly 5–15% of CRCs (1–3, 17), and are generally mutually exclusive with KRAS mutations (1). In fact, a recent report highlighted gene expression similarities in these two genetically distinct MT CRCs, underscoring the overlap in signaling downstream from these mutant oncogenes (18). Single-agent BRAF inhibitors have been largely ineffective in BRAF MT CRCs (19), despite activity in BRAF MT melanomas (20). However, some laboratory models of BRAF mutant CRCs are sensitive to the combination of BRAF and receptor tyrosine kinase inhibitors, particularly EGFR, and this approach is currently under evaluation in the clinic (21, 22). While some of these novel therapeutic strategies for KRAS and BRAF MT CRCs being explored in clinical trials will hopefully demonstrate some activity, it is very likely that clinical resistance will emerge, necessitating additional treatment strategies. Thus, there continues to be an urgent need to develop additional targeted therapies for KRAS MT as well as BRAF MT CRCs.
We sought to uncover targeted therapy strategies that demonstrate specificity towards KRAS or BRAF MT CRCs compared to their WT counterparts. We leveraged the results from a high-throughput screen that assessed the sensitivity of over 1,000 cell lines to more than 130 drugs (23). Since the induction of both apoptosis and growth arrest is a hallmark of many successful targeted therapy approaches (24–26), we built upon the screen results and further mechanistic insights to establish a combination strategy producing these biological effects.
Results
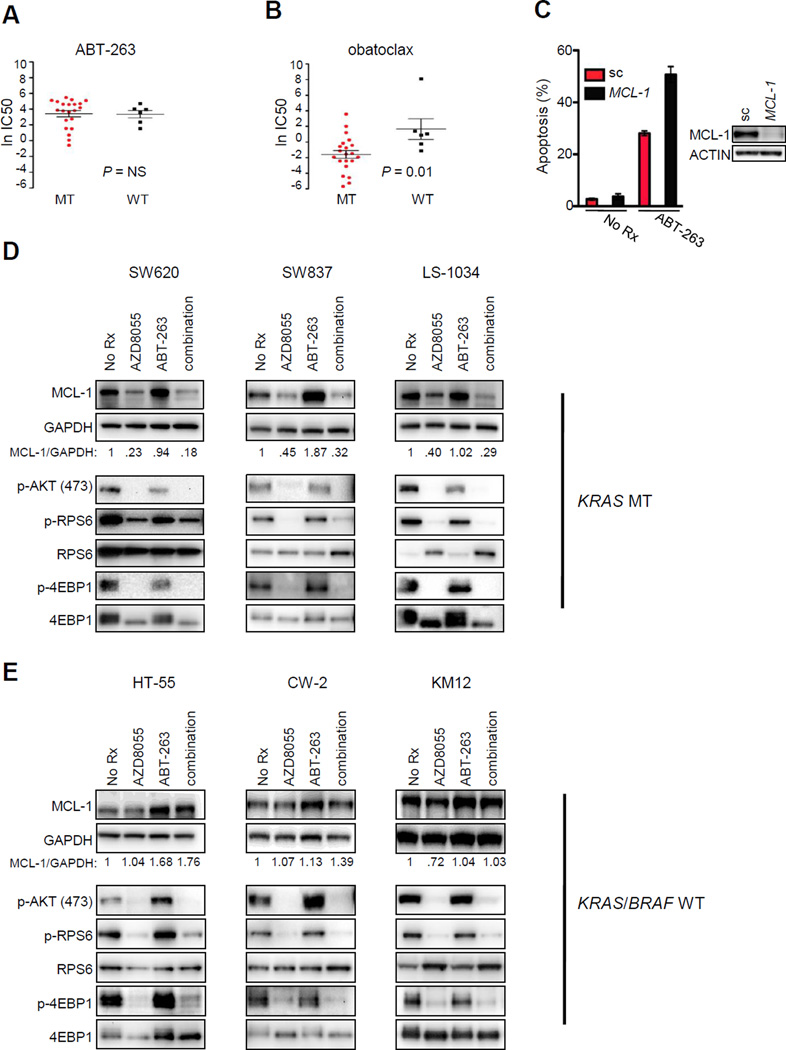
Data obtained from our recently described high-throughput drug screen (23, 27) allowed us to compare the efficacy of drugs between KRAS MT and BRAF MT human CRCs versus WT human CRCs. Included among the large number of compounds in the drug screen was ABT-263, a BCL-2/XL inhibitor (BH3 mimetic) that has demonstrated pre-clinical efficacy in some tumors (28, 29) and is under clinical evaluation as a single agent or in combination with chemotherapy (30, 31). In this study, we found that ABT-263 had similar activity in KRAS and BRAF MT compared to WT CRCs (Fig. 1A). In contrast to ABT-263, a different BH3 mimetic, obatoclax, neutralizes another BCL-2 family member, MCL-1, in addition to BCL-2 and BCL-XL (32). Unlike ABT-263, obatoclax was more effective in both KRAS and BRAF MT CRCs than in WT CRCs (Fig. 1B). The selectivity of obatoclax for MT CRCs was notable as many common chemotherapies and experimental therapies did not discriminate between the MT and WT CRCs (Sup. Fig.1, P=NS for all comparisons). The differential sensitivity to obatoclax was not explained simply by expression levels of either MCL-1 or other BCL-2 family members (Sup. Fig. 2A, 2B). Consistent with the increased sensitivity of MT CRCs to obatoclax, RNAi knockdown of MCL-1 sensitized MT CRCs, but not WT CRCs, to ABT-263 (Fig. 1C, Sup. Fig. 2C). In total, these findings suggest that, in comparison to their WT counterparts, MT cells have a heightened sensitivity to combined inhibition of MCL-1, BCL-XL, and BCL-2.
Figure 1. KRAS and BRAF mutant colorectal cancers have increased sensitivity to obatoclax compared to their wild-type counterparts and also have MCL-1 expression under the regulation of TORC1/2.
KRAS mutant (MT), BRAF MT, and wild-type (WT) colorectal cells were treated with increasing concentrations of (A) the BCL-2/XL inhibitor ABT-263 or (B) the BCL-2/XL/MCL-1 inhibitor obatoclax for 72 hours; cell number was determined, and IC50s were calculated (and converted to natural log, y axis). Student’s t tests were performed between the two groups (KRAS or BRAF MT versus WT) and P values were determined (NS=not significant) (n=27). (C) KRAS MT SW620 cells were transfected with 50 nM scrambled (sc) or 50 nM MCL-1 siRNA for 24 hours. An aliquot of transfected cells was used to prepare lysates for Western blot analyses with the indicated antibodies (lower panels) or re-seeded and the next day treated for 48 hours with vehicle (no Rx) or 1 µM ABT-263; percent of apoptotic cells was quantified by FACS (n=3). (D) KRAS MT or (E) KRAS/BRAF WT colorectal cell lines were treated for 16 hours with no drug (No Rx), 500 nM AZD8055, 1 µM ABT-263, or combination AZD8055/ABT-263, and proteins from lysates were subjected to Western blot analyses with the indicated antibodies. The numbers in the panels represent the amount of MCL-1 normalized to GAPDH. The detection of MCL-1 and GAPDH were performed on the same membrane.
These data indicated that targeting BCL-2, BCL-XL, and MCL-1 may be an effective therapeutic strategy in MT CRCs. However, obatoclax has significant toxicity, possibly off-target, limiting its clinical utility (33–35). Furthermore, many effective targeted therapies lead to both apoptosis and growth arrest, and inhibition of BCL-2 family members might be expected to induce apoptosis (29), but fail to strongly impair proliferation. Thus, we sought an alternative approach to both induce apoptosis and concomitantly block proliferation. Previous studies had demonstrated that efficient translation of MCL-1 mRNA requires cap-dependent translation in some cancers (36–38). TORC1 inhibition, via loss of 4EBP1 (also referred to as EIF4EBP1) phosphorylation, results in a decrease of MCL-1 protein expression in these cancers (37–40). Consistent with those results, we found that treatment of KRAS MT and BRAF MT CRCs with the TORC1/2 inhibitor, AZD8055 (41), led to a decrease in MCL-1 expression (Fig. 1D, Sup. Fig. 2D and Sup. Fig 3). Interestingly, AZD8055 did not substantially suppress MCL-1 expression in KRAS and BRAF WT cells (Fig. 1E). Indeed, the regulation of MCL-1 expression by TORC1/2 appeared to be different between WT and MT CRCs since AZD8055 effectively suppressed TORC1 and TORC2 in both genetic backgrounds as measured by its effect on phospho-RPS6, phosho-4EBP1 and phospho-AKT at residue 473 (Fig. 1D,E). Consistent with this, treatment with AZD8055 led to a G1 arrest in both WT and MT CRC, and thus MT and WT CRCs had similar sensitivities to single-agent AZD8055 (Sup. Fig. 4A,B). The suppression of MCL-1 protein expression by AZD8055 in MT cells was not associated with a decrease in MCL-1 RNA levels (Sup. Fig. 5A), consistent with an effect on translation of the MCL-1 mRNA in these cancers. Furthermore, MCL-1 mRNA levels only slightly increased in both MT and WT cells following AZD8055 treatment, suggesting there is no differential feedback of MCL-1 transcription contributing to differences in MCL-1 protein expression between the cancer types (Sup. Fig 5A). A time-course of AZD8055 treatment in MT cells demonstrated clear suppression of MCL-1 protein expression within two hours of treatment (Sup. Fig. 5B). In contrast, MCL-1 expression was not diminished in WT cells at any time point examined (Sup. Fig 5B).
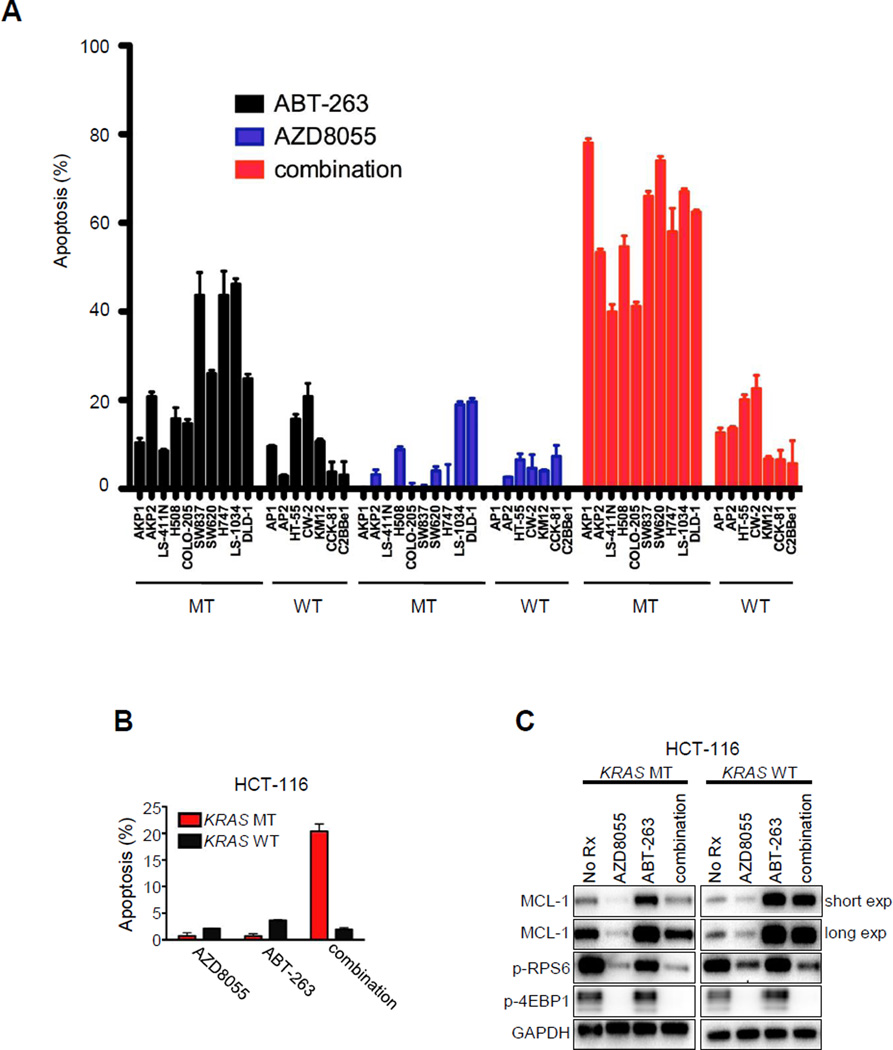
Since TORC1/2 inhibition effectively suppressed MCL-1 expression in KRAS or BRAF MT cells, we explored whether TORC1/2 inhibitors would combine with ABT-263 to induce apoptosis specifically in these genetic backgrounds (analogous to MCL-1 siRNA sensitizing to ABT-263, Fig. 1C). It is notable that ABT-263 usually induces expression of MCL-1, and that the differential effects of AZD8055 on the expression of MCL-1 between MT and WT cells were often more dramatic when AZD8055 was combined with ABT-263 (Fig. 1D, E, Sup. Fig. 2D and Sup. Fig. 3). We assessed the induction of apoptosis across a panel of human MT and WT CRC cell lines. In addition to human cancer cell lines, we also included ex vivo mouse cell lines derived from CRCs induced in KRAS MT (KRAS/APC/p53) and WT (APC/p53) genetically-engineered mouse models (GEMMs) of CRC (42, 43). The combination of ABT-263 and AZD8055 induced substantial apoptosis in the MT human and murine CRCs (average apoptosis 59.4 ± 4.0%) (Fig. 2A). In contrast, marked apoptosis was not observed in any of the WT CRC cell lines (average apoptosis 12.5 ± 2.6%) (Fig. 2A). We also compared the effect of this combination on a pair of isogenic HCT-116 cell lines with and without KRAS mutations(44) We observed that the KRAS MT HCT-116 cells had approximately a seven-fold increase in apoptosis following combination therapy compared to the KRAS WT HCT-116 cells (Fig. 2B). Of note, MCL-1 levels decreased significantly more following AZD8055 (and the AZD8055/ABT-263 combination) in MT cells compared to the WT cells (Fig. 2C, ten-fold versus two-fold, respectively). In total, these findings support the notion that MT CRCs may be particularly more sensitive to this combination than their WT counterparts.
Figure 2. Increased induction of apoptosis in KRAS and BRAF mutant colorectal cancers treated with combined ABT-263/AZD8055 compared to KRAS/BRAF wild-type colorectal cancers.
(A) The indicated cell lines were treated with no drug, 500 nM AZD8055, 1 µM ABT-263, or combination AZD8055/ABT-263 for 72 hours. Percent of apoptotic cells was quantified by FACS. Each bar graph represents the amount of apoptosis induced by treatment relative to no drug treatment. Error bars are the S.D. (n=3). Mutant (MT) cell lines consist of KRAS or BRAF MT human CRC cell lines, as well as CRC cell lines derived from a KRAS MT genetically-engineered mouse model (GEMM) (AKP1 and AKP2). Wild-type (WT) cells consist of KRAS/BRAF WT human CRC cell lines as well as CRC cell lines derived from a KRAS WT GEMM (AP1 and AP2). (B and C) Isogenic HCT-116 cells that were either KRAS MT or WT were treated with no drug (No Rx), 500 nM AZD8055, 1 µM ABT-263, or the combination of ABT-263/AZD8055 (263/8055) for (B) 72 hours and the percent of apoptotic cells was quantified by FACS, or (C) overnight and proteins from lysates were subjected to Western blot analyses with the indicated antibodies. For (B), each bar graph represents the amount of apoptosis induced by treatment relative to no drug treatment. Error bars are the S.D. (n=3).
These findings prompted us to further examine the regulation of MCL-1 in KRAS MT cancers and the mechanism by which its inhibition synergized with ABT-263 to promote apoptosis. Knockdown of KRAS using inducible shRNA was sufficient to suppress 4EBP1 phosphorylation, and accordingly, MCL-1 in KRAS MT SW620 cells (Fig. 3A). However, MEK inhibition alone did not suppress phosphorylation of 4EBP1, and accordingly, it did not decrease expression of MCL-1 (Fig. 3B). This finding is consistent with a report suggesting that other KRAS-regulated pathways, in addition to MEK-ERK, contribute to phosphorylation of 4EBP1(45). In contrast, KRAS knockdown did not decrease MCL-1 levels in KRAS WT CW-2 cells (Sup. Fig. 5C).
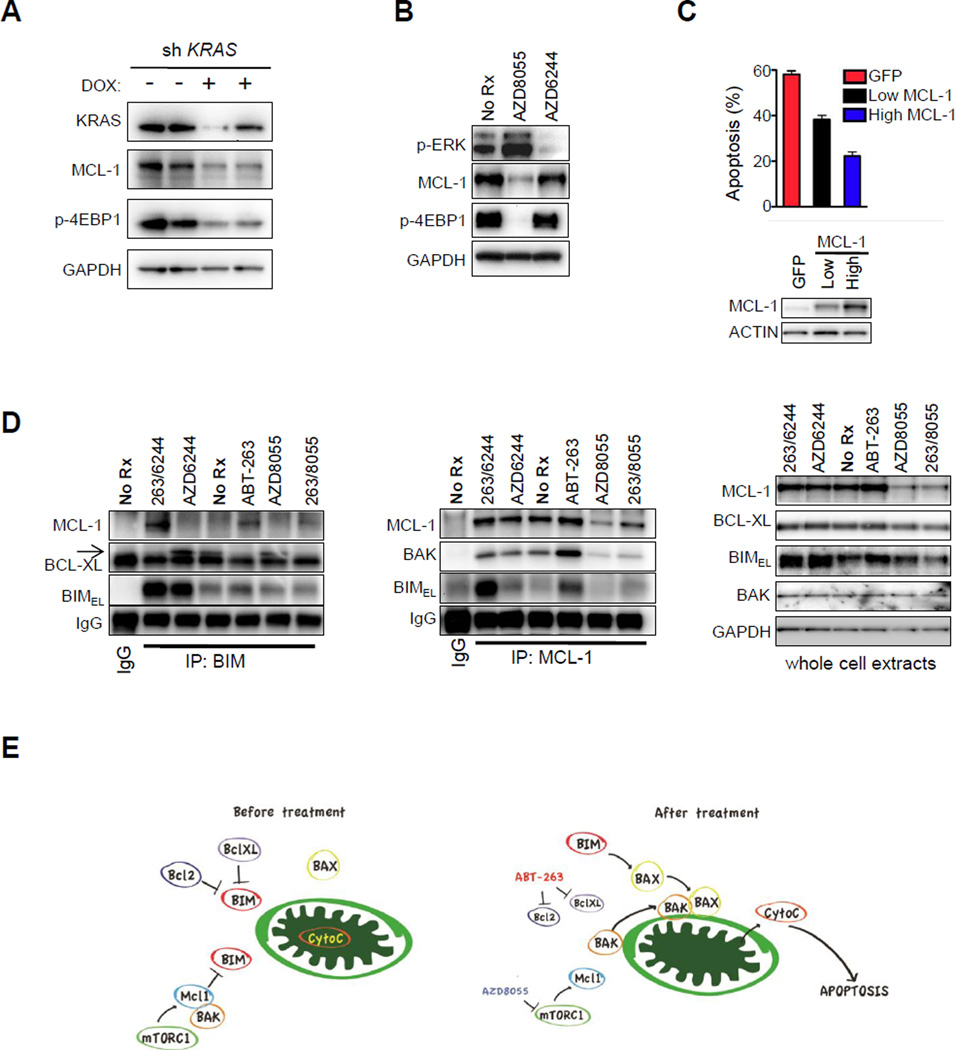
Figure 3. TORC inhibition, but not MEK inhibition, downregulates MCL-1 and disrupts MCL-1:BIM complexes following ABT-263 therapy, leading to enhanced apoptosis.
(A) KRAS MT SW620 colorectal cells expressing doxycycline-inducible KRAS shRNA vectors were treated with or without 50 ng/mL doxycycline (DOX) for 72 hours, and lysates were subjected to Western blot analyses with the indicated antibodies. (B) KRAS MT SW620 colorectal cells were treated for 16 hours with no drug (no Rx), 500 nM AZD8055, or 1 µM AZD6244, and lysates were subjected to Western blot analyses with the indicated antibodies. (C) Stable clones of SW620 cells expressing GFP alone or a GFP-IRES-MCL-1 were sorted by FACS by fluorescence intensity and the populations of cells were separated. Western blot analyses of the resultant cell lines are shown in the lower panel. Cells were treated for 72 hours with the ABT-263/AZD8055 combination therapy and apoptosis was determined by FACS. Each bar graph represents the amount of apoptosis induced by treatment relative to no drug treatment. Error bars are the S.D. (n=3). (D) SW620 parental cells underwent immunoprecipitation (IP) with antibodies targeting BIM (left panel), MCL-1 (middle panel), or IgG (left and middle panels) following six hours of no drug (no Rx), 500 nM AZD8055, 1 µM ABT-263, combination therapy (263/8055), 1 µM AZD6244, or combination of AZD6244 and ABT-263 (263/6244). Precipitates were analyzed by Western blot analyses with the indicated antibodies. Arrow indicates BCL-XL that migrates slower than the light chain. Whole cell lysates were set aside prior to IP and were subjected to Western blot analyses with the indicated antibodies (right panel). (E) The schematic displays how ABT-263 and AZD8055 combination therapy affect the balance of BCL-2 family proteins to induce apoptosis in KRAS MT cells. Mitochondria are depicted in green.
We further interrogated the molecular mechanisms by which the combination of TORC1/2 and BCL-2/XL inhibitors led to apoptosis in the MT cancers. To determine if the induction of apoptosis by the AZD8055/ABT-263 combination was indeed dependent on the suppression of MCL-1 expression, we expressed exogenous MCL-1 via lentiviral gene delivery. As expected, the exogenous expression of MCL-1 suppressed the induction of apoptosis (Fig. 3C, Sup. Fig. 6A). To understand how the drug combination impacted the protein complexes of the BCL-2 family, we performed immunoprecipitations in the absence and presence of inhibitors. Previous studies have shown that BIM plays a critical role in BH3 mimetic (including ABT-263)-induced apoptosis (46, 47). Analysis of immunoprecipitated BIM and MCL-1 complexes revealed that ABT-263 treatment abrogated the interaction between BIM and BCL-XL. However, ABT-263 treatment led to a proportional increase in the association between MCL-1 and BIM (Fig. 3D). These findings suggest that the BIM released from BCL-2 and BCL-XL in response to ABT-263 was bound to MCL-1, preventing apoptosis. Adding AZD8055 to ABT-263 resulted in loss of BIM/MCL-1 complexes (Fig. 3D) corresponding to decreases of MCL-1 expression in the whole cell extracts (Fig. 3D). AZD8055 treatment also led to reduced complexes between MCL-1 and BAK due to loss of MCL-1 (Fig. 3D, middle panel), likely contributing to the amount of apoptosis induced by the combination. In KRAS WT cells, analysis of immunoprecipitated BIM complexes revealed that, similar to the KRAS MT cells, ABT-263 treatment blocked the interaction between BIM and BCL-XL, and that treatment led to a proportional increase in the association between MCL-1 and BIM. However, in contrast to the MT cells, AZD8055 treatment did not reduce BIM/MCL-1 complexes (Sup. Fig. 6B), consistent with the lack of reduction of MCL-1 following AZD8055 treatment in the total cell lysates (Sup. Fig. 6B) and the lack of efficacy of the combination (Fig. 2A). A model depicting the impact of the combination on the BCL-2 family is shown in Fig. 3E. Of note, this mechanism of apoptosis is distinct from the recently described combination of ABT-263 and MEK inhibitor(16), AZD6244. Unlike AZD8055, the MEK inhibitor does not decrease levels of MCL-1, but rather increases levels of BIM (Fig. 3D).
Next, we asked whether our findings in MT CRCs extended to another type of KRAS mutant cancer, KRAS MT non-small cell lung cancers (NSCLCs). In contrast to KRAS MT CRC, AZD8055 failed to markedly downregulate MCL-1 in KRAS MT NSCLC cell lines (Sup. Fig. 7A), and accordingly, it failed to substantially increase the amount of apoptosis induced by ABT-263 in the NSCLC cell lines (Sup. Fig. 7B). However, NSCLC cells were highly sensitive to ABT-263 treatment following MCL-1 knockdown (Sup. Fig. 7C), suggesting KRAS MT cancers in general may be sensitive to simultaneous BCL-2, BCL-XL and MCL-1 inhibition, but that TORC1 regulation of MCL-1 may be different between KRAS MT CRC and KRAS MT NSCLC. In support of this hypothesis, AZD8055 treatment led to a modest increase in the level of MCL-1 transcript in KRAS MT NSCLCs (Sup. Fig. 7D), similar to KRAS MT CRCs and KRAS WT CRCs (Sup. Fig 5A). Furthermore, KRAS knockdown in KRAS mutant H2009 and H358 cells failed to significantly downregulate MCL-1 expression (Sup. Fig. 7E) in contrast to KRAS knockdown in KRAS mutant SW620 cells (Fig. 3A). Of note, we did not detect any differences between the stability of MCL-1 protein between KRAS MT CRC and KRAS MT NSCLC cell lines, as determined by Western blotting of MCL-1 following a short-term time-course of treatment with the protein translational inhibitor, cycloheximide (Sup. Fig. 8). Altogether, these data suggest that MCL-1 may be an important survival signal for KRAS MT cancers in general, however, the regulation of MCL-1 expression by TORC1/2 is not equivalent in KRAS MT NSCLCs.
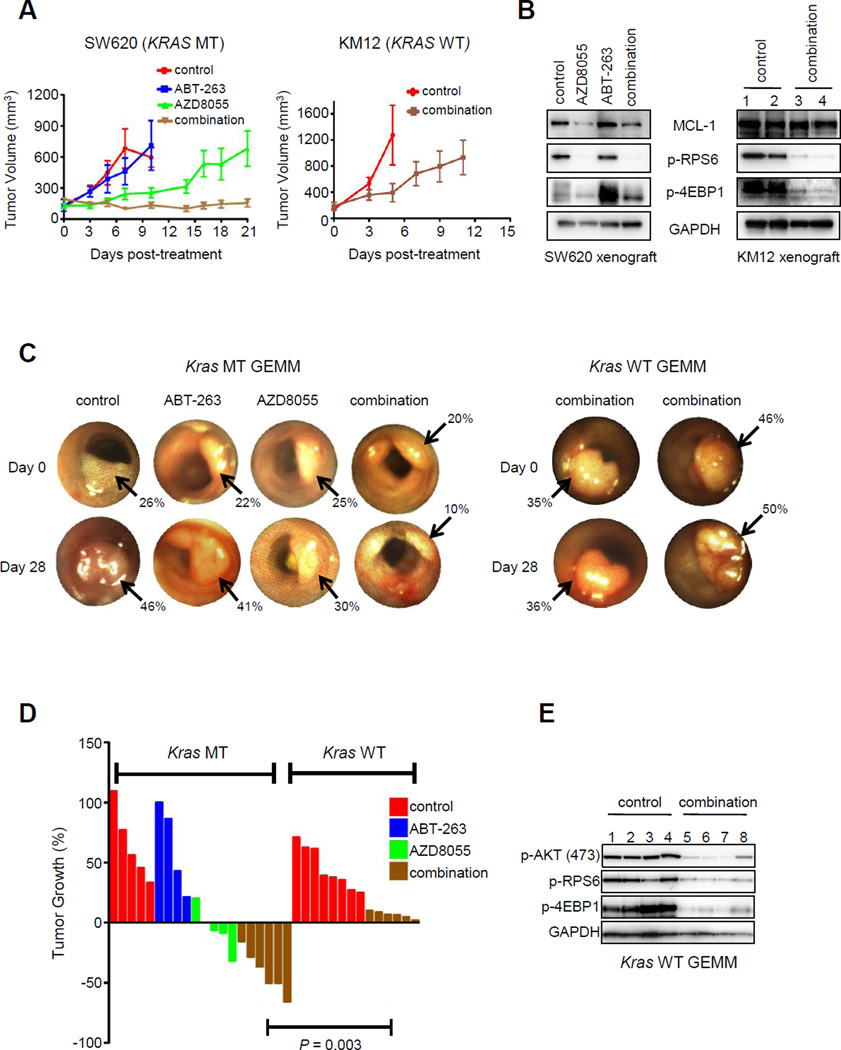
Since the ABT-263/AZD8055 combination in KRAS MT CRCs induces both apoptosis (Fig. 2A) and growth arrest (Sup. Fig. 4A), we evaluated this therapeutic strategy in vivo against established KRAS MT CRCs. In a SW620 KRAS MT CRC xenograft tumor model, combination treatment resulted in marked tumor growth impairment, and even some regressions (Fig. 4A, left panel). AZD8055 downregulated MCL-1 and there was a marked induction of cell death following combination therapy in vivo (Fig. 4B, Sup. Fig. 9A). Notably, SW620 tumors have been resistant to several other combination approaches in clinical development, highlighting the potential of the current approach (Sup. Fig. 9B). As expected, the combination of AZD8055/ABT-263 failed to significantly prevent tumor growth or suppress MCL-1 expression in a KRAS WT xenograft tumor model (Fig. 4A, right panel, Fig. 4B). Thus, this combination demonstrated more impressive activity against the KRAS mutant CRC in vivo, consistent with the in vitro data.
Figure 4. The combination of AZD8055/ABT-263 has in vivo efficacy in KRAS mutant CRC.
(A) Cohorts of KRAS MT SW620 xenograft tumors (left panel) were treated with no drug (control), AZD8055 (16 mg/kg/qd), ABT-263 (80 mg/kg/qd), or the combination (16 mg/kg/qd AZD8055, 80 mg/kg/qd ABT-263). Average tumor sizes for each treatment group are plotted. Cohorts of KRAS wild-type KM12 xenograft tumors (right panel) were treated with vehicle control or AZD8055/ABT-263 combination, and average tumor sizes are plotted. (B) Approximately three hours following the final drug treatment, mice were euthanized and tumors were harvested. Protein lysates were prepared from SW620 (left) and KM12 (right) tumors and were subjected to Western blot analyses with the indicated antibodies. (C) Representative images of tumors taken from serial colonoscopies of KRAS MT and KRAS WT GEMMs as treated in (A). Arrows depict each tumor. Tumor size was calculated by determining the percent occlusion of the lumen as described in Materials and Methods. (D) Colorectal cancers in KRAS MT and KRAS WT GEMMs were treated as in (A) for 28 days and changes in tumor volume for each mouse are shown by a waterfall plot. (E) Lysates from KRAS WT GEMM tumors were harvested approximately three hours following the last drug treatment of the experiment, and protein lysates were subjected to Western blotting analyses with the indicated antibodies. Each number represents a different tumor.
We next tested this combination in genetically engineered mouse models (GEMMs) of KRAS MT and KRAS WT CRC. These GEMMs are valuable preclinical models. In contrast to xenograft cancer models, the GEMM tumors develop in the physiologically relevant microenvironment of the colon and do not undergo growth selection in vitro prior to in vivo testing (42). Furthermore, the ability to monitor tumor growth and/or regressions using optical colonoscopy allows us to follow the dynamic tumor responses to treatment in real time, making this model particularly valuable and effective for evaluating experimental therapeutics. We examined the efficacy of the combination regimen on mice with colonic tumors that were APC mutant (null) (48) with or without KRAS mutation (42). The growth or regressions of individual tumors were monitored weekly by serial colonoscopies (42). The KRAS MT CRC model treated with combination therapy exhibited shrinkage of all treated tumors (Fig. 4C,D). In contrast, the combination failed to induce tumor regressions in KRAS WT GEMMs (Fig. 4C,D), despite effective inhibition of TORC1/2 signaling in vivo (Fig. 4E). Therefore, GEMMs of CRC reflect our findings in human CRC cell lines assessed in vitro and as xenografts in vivo, further supporting the efficacy and specificity of this regimen for KRAS MT CRCs.
Discussion
Advanced CRCs with KRAS or BRAF mutations require new therapeutic options. Here, we show that combining AZD8055 with ABT-263 induces robust apoptosis in KRAS and BRAF MT human CRC cell lines. Further, this combination produced tumor regressions in KRAS MT CRCs in vivo both in human xenografts and in GEMMs, increasing the likeliness that this combination may have efficacy in clinical trials of KRAS MT CRCs. While our initial observation that KRAS and BRAF MT CRCs had enhanced sensitivity to single-agent obatoclax compared to their WT counterparts, we believe that this combination is likely to be preferable to obatoclax for two main reasons: 1) BH3 mimetics alone induce apoptosis but do not appear to induce growth arrest, and 2) clinical trial data suggest that the dose-limiting toxicities of obatoclax may preclude sufficient target inhibition, in contrast to ABT-263 (30, 34, 35). Furthermore, we are optimistic that this combination may have an attractive therapeutic index. Doses of AZD8055 as low as ~17nM effectively reduced MCL-1 levels and sensitized the cells to ABT-263 (Sup. Fig. 10A,B). Even when we decreased the concentrations of AZD8055 in vivo from 16mg/kg to 2mg/kg, significant tumor inhibition in the SW620 model was still achieved (Sup. Fig.10C).
Recently, there have been a number of studies that have sought effective approaches to treat KRAS mutant cancers. These therapies include MEK inhibition in combination with BCL-2/XL inhibition (16), STK33 inhibition (13), TAK1 inhibition (14), MEK inhibition in combination with PI3K inhibition (11), GATA2 inhibition (15, 49), CDC6 inhibition (49), APC/C inhibition (50) and PLK1 inhibition (50). At this point, it is difficult to predict which approaches will ultimately emerge as effective therapies in the clinic, and potential therapeutic indices for each of these therapies have yet to be established. This will likely have a large impact on determining which approaches have greatest clinical value. In addition, there may be different subsets of KRAS mutant cancers that are susceptible to a particular combination. As an example, the combination of a MEK inhibitor with a BCL-2/XL inhibitor demonstrated efficacy across a large swath of KRAS mutant cancers, including KRAS mutant lung cancers, but tended to be less effective in KRAS mutant cancers with mesenchymal features (16). In contrast, the combination of AZD8055 with a BCL-2/XL inhibitor appeared less active against KRAS MT lung cancers, but demonstrated potency against all KRAS MT CRCs examined.
The current study offers some unique components that may facilitate drug development. Most published studies utilized RNA interference approaches to identify potential therapeutic strategies, and thus further drug development efforts will be necessary to achieve clinical translation. The current study differs in that we utilized data from a high- throughput drug screen involving clinically relevant inhibitors to identify a treatment strategy specific for KRAS MT CRC. Although our current approach restricts the number of targets being explored (compared to comprehensive RNA interference screens), it is rather straightforward to examine the efficacy of this drug combination in the clinic relatively quickly.
The combination developed in this study for KRAS MT CRCs is less active in KRAS WT CRCs. The activity of this combination centers on the sensitization of these cancers to ABT-263 by suppression of MCL-1 expression upon treatment with TORC1/2 inhibitors. In contrast to the KRAS mutant CRCs, the WT CRCs were not sensitized to ABT-263 by downregulation of MCL-1 nor did they have MCL-1 expression under the regulation of TORC. Although further studies will be required to determine the activity of this combination in other subsets of cancer, we speculate that this combination may be effective for cancers that have MCL-1 expression under strict regulation of the TORC1 pathway and are sensitive to simultaneous BCL-2/BCL-XL/MCL-1 disruption.
Overall, we believe that the combination of TORC inhibitors with BCL-2/XL inhibitors are mechanistically designed to induce apoptosis and growth arrest in the subset of CRCs with KRAS or BRAF mutations and warrants further investigation as a potential clinical treatment for these recalcitrant cancers.
MATERIALS AND METHODS
Cell Lines
All of the cell lines used in this study were provided by the Center for Molecular Therapeutics (CMT) at Massachusetts General Hospital, with the exception of the genetically engineered mouse model cell lines (42) and the HCT-116 isogenic cell lines (44). The cell lines from CMT routine undergo cell line authentication testing by SNP and STR analysis. These cell lines have been acquired over the past five years. Limited genotyping of human colorectal cell lines used in the drug screen is available in Supp. Table 1. The human colorectal cell lines used in Western blotting, apoptosis assays and cell cycle assays were grown in DMEM with 10% FBS in the presence of 1ug/ml penicillin and streptomycin, expect the BRAF mutant COLO-205 and LS-411N cell lines which were grown in RPMI with 10% FBS. The KRAS mutant lung cancer cell lines (H2009, H460, H2030 and H358) were grown in RPMI with 10% FBS.
Drug Screen
The drug screen has previously been described (23) as has the website for publically accessible drug-sensitivity data (27) (www.cancerRxgene.org). Data analysis was performed using data sets available in early 2013 and these data are recorded in Supplemental Table 1.
Antibodies and Reagents
The following antibodies were used for Western blot analyses: phospho-AKT (473) (cat #9271), phospho-RPS6 (240/244) (cat #2211), phospho-4EBP1 (cat #2855), phospho-ERK (cat #9101), total RPS6 (cat #2217), phospho-4EBP1 (cat #2855), BIM (cat #2819), BAK (cat #3814) and BCL-XL (cat #2762) were from Cell Signaling (Beverly, MA). MCL-1 (S-19) antibody was from Santa Cruz Biotechnology (Santa Cruz, CA). Antibody to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was purchased from Chemicon. Annexin-Cy5 was from Biosource International, Camarillo, CA. Propidium iodide was from Sigma-Aldrich. Opti-MEM was from Invitrogen (Carlsbad, CA). Protein A sepharose beads were from Amersham (GE Healthcare).
Western Blotting
For Western blotting of cellular lystates, cells were resuspended in lysis buffer (20 mM Tris, 150 mM NaCl, 1% Nonidet P-40, 0.1 mM EDTA, and protease and phosphatase inhibitors), incubated on ice for 10 m and centrifuged at 14,000 rpm for 10 minutes. For whole tumor lysates, these were prepared by homogenization with a TH tissue homogenizer (Omni International) in tissue lysis buffer (25mM Tris-HCl (pH 7.6), 150mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS, 1mM DTT, and protease and phosphatase inhibitors), incubated for 10 minutes on ice, vigorously shaken for 30 minutes at 4 °C at 1400 rpm in a thermomixer and centrifuged at 14,000 rpm for 10 minutes at 4 °C. Protein concentration was determined by BCA Protein Assay (Pierce). Proteins were resolved using the NuPAGE® Novex® Midi Gel system on 4% to 12% Bis-Tris Gels (Invitrogen), transferred to nylon membranes and probed with the antibodies listed above. Representative blots are shown from several experiments. Chemiluminescence was detected with the Syngene G:Box camera (Synoptics). All measurements were performed in the linear range without saturation and were normalized to GAPDH or ACTIN loading control.
Apoptosis
Cells were seeded at roughly 30–40% confluency and cells were treated for the indicated times with the indicated treatments, with no-treatment controls in parallel. Treatments were in triplicate where indicated. Apoptosis experiments were performed as previously described (24) and analyzed on a BD LSR III (Becton Dickenson). The number of cells in quadrants II and IV (Annexin positive) were counted as apoptotic.
Cell Cycle
Cells were seeded at roughly 30–40% confluency and were treated overnight with the indicated treatments in triplicate, with no-treatment controls in parallel and the cells were prepared for cell cycle analysis as previously described (24) and analyzed on a BD LSR III (Becton Dickenson).
siRNA Experiments
For the siRNA experiments, MCL-1 and scrambled control oligos (Dharmacon smart pool) were used at a concentration of 50nM and transfected with Hiperfect reagent (Qiagen). Briefly, cells were plated to achieve next day confluency of roughly 60% confluency. 75uL of Hiperfect was added to 750uL of OPTI-MEM in a 1.5mL eppendorf tube. In parallel, 10nM of siRNA was added to 750uL of Opti-MEM in a separate 1.5mL eppendorf tube. After ten minutes, the tubes were combined and mixed gently. Following ten more minutes, the Hiperfect-siRNA mix was then drop-wise added to cells, and gently shaken for five minutes. 24 hours later, the cells were either re-seeded in 6-well plates and treated with the appropriate drug the following day, or lysed for Western blot analysis to determine the efficiency of the knockdown.
shRNA Experiments
For the shRNA experiments, cells were transduced with tetracycline-inducible shKRAS vectors as previously described (9). Following selection in 2ug/ml puromycin, cells were grown in Tet-approved FBS and knockdown was performed in the presence of 50ng/ml of doxycycline for 72 hours.
Protein Complex Immunoprecipitation
Following indicated drug treatments, cells were lysed in the same buffer as used for Western blotting. 20uL of protein A sepharose beads were added to 250–500µg of protein, and 5µg of either MCL-1 antibody (S-19, Santa Cruz technology) or BIM antibody (cat# 2819, Cell Signaling) were added to the lysates. Following overnight incubation with motion, the supernatant was collected and the cell pellets were washed three times with PBS.
Xenograft Studies
KrasG12D genetically engineered mouse model
Homozygous mutant APC and heterozygous mutant KRAS G12D mice were generated as previously described1.
Adenoviral Infection of Colonic Epithelium
Tumors were induced in the colons of Apc−/− KrasG12D/+ mice by adenovirus expressing cre recombinase and followed by optical colonoscopy as previously described (42).
In vivo treatments
For the human xenografts, KM-12 (n=5 for treatment group) or SW620 cells (n =4 for each cohort) were injected (5 × 106 cells per mouse) into the rear right flanks of athymic nude mice (Charles River Laboratories). When tumors reached approximately 100mm3 to 200mm3, treatments began. All drugs were administered directly to the stomach daily by oral gavage, unless otherwise indicated in the figure legend describing the data. For xenograft experiments, tumors were measured by electronic caliper two to three times a week, in two-dimensions (length and width), and with the formula : v = l × (w)2 (π/6), where v = tumor volume, l = length, and w =width. For GEMMs, tumor size was determined by the Tumor Size Index (TSI) as calculated by (tumor area/colonic lumen area)×100 (%) (42).
All animal experiments were performed in accordance with Massachusetts General Hospital Animal Care or Tufts Institutional Animal Care and Use Committee.
Histopathology and immunohistochemistry
Tissue was fixed in 10% paraformaldehyde overnight, embedded in paraffin, and sectioned at the Department of Pathology, Massachusetts General Hospital. Cell death was determined by cleaved caspase 3 (CC3) positivity.
Statistical Considerations
IC50s for cell lines were calculated as previously described in the drug screens6. The Statistical tests used in this study were Student’s t-test (two-sided) and Wilcoxon rank sum tests performed using Graphpad Prism Software or R version 2.14.1. Differences were considered statistically different if P<0.05. The average amount of apoptosis between all the mutant groups and all the wild-type groups, as described in the text, is +/− S.E.M.
Supplementary Material
Significance.
Effective targeted therapies directed against CRC with activating mutations in KRAS remains elusive. We have leveraged drug screen data from a large panel of human CRCs to uncover an effective, rational targeted therapy strategy that has preferential activity in CRCs with KRAS or BRAF mutations. This combination may be developed for clinical testing.
Acknowledgments
Financial Support: This was work was supported by DF/HCC Gastrointestinal Cancer SPORE P50 CA127003 (J.A. Engelman), grants from the National Institute of Health: R01CA140594 (J.A. Engelman), R01CA137008 (J.A. Engelman), 1U54HG006097-01 (C.H. Benes), a grant from the Wellcome Trust (086357; C.H. Benes), awards from the Burroughs Wellcome Fund and the Howard Hughes Medical Institute (M.N.R.), and an American Cancer Society Postdoctoral Fellowship (A.C. Faber).
Footnotes
There are no conflicts of interest to disclose
REFERENCES
- 1.Artale S, Sartore-Bianchi A, Veronese SM, Gambi V, Sarnataro CS, Gambacorta M, et al. Mutations of KRAS and BRAF in primary and matched metastatic sites of colorectal cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2008;26:4217–4219. doi: 10.1200/JCO.2008.18.7286. [DOI] [PubMed] [Google Scholar]
- 2.Vaughn CP, Zobell SD, Furtado LV, Baker CL, Samowitz WS. Frequency of KRAS, BRAF, and NRAS mutations in colorectal cancer. Genes, chromosomes & cancer. 2011;50:307–312. doi: 10.1002/gcc.20854. [DOI] [PubMed] [Google Scholar]
- 3.Tian S, Simon I, Moreno V, Roepman P, Tabernero J, Snel M, et al. A combined oncogenic pathway signature of BRAF, KRAS and PI3KCA mutation improves colorectal cancer classification and cetuximab treatment prediction. Gut. 2013;62:540–549. doi: 10.1136/gutjnl-2012-302423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yeh JJ, Routh ED, Rubinas T, Peacock J, Martin TD, Shen XJ, et al. KRAS/BRAF mutation status and ERK1/2 activation as biomarkers for MEK1/2 inhibitor therapy in colorectal cancer. Molecular cancer therapeutics. 2009;8:834–843. doi: 10.1158/1535-7163.MCT-08-0972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karapetis CS, Khambata-Ford S, Jonker DJ, O'Callaghan CJ, Tu D, Tebbutt NC, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. The New England journal of medicine. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 6.Laurent-Puig P, Cayre A, Manceau G, Buc E, Bachet JB, Lecomte T, et al. Analysis of PTEN, BRAF, and EGFR status in determining benefit from cetuximab therapy in wild-type KRAS metastatic colon cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27:5924–5930. doi: 10.1200/JCO.2008.21.6796. [DOI] [PubMed] [Google Scholar]
- 7.Siena S, Sartore-Bianchi A, Di Nicolantonio F, Balfour J, Bardelli A. Biomarkers predicting clinical outcome of epidermal growth factor receptor-targeted therapy in metastatic colorectal cancer. Journal of the National Cancer Institute. 2009;101:1308–1324. doi: 10.1093/jnci/djp280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di Nicolantonio F, Martini M, Molinari F, Sartore-Bianchi A, Arena S, Saletti P, et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2008;26:5705–5712. doi: 10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- 9.Ebi H, Corcoran RB, Singh A, Chen Z, Song Y, Lifshits E, et al. Receptor tyrosine kinases exert dominant control over PI3K signaling in human KRAS mutant colorectal cancers. The Journal of clinical investigation. 2011;121:4311–4321. doi: 10.1172/JCI57909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nature medicine. 2008;14:1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halilovic E, She QB, Ye Q, Pagliarini R, Sellers WR, Solit DB, et al. PIK3CA mutation uncouples tumor growth and cyclin D1 regulation from MEK/ERK and mutant KRAS signaling. Cancer research. 2010;70:6804–6814. doi: 10.1158/0008-5472.CAN-10-0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scholl C, Frohling S, Dunn IF, Schinzel AC, Barbie DA, Kim SY, et al. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell. 2009;137:821–834. doi: 10.1016/j.cell.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 14.Singh A, Sweeney MF, Yu M, Burger A, Greninger P, Benes C, et al. TAK1 inhibition promotes apoptosis in KRAS-dependent colon cancers. Cell. 2012;148:639–650. doi: 10.1016/j.cell.2011.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar MS, Hancock DC, Molina-Arcas M, Steckel M, East P, Diefenbacher M, et al. The GATA2 transcriptional network is requisite for RAS oncogene-driven non-small cell lung cancer. Cell. 2012;149:642–655. doi: 10.1016/j.cell.2012.02.059. [DOI] [PubMed] [Google Scholar]
- 16.Corcoran RBCK, Ebi H, Hata A, Faber AC, Coffee EM, Greninger P, Brown RD, Godfrey JT, Cohoon TJ, Song Y, Lifshits E, Hung KE, Shioda T, Dias-Santagata D, Singh A, Settleman J, Benes CH, Mino-Kenudson M, Wong KK, Engelman JA. Synthetic lethal interaction of combined BCL-XL and MEK inhibition promotes tumor regressions in KRAS mutant cancer models. doi: 10.1016/j.ccr.2012.11.007. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 18.Popovici V, Budinska E, Tejpar S, Weinrich S, Estrella H, Hodgson G, et al. Identification of a poorprognosis BRAF-mutant-like population of patients with colon cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2012;30:1288–1295. doi: 10.1200/JCO.2011.39.5814. [DOI] [PubMed] [Google Scholar]
- 19.Tol J, Nagtegaal ID, Punt CJ. BRAF mutation in metastatic colorectal cancer. The New England journal of medicine. 2009;361:98–99. doi: 10.1056/NEJMc0904160. [DOI] [PubMed] [Google Scholar]
- 20.Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. The New England journal of medicine. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–103. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 22.Corcoran RB, Ebi H, Turke AB, Coffee EM, Nishino M, Cogdill AP, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer discovery. 2012;2:227–235. doi: 10.1158/2159-8290.CD-11-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Faber AC, Corcoran RB, Ebi H, Sequist LV, Waltman BA, Chung E, et al. BIM expression in treatment-naive cancers predicts responsiveness to kinase inhibitors. Cancer discovery. 2011;1:352–365. doi: 10.1158/2159-8290.CD-11-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Faber AC, Ebi H, Costa C, Engelman JA. Apoptosis In Targeted Therapy Responses: The Role of BIM. Adv Pharmacol. 2012;65:519–542. doi: 10.1016/B978-0-12-397927-8.00016-6. [DOI] [PubMed] [Google Scholar]
- 26.Brachmann SM, Hofmann I, Schnell C, Fritsch C, Wee S, Lane H, et al. Specific apoptosis induction by the dual PI3K/mTor inhibitor NVP-BEZ235 in HER2 amplified and PIK3CA mutant breast cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:22299–22304. doi: 10.1073/pnas.0905152106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang W, Soares J, Greninger P, Edelman EJ, Lightfoot H, Forbes S, et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic acids research. 2013;41:D955–D961. doi: 10.1093/nar/gks1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tse C, Shoemaker AR, Adickes J, Anderson MG, Chen J, Jin S, et al. ABT-263: a potent and orally bioavailable Bcl-2 family inhibitor. Cancer research. 2008;68:3421–3428. doi: 10.1158/0008-5472.CAN-07-5836. [DOI] [PubMed] [Google Scholar]
- 29.Shoemaker AR, Mitten MJ, Adickes J, Ackler S, Refici M, Ferguson D, et al. Activity of the Bcl-2 family inhibitor ABT-263 in a panel of small cell lung cancer xenograft models. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14:3268–3277. doi: 10.1158/1078-0432.CCR-07-4622. [DOI] [PubMed] [Google Scholar]
- 30.Gandhi L, Camidge DR, Ribeiro de Oliveira M, Bonomi P, Gandara D, Khaira D, et al. Phase I study of Navitoclax (ABT-263), a novel Bcl-2 family inhibitor, in patients with small-cell lung cancer and other solid tumors. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29:909–916. doi: 10.1200/JCO.2010.31.6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roberts AW, Seymour JF, Brown JR, Wierda WG, Kipps TJ, Khaw SL, et al. Substantial susceptibility of chronic lymphocytic leukemia to BCL2 inhibition: results of a phase I study of navitoclax in patients with relapsed or refractory disease. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2012;30:488–496. doi: 10.1200/JCO.2011.34.7898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen M, Marcellus RC, Roulston A, Watson M, Serfass L, Murthy Madiraju SR, et al. Small molecule obatoclax (GX15-070) antagonizes MCL-1 and overcomes MCL-1-mediated resistance to apoptosis. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:19512–19517. doi: 10.1073/pnas.0709443104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paik PK, Rudin CM, Pietanza MC, Brown A, Rizvi NA, Takebe N, et al. A phase II study of obatoclax mesylate, a Bcl-2 antagonist, plus topotecan in relapsed small cell lung cancer. Lung Cancer. 2011;74:481–485. doi: 10.1016/j.lungcan.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hwang JJ, Kuruvilla J, Mendelson D, Pishvaian MJ, Deeken JF, Siu LL, et al. Phase I dose finding studies of obatoclax (GX15-070), a small molecule pan-BCL-2 family antagonist, in patients with advanced solid tumors or lymphoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2010;16:4038–4045. doi: 10.1158/1078-0432.CCR-10-0822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schimmer AD, O'Brien S, Kantarjian H, Brandwein J, Cheson BD, Minden MD, et al. A phase I study of the pan bcl-2 family inhibitor obatoclax mesylate in patients with advanced hematologic malignancies. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14:8295–8301. doi: 10.1158/1078-0432.CCR-08-0999. [DOI] [PubMed] [Google Scholar]
- 36.Wendel HG, Silva RL, Malina A, Mills JR, Zhu H, Ueda T, et al. Dissecting eIF4E action in tumorigenesis. Genes & development. 2007;21:3232–3237. doi: 10.1101/gad.1604407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsieh AC, Costa M, Zollo O, Davis C, Feldman ME, Testa JR, et al. Genetic dissection of the oncogenic mTOR pathway reveals druggable addiction to translational control via 4EBP-eIF4E. Cancer cell. 2010;17:249–261. doi: 10.1016/j.ccr.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mills JR, Hippo Y, Robert F, Chen SM, Malina A, Lin CJ, et al. mTORC1 promotes survival through translational control of Mcl-1. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:10853–10858. doi: 10.1073/pnas.0804821105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Faber AC, Li D, Song Y, Liang MC, Yeap BY, Bronson RT, et al. Differential induction of apoptosis in HER2 and EGFR addicted cancers following PI3K inhibition. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:19503–19508. doi: 10.1073/pnas.0905056106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schatz JH, Oricchio E, Wolfe AL, Jiang M, Linkov I, Maragulia J, et al. Targeting cap-dependent translation blocks converging survival signals by AKT and PIM kinases in lymphoma. The Journal of experimental medicine. 2011;208:1799–1807. doi: 10.1084/jem.20110846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chresta CM, Davies BR, Hickson I, Harding T, Cosulich S, Critchlow SE, et al. AZD8055 is a potent, selective, and orally bioavailable ATP-competitive mammalian target of rapamycin kinase inhibitor with in vitro and in vivo antitumor activity. Cancer research. 2010;70:288–298. doi: 10.1158/0008-5472.CAN-09-1751. [DOI] [PubMed] [Google Scholar]
- 42.Hung KE, Maricevich MA, Richard LG, Chen WY, Richardson MP, Kunin A, et al. Development of a mouse model for sporadic and metastatic colon tumors and its use in assessing drug treatment. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:1565–1570. doi: 10.1073/pnas.0908682107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin ES, Belmont PJ, Sinnamon MJ, Richard LG, Yuan J, Coffee EM, et al. Development of a colon cancer GEMM-derived orthotopic transplant model for drug discovery and validation. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19:2929–2940. doi: 10.1158/1078-0432.CCR-12-2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shirasawa S, Furuse M, Yokoyama N, Sasazuki T. Altered growth of human colon cancer cell lines disrupted at activated Ki-ras. Science. 1993;260:85–88. doi: 10.1126/science.8465203. [DOI] [PubMed] [Google Scholar]
- 45.She QB, Halilovic E, Ye Q, Zhen W, Shirasawa S, Sasazuki T, et al. 4E-BP1 is a key effector of the oncogenic activation of the AKT and ERK signaling pathways that integrates their function in tumors. Cancer cell. 2010;18:39–51. doi: 10.1016/j.ccr.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Del Gaizo Moore V, Brown JR, Certo M, Love TM, Novina CD, Letai A. Chronic lymphocytic leukemia requires BCL2 to sequester prodeath BIM, explaining sensitivity to BCL2 antagonist ABT-737. The Journal of clinical investigation. 2007;117:112–121. doi: 10.1172/JCI28281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sale MJ, Cook SJ. The BH3 mimetic ABT-263 synergizes with the MEK1/2 inhibitor selumetinib/AZD6244 to promote BIM-dependent tumour cell death and inhibit acquired resistance. The Biochemical journal. 2013;450:285–294. doi: 10.1042/BJ20121212. [DOI] [PubMed] [Google Scholar]
- 48.Kuraguchi M, Wang XP, Bronson RT, Rothenberg R, Ohene-Baah NY, Lund JJ, et al. Adenomatous polyposis coli (APC) is required for normal development of skin and thymus. PLoS genetics. 2006;2:e146. doi: 10.1371/journal.pgen.0020146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steckel M, Molina-Arcas M, Weigelt B, Marani M, Warne PH, Kuznetsov H, et al. Determination of synthetic lethal interactions in KRAS oncogene-dependent cancer cells reveals novel therapeutic targeting strategies. Cell research. 2012;22:1227–1245. doi: 10.1038/cr.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, Westbrook TF, et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137:835–848. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.