Abstract
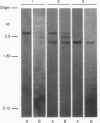
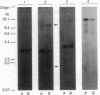
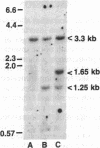
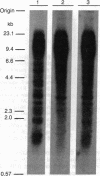
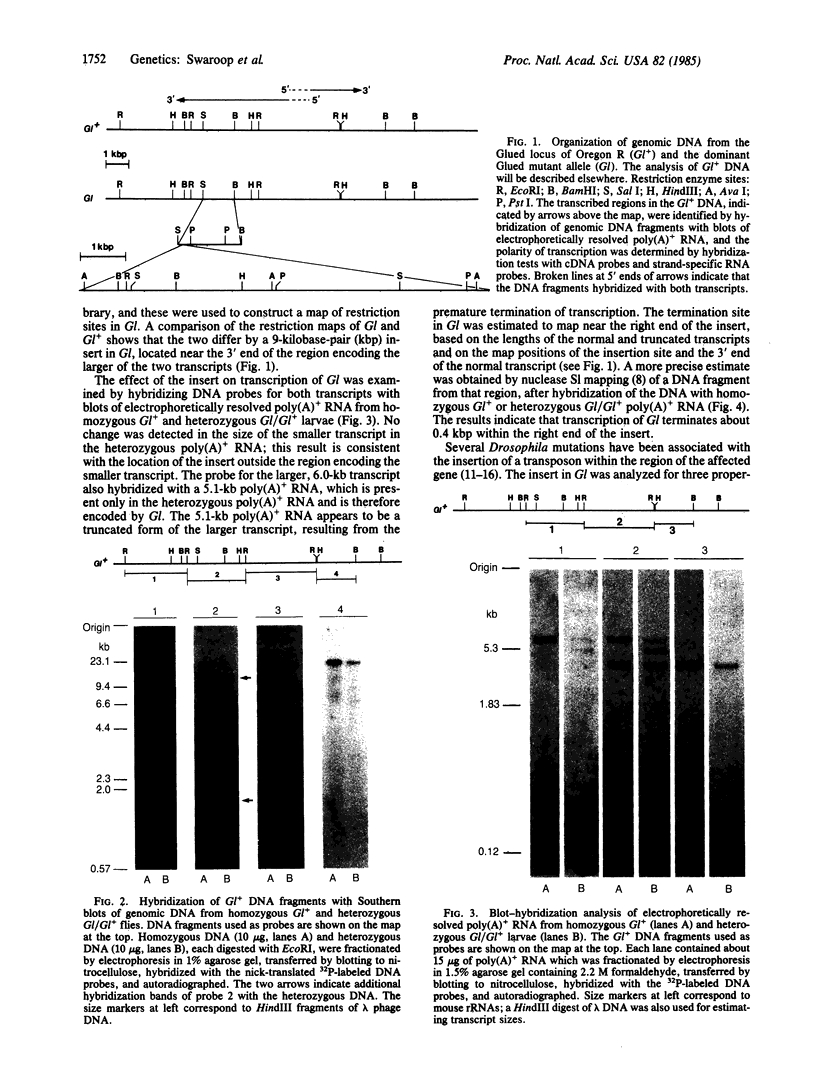
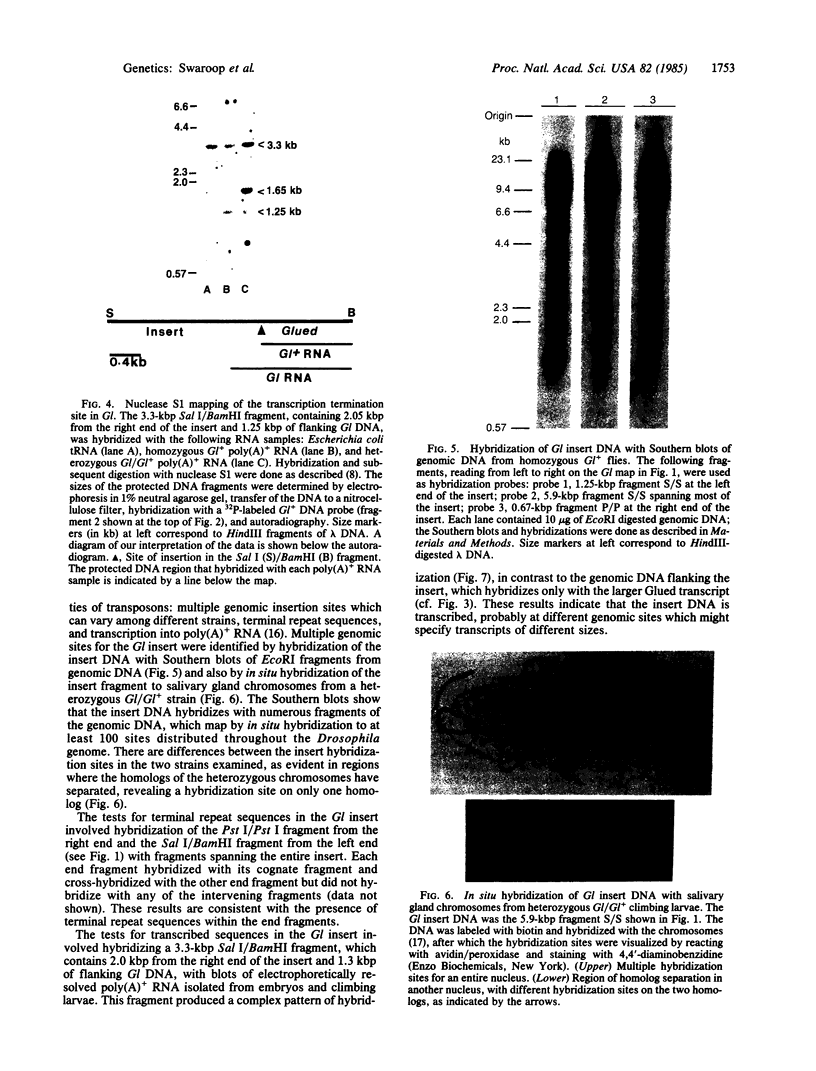
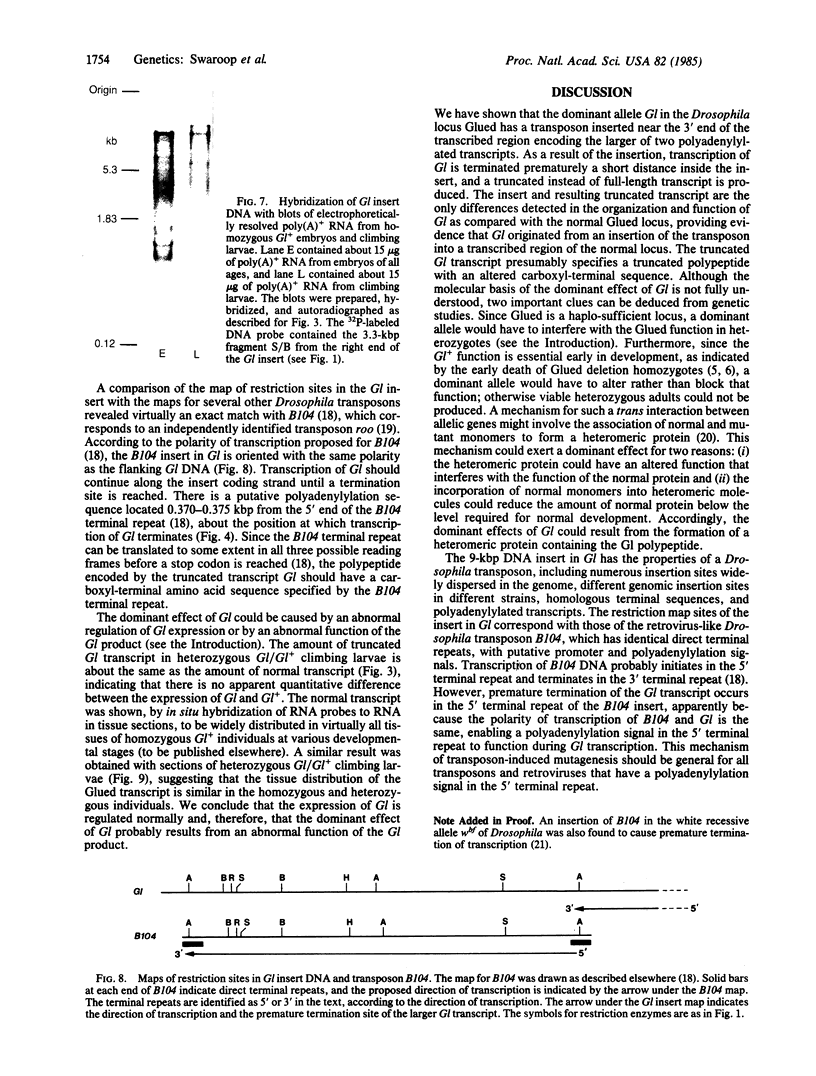
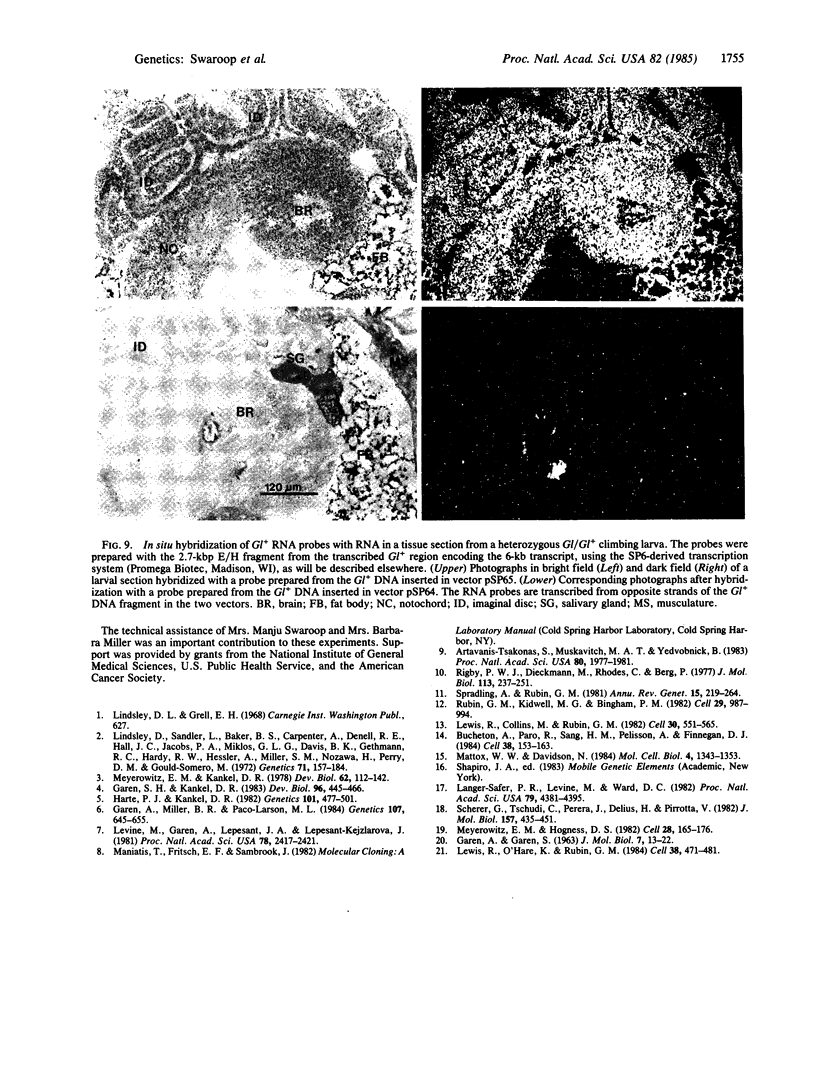
The organization of the Drosophila locus Glued containing the dominant allele Gl was shown to differ from that of the normal locus by an insertion of a 9-kilobase-pair DNA segment near the 3' end of a transcribed region. The insertion causes the formation of a truncated polyadenylylated transcript of 5.1 kilobases instead of the normal 6.0 kilobases. The inserted DNA segment has the properties of a transposon and was identified by its corresponding restriction map as B104, which is a retrovirus-like transposon with direct terminal repeats. B104 appears to be oriented in Gl with the same polarity of transcription as Gl. The truncated Gl transcript terminates prematurely inside the 5' terminal repeat of B104, in the region of a putative polyadenylylation signal. We discuss the general implications of this finding for transposon- and retrovirus-induced mutagenesis and for the origin of dominant mutations.
Full text
PDF
Images in this article
Selected References
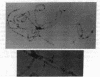
These references are in PubMed. This may not be the complete list of references from this article.
- Artavanis-Tsakonas S., Muskavitch M. A., Yedvobnick B. Molecular cloning of Notch, a locus affecting neurogenesis in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1977–1981. doi: 10.1073/pnas.80.7.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucheton A., Paro R., Sang H. M., Pelisson A., Finnegan D. J. The molecular basis of I-R hybrid dysgenesis in Drosophila melanogaster: identification, cloning, and properties of the I factor. Cell. 1984 Aug;38(1):153–163. doi: 10.1016/0092-8674(84)90536-1. [DOI] [PubMed] [Google Scholar]
- GAREN A., GAREN S. Complementation in vivo between structural mutants of alkaline phosphatase from E. coli. J Mol Biol. 1963 Jul;7:13–22. doi: 10.1016/s0022-2836(63)80015-7. [DOI] [PubMed] [Google Scholar]
- Garen A., Miller B. R., Paco-Larson M. L. Mutations affecting functions of the Drosophila gene glued. Genetics. 1984 Aug;107(4):645–655. doi: 10.1093/genetics/107.4.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garen S. H., Kankel D. R. Golgi and genetic mosaic analyses of visual system mutants in Drosophila melanogaster. Dev Biol. 1983 Apr;96(2):445–466. doi: 10.1016/0012-1606(83)90182-3. [DOI] [PubMed] [Google Scholar]
- Harte P. J., Kankel D. R. Genetic analysis of mutations at the Glued locus and interacting loci in Drosophila melanogaster. Genetics. 1982 Jul-Aug;101(3-4):477–501. doi: 10.1093/genetics/101.3-4.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer-Safer P. R., Levine M., Ward D. C. Immunological method for mapping genes on Drosophila polytene chromosomes. Proc Natl Acad Sci U S A. 1982 Jul;79(14):4381–4385. doi: 10.1073/pnas.79.14.4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M., Garen A., Lepesant J. A., Lepesant-Kejzlarova J. Constancy of somatic DNA organization in developmentally regulated regions of the Drosophila genome. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2417–2421. doi: 10.1073/pnas.78.4.2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis R., Collins M., Rubin G. M. FB elements are the common basis for the instability of the wDZL and wC Drosophila mutations. Cell. 1982 Sep;30(2):551–565. doi: 10.1016/0092-8674(82)90252-5. [DOI] [PubMed] [Google Scholar]
- Levis R., O'Hare K., Rubin G. M. Effects of transposable element insertions on RNA encoded by the white gene of Drosophila. Cell. 1984 Sep;38(2):471–481. doi: 10.1016/0092-8674(84)90502-6. [DOI] [PubMed] [Google Scholar]
- Lindsley D. L., Sandler L., Baker B. S., Carpenter A. T., Denell R. E., Hall J. C., Jacobs P. A., Miklos G. L., Davis B. K., Gethmann R. C. Segmental aneuploidy and the genetic gross structure of the Drosophila genome. Genetics. 1972 May;71(1):157–184. doi: 10.1093/genetics/71.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattox W. W., Davidson N. Isolation and characterization of the Beadex locus of Drosophila melanogaster: a putative cis-acting negative regulatory element for the heldup-a gene. Mol Cell Biol. 1984 Jul;4(7):1343–1353. doi: 10.1128/mcb.4.7.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerowitz E. M., Hogness D. S. Molecular organization of a Drosophila puff site that responds to ecdysone. Cell. 1982 Jan;28(1):165–176. doi: 10.1016/0092-8674(82)90386-5. [DOI] [PubMed] [Google Scholar]
- Meyerowitz E. M., Kankel D. R. A genetic analysis of visual system development in Drosophilia melanogaster. Dev Biol. 1978 Jan;62(1):112–142. doi: 10.1016/0012-1606(78)90096-9. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Rubin G. M., Kidwell M. G., Bingham P. M. The molecular basis of P-M hybrid dysgenesis: the nature of induced mutations. Cell. 1982 Jul;29(3):987–994. doi: 10.1016/0092-8674(82)90462-7. [DOI] [PubMed] [Google Scholar]
- Scherer G., Tschudi C., Perera J., Delius H., Pirrotta V. B104, a new dispersed repeated gene family in Drosophila melanogaster and its analogies with retroviruses. J Mol Biol. 1982 May 25;157(3):435–451. doi: 10.1016/0022-2836(82)90470-3. [DOI] [PubMed] [Google Scholar]
- Spradling A. C., Rubin G. M. Drosophila genome organization: conserved and dynamic aspects. Annu Rev Genet. 1981;15:219–264. doi: 10.1146/annurev.ge.15.120181.001251. [DOI] [PubMed] [Google Scholar]