Abstract
DNA sequencing by synthesis on a solid surface offers new paradigms to overcome limitations of electrophoresis-based sequencing methods. Here we report DNA sequencing by synthesis using photocleavable (PC) fluorescent nucleotides [dUTP-PC-4,4-difluoro-4-bora-3α,4α-diaza-s-indacene (Bodipy)-FL-510, dCTP-PC-Bodipy-650, and dUTP-PC-6-carboxy-X-rhodamine (ROX)] on a glass chip constructed by 1,3-dipolar azide-alkyne cycloaddition coupling chemistry. Each nucleotide analogue consists of a different fluorophore attached to the base through a PC 2-nitrobenzyl linker. We constructed a DNA microarray by using the 1,3-dipolar cycloaddition chemistry to site-specifically attach azido-modified DNA onto an alkyne-functionalized glass chip at room temperature under aqueous conditions. After verifying that the polymerase reaction could be carried out successfully on the above-described DNA array, we then performed a sequencing reaction on the chip by using a self-primed DNA template. In the first step, we extended the primer using DNA polymerase and dUTP-PC-Bodipy-FL-510, detected the fluorescent signal from the fluorophore Bodipy-FL-510, and then cleaved the fluorophore using 340 nm UV irradiation. This process was followed by extension of the primer with dCTP-PC-Bodipy-650 and the subsequent detection of the fluorescent signal from Bodipy-650 and its photocleavage. The same procedure was also performed by using dUTP-PC-ROX. The entire process was repeated five times by using the three fluorescent nucleotides to identify 7 bases in the DNA template. These results demonstrate that the PC nucleotide analogues can be incorporated accurately into a growing DNA strand during polymerase reaction on a chip, and the fluorophore can be detected and then efficiently cleaved using near-UV irradiation, thereby allowing the continuous identification of the template sequence.
Keywords: 1,3-dipolar azide-alkyne cycloaddition; DNA chip; DNA sequencing by synthesis; 2-nitrobenzyl linker; photocleavage
The completion of the Human Genome Project has set the stage for screening genetic mutations to identify disease genes on a genome-wide scale (1). Accurate high-throughput methods for resequencing the intron/exon regions of candidate genes are needed to explore the complete human genome sequence for disease-gene discovery. Recent studies also have demonstrated that an important route for identifying functional elements in the human genome is to sequence the genomes of many species that represent a wide sampling of the evolutionary tree (2). To overcome the limitations of the current sequencing technology based on electrophoresis using laser-induced fluorescence detection (3–5), a variety of new DNA-sequencing methods have been explored. Such techniques include pyrosequencing (6), MS sequencing (7–9), sequencing by hybridization (10), sequence-specific detection of single-stranded DNA using engineered nanopores (11), and sequencing of single DNA molecules (12) and polymerase colonies (13).
Recently, new DNA-sequencing approaches based on sequencing by synthesis (DSS) on a solid surface have attracted much attention, because they have the potential to offer an efficient method to decipher the genome and screen genetic mutations. However, only limited success with such approaches has been reported thus far. The key requirement for these methods to succeed is the ability to continuously determine the identity of each nucleotide immediately after its incorporation into a growing DNA strand in a polymerase reaction and to subsequently remove the reporter signal such as a fluorophore in a rapid and efficient manner after its detection and before the incorporation of the next nucleotide. To this end, we have designed a parallel DNA-sequencing chip system based on DSS (14). This system involves the construction of a chip with immobilized single-stranded DNA templates that can self-prime for the generation of a complementary DNA strand in a polymerase reaction using four unique fluorescently labeled nucleotide analogues. Each of the four nucleotide analogues consists of a fluorophore attached to the base through a photocleavable (PC) linker and a small chemically cleavable moiety to cap the 3′-OH group to allow temporary termination of the DNA polymerase reaction after the incorporation of each nucleotide. A four-color fluorescence imager is used to identify the incorporated nucleotide on each spot of the chip. After removing the fluorophore photochemically and cleaving the 3′-OH capping group, the polymerase reaction is allowed to proceed with the incorporation of the next nucleotide analogue and the detection of the subsequent base on the template sequence. A significant advantage offered by the photochemical cleavage of the fluorescent dye is that no additional chemical reagents are required to be introduced into the system, and clean products can be generated with no requirement for subsequent purification. We demonstrated previously that a 2′-deoxyuridine 5′-triphosphate bearing a fluorophore on its 5 position by a PC 2-nitrobenzyl linker [dUTP-PC-4,4-difluoro-4-bora-3α,4α-diaza-s-indacene (Bodipy)-FL-510] is an excellent substrate for the DNA polymerase Thermo Sequenase in a solution-phase DNA-extension reaction (15). The fluorophore was shown to be cleaved completely by near-UV irradiation (λ ≈ 340 nm) after its incorporation into a growing DNA strand. Subsequently, Mitra et al. (13) also demonstrated the use of the similar PC fluorescent nucleotides for in situ DNA sequencing on polymerase colonies. These results firmly established the feasibility of using PC fluorescent nucleotide analogues to perform DSS.
Alternative methods were also explored to remove the fluorescence signal from a fluorescent nucleotide in the DSS approach. Braslavsky et al. (12) reported the use of photobleaching to eliminate the fluorescent signal from an incorporated nucleotide before the addition of the subsequent nucleotide in polymerase reaction, but in this method, the photobleached fluorophores remain with the DNA template and interfere with the polymerase activity for the incorporation of the subsequent nucleotide. Mitra et al. (13) reported the utilization of a disulfide group as a chemically cleavable linker to attach a fluorophore to a deoxynucleotide and the use of 2-mercaptoethanol to remove the fluorophore after the nucleotide incorporation and detection. However, the disulfide bond can be reversed and destabilized under certain conditions (16, 17).
Another crucial requirement for the development of a sequencing approach based on DSS is the construction of a chip with an immobilized self-primed DNA moiety where the primer cannot be washed away from the DNA template. The development of a chemoselective coupling chemistry for the immobilization of DNA on a solid surface is essential for accurate gene-expression measurement (18) and polymorphism or mutation detection (19, 20). Because covalent coupling chemistries have been shown to typically lead to more stable DNA arrays than noncovalent chemistries, a variety of covalent coupling methods have been used for DNA immobilization on a solid surface (21–23). However, additional improvement of the coupling chemistry for immobilizing DNA on a surface is required to achieve high selectivity and coupling efficiency. One ideal property required for the functional groups to be coupled (one from the DNA and the other from the surface) is the stability of the groups in aqueous conditions, which are typically needed to perform the coupling. We previously explored the use of the 1,3-dipolar cycloaddition click chemistry between an azide and alkyne for coupling a f luorophore [5-carboxyf luorescein (FAM)] to single-stranded DNA (24). This chemistry was shown to chemoselectively produce FAM-labeled DNA in a quantitative yield under aqueous conditions at 80°C. Recently it was reported that this reaction can be conducted at room temperature in the presence of Cu(I) catalyst to produce only 1,4-regioisomeric triazoles (25, 26). Because of its mild reaction conditions and high selectivity and efficiency, this ligation reaction has also been applied for the modification of virus particles with fluorescent dyes (27), cell surface labeling (28), the profiling of enzyme activity in proteomes (29), and the attachment of oligosaccharides to microtiter plates for biological assays (30). We report here the use of this Cu(I)-catalyzed [3+2] azide-alkyne cycloaddition to immobilize DNA on a glass chip for DSS using PC fluorescent nucleotides. From the results of this study, we demonstrated that the nucleotide analogues could be incorporated successively and accurately into a growing DNA strand as substrates during a polymerase reaction on a glass chip, and the fluorescent dye could be detected and cleaved with high efficiency by using UV irradiation at 340 nm, thereby allowing the continuous identification of the template sequence.
Materials and Methods
All chemicals were purchased from Sigma–Aldrich unless otherwise indicated. 1H and 13C NMR spectra were recorded on Bruker (Billerica, MA) 400 and 300 spectrometers, respectively. High-resolution MS (HRMS) data were obtained by using a JEOL JMS HX 110A mass spectrometer. Mass measurement of DNA was made on a Voyager DE matrix-assisted laser desorption ionization–time-of-flight (MALDI-TOF) mass spectrometer (Applied Biosystems). A home-made contact-angle goniometer was used for the water contact-angle measurement, and the atomic force microscopy images were acquired by using a NanoScope multimode scanning probe microscope in the tapping mode. Photolysis was performed at 340 nm (10-nm band pass) by using a 450-W high-pressure xenon lamp (Thermo Oriel, Stratford, CT) in conjunction with a 340-nm UV interference filter (CVI Laser, Albuquerque, NM) with a light intensity of 6 mW/cm2. The scanned fluorescence images were obtained by using a ScanArray Express scanner (Perkin–Elmer Life Sciences), which was equipped with four lasers with excitation wavelengths of 488, 543, 594, and 633 nm and emission filters centered at 522, 570, 614, and 670 nm. All NMR data for newly synthesized molecules are reported in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
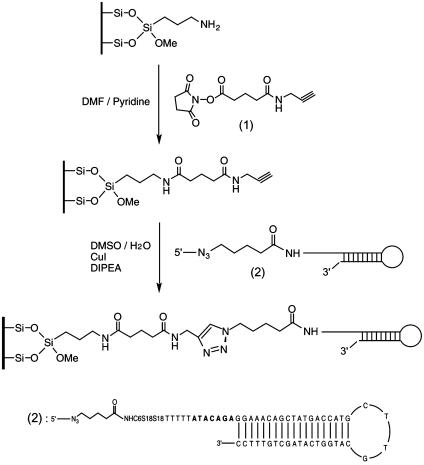
Construction of the DNA Chip by Using 1,3-Dipolar Azide-Alkyne Cycloaddition Coupling Chemistry. The DNA chip was constructed as shown in Scheme 1 and involved the following three steps.
Scheme 1.
Construction of the DNA chip using 1,3-dipolar azide-alkyne cycloaddition coupling chemistry. A glass chip is first functionalized to contain an alkynyl group, which forms a covalent bond with the 5′-azido-modified DNA.
I. Synthesis of the crosslinker succinimidyl N-propargyl glutariamidate (1). Glutaric anhydride (2.59 g, 22.7 mmol), propargylamine (1.27 g, 23 mmol), and triethylamine (2.33 g, 23 mmol) were dissolved in CH2Cl2 (50 ml) and stirred for 12 h at room temperature. After the CH2Cl2 solvent was evaporated, the residue was acidified by adding 1 M HCl solution (5 ml). The solvent was removed under vacuum, and the crude mixture was purified by silica gel chromatography (CHCl3/CH3OH = 5:1, Rf = 0.45) to yield 2.77 g of pure N-propargyl glutariamidic acid as a yellow solid (72% yield). HRMS [fast atom bombardment (FAB)+] m/z: calcd for C8H12O3N(M + H+), 170.0817; found, 170.0823. To a solution of N-propargyl glutariamidic acid (0.40 g, 2.36 mmol) in CH2Cl2 (25 ml) was added N-hydroxysuccinimide (NHS) (0.28 g, 2.4 mmol), followed by the addition of 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (0.46 g, 2.4 mmol) at room temperature. After stirring for 8 h, the mixture was washed with H2O(2 × 20 ml) and the aqueous layer was extracted with CH2Cl2 (30 ml). The combined organic layers were washed with brine, dried over Na2SO4, and concentrated in vacuo to yield 0.47 g of the crosslinker succinimidyl N-propargyl glutariamidate 1 as a yellow solid (75% yield). HRMS (FAB+) m/z: calcd for C12H15O5N2 (M + H+), 267.0981; found, 267.0995.
II. Synthesis of the azido-labeled DNA (2). The amino-labeled hairpin DNA (sequence shown in Scheme 1) was prepared by phosphoramidite chemistry on a DNA synthesizer. The self-primed DNA moiety consisted of a loop with a sequence [G(CTTG)C] and a stem formed by using the M13-28 reverse primer sequence. The stem was followed by a 7-bp sequence at the 5′ end with an “A” nucleotide inserted alternately (AGACATA). Five T nucleotides, two spacer 18 phosphoramidites (S18), and one 5′-amino modifier C6 phosphoramidite (Glen Research, Sterling, VA) were added subsequently. Azido-labeling of this DNA molecule was achieved by reacting the above-described 5′-amino-modified DNA with succinimidyl 5-azidovalerate in 0.25 M Na2CO3/NaHCO3 buffer (pH 9.0) for 12 h at room temperature. The resulting azido-labeled DNA was purified by size-exclusion chromatography and desalted with an oligonucleotide purification cartridge (24). The DNA product was analyzed by MALDI-TOF MS by using the amino-modified DNA as an internal standard. The theoretical mass difference between the amino-modified DNA and the azido-labeled DNA was calculated as 125 Da, and the observed mass difference was 119 Da, which is typically within the error range of MALDI-TOF MS measurement for DNA.
III. DNA immobilization on a glass surface. The amino-modified glass slide (Sigma) was cleaned by immersion into a basic solution [dimethylformamide (DMF)/N,N-diisopropyl-ethylamine (DIPEA), 9:1 (vol/vol)] for 1 h, sonicated for 5 min, washed with DMF and ethanol, and then dried under argon. The precleaned glass surface was functionalized to contain a terminal alkyne group by immersing it into the alkyne crosslinker solution [20 mM succinimidyl N-propargyl glutariamidate 1 in DMF/pyridine, 9:1 (vol/vol)] for 5 h at room temperature. After sonication for 5 min, the glass surface was washed with DMF and ethanol and dried under argon. The azido-labeled DNA was dissolved in DMSO/H2O [1:2 (vol/vol)] to obtain a 20 μM solution. This DNA solution then was spotted onto the alkynyl-functionalized glass surface in the form of 4-μl drops, followed by the addition of CuI (400 pmol, 5 eq) and DIPEA (400 pmol, 5 eq) solution. The glass slide was incubated in a humid chamber at room temperature for 12 h, then washed with deionized water (dH2O) and SPSC buffer (0.25 M sodium phosphate/2.5 M NaCl, pH 6.5) extensively for 1 h to remove nonspecifically bound DNAs (31), and finally rinsed with dH2O and ethanol. Atomic force microscopy and water contact-angle measurement were used to characterize the change on the surface after each step in the immobilization process.
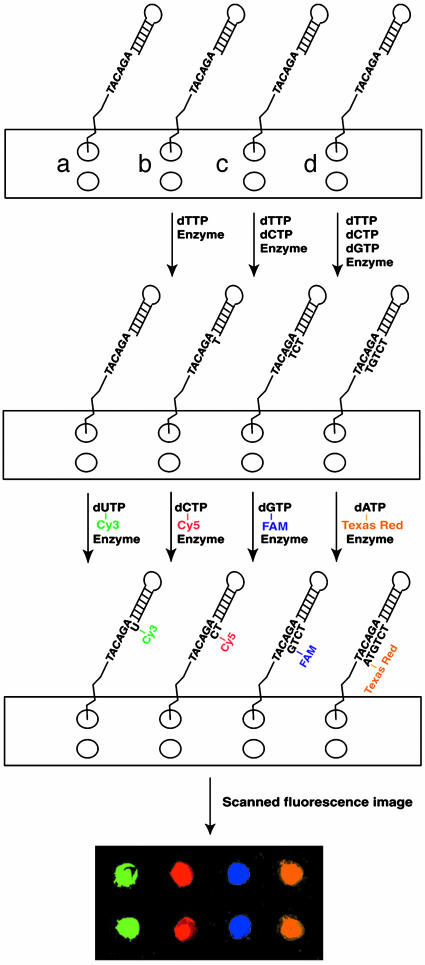
Polymerase-Extension Reaction on the DNA Chip Constructed by 1,3-Dipolar Azide-Alkyne Cycloaddition Chemistry. The overall procedure for the reaction is shown in Scheme 2. Each area (a–d) of the chip had two identical spots, which were spotted with the azido-modified DNA as described above. The extension reaction conditions are described in Supporting Materials and Methods.
Scheme 2.
Polymerase-extension reaction using four fluorescent nucleotides on a DNA chip constructed by using the 1,3-dipolar cycloaddition chemistry. Each of the four colors in the fluorescence image matched with the fluorescence emissions from the four fluorophores used to label the corresponding nucleotides (green, Cy3; red, Cy5; blue, FAM; orange, Texas red).
Synthesis of a PC Fluorescent Nucleotide: dCTP-PC-Bodipy-650. PC-Bodipy-650 was prepared by using Bodipy-650-NHS ester (Molecular Probes) and 1-[5-(aminomethyl)-2-nitrophenyl]ethanol following the procedures reported previously for the synthesis of PC-Bodipy-FL-510 (15). HRMS (FAB+) m/z: calcd for C38H38BF2N5O6S (M + H+), 741.2611; found, 741.2583.
PC-Bodipy-650-NHS ester was prepared by using the PC-Bodipy-650 prepared above and N,N′-disuccinimidyl carbonate following the procedures reported previously for the synthesis of PC-Bodipy-FL-510-NHS (15). HRMS (FAB+) m/z: calcd for C43H41BF2N6O10S (M + H+), 882.2674; found, 882.2697.
dCTP-PC-Bodipy-650 was synthesized from the coupling reaction between PC-Bodipy-650-NHS ester and 5-aminoallyl-2′-deoxycytidine-5′-triphosphate (dCTP-NH2) (TriLink BioTechnologies, San Diego). To a solution of dCTP-NH2 (1 mg, 2 μmol) in 300 μl of 0.1 M Na2CO3/NaHCO3 buffer (pH 8.5) was added PC-Bodipy-650-NHS ester (3 mg, 3.5 μmol) in 300 μl of acetonitrile and then stirred at room temperature for 5 h. Preparative TLC was used to remove the unreacted PC-Bodipy-650-NHS (CHCl3/CH3OH = 4:1), and the fractions containing dCTP-PC-Bodipy-650 were collected and purified further by reverse-phase HPLC by using similar conditions as described (15). The identity of the purified compound dCTP-PC-Bodipy-650 was confirmed by using it to generate a DNA-extension product, 5′-TCAAGGACGTACCCGCC(PC-Bodipy-650)G-biotin-3′, that was characterized by MALDI-TOF MS (calcd, 6,669; found, 6,668).
Synthesis of dUTP-PC-6-carboxy-X-rhodamine. dUTP-PC-6-carboxy-X-rhodamine (ROX) was synthesized and characterized following a similar procedure as described above (MALDI-TOF MS characterization of the DNA-extension product 5′-AGAGGATCCAACCGAGACU(PC-ROX)G-biotin-3′ containing dUTP-PC-ROX: calcd, 7,308; found, 7,307).
The detailed procedure for characterizing the above-described two PC nucleotide analogues by using DNA extension and photocleavage is described in Supporting Materials and Methods.
DSS on a Chip. One microliter of a solution consisting of dUTP-PC-Bodipy-FL-510 (20 pmol), Thermo Sequenase (2 units), and 1× reaction buffer was spotted on the area of the glass surface immobilized with the self-primed DNA, and the nucleotide analogue was allowed to incorporate into the primer at 72°C for 5 min. After washing with dH2O, SPSC buffer, 0.1% SDS, dH2O, and ethanol, the surface was scanned with the ScanArray Express microarray scanner to detect the fluorescence signal. To perform the photocleavage reaction, the glass chip was irradiated by UV light (λ ≈ 340 nm) for 10 min in acetonitrile/water [1:1 (vol/vol)] solution. After washing with dH2O, SPSC buffer, 0.1% SDS, and ethanol, the surface was scanned again to compare the intensity of fluorescence after photocleavage with the original fluorescence intensity. This process was followed by the incorporation of dCTP-PC-Bodipy-650, with the subsequent washing, fluorescence detection, and photocleavage processes performed as described above. The next three steps involved primer extension first by dUTP-PC-ROX, then by dGTP and dUTP-PC-Bodipy-FL-510, and finally by dATP and dUTP-PC-ROX, with the washing, detection, and photocleavage steps repeated between every successive incorporation. The absorption and emission maxima for each fluorophore used to construct the PC fluorescent nucleotides are as follows: Bodipy-FL-510, λabs = 502 nm and λem = 510 nm; Bodipy-650, λabs = 630 nm and λem = 650 nm; ROX, λabs = 575 nm and λem = 602 nm.
Results and Discussion
The DNA chip was constructed as shown in Scheme 1 by using the 1,3-dipolar cycloaddition coupling chemistry to attach the azido-labeled DNA onto the alkyne-modified glass surface in the presence of a Cu(I) catalyst. This cycloaddition was carried out under mild reaction conditions in an aqueous solution with high selectivity and efficiency. In addition, the two functional groups (azido and alkynyl) to be coupled are very stable in aqueous conditions. Thus, the 1,3-dipolar cycloaddition coupling chemistry provides an ideal coupling reaction to immobilize DNA covalently on a surface to construct a DNA chip. During the immobilization process, the change in topography of the solid surface was monitored and characterized by atomic force microscopy (Fig. 5, which is published as supporting information on the PNAS web site) and contact-angle measurement.
To evaluate the functionality, accessibility, and stability of the surface-bound DNA, a polymerase-extension reaction was carried out by using conventional dNTPs (Scheme 2). The DNA moiety contained a hairpin structure that formed an entity capable of self-priming in a polymerase reaction. The self-primed DNA moiety formed by the specific loop sequence [G(CTTG)C] has been shown to be thermally stable with a melting temperature of 86°C (32). Because of the specificity of self-priming, the possibility of mispriming was reduced. The sequence beyond the priming site was chosen as AGACATA, consisting of A nucleotides alternating with other nucleotides. This sequence was followed by two spacers and an amino linker that facilitated the attachment of DNA on the glass surface by reducing the effect of steric hindrance. The azido-modified loop DNA was spotted on the chip surface at areas designated as a–d (two spots were made for each label to demonstrate the reproducibility of the data). After DNA immobilization, the glass chip was incubated at 94°C for 10 min and 48°C for 10 min to ensure the complete denaturation and subsequent renaturation of the hairpin structure. In the first step of the extension reaction, unmodified dNTPs (none for spots a, dTTP for spots b, dTTP and dCTP for spots c, and dTTP, dCTP, and dGTP for spots d) and Thermo Sequenase DNA polymerase were added to each spot in accordance to the 7-bp sequence mentioned above. After the primer-extension reaction, the four spots (a–d) each had the primer extended by zero, one, three and five bases, generating DNA moieties with free A, G, C, and T bases, respectively, immediately after the priming site on the template. Each of four dye-labeled dNTPs (Cy3-dUTP for a, Cy5-dCTP for b, FAM-dGTP for c, and Texas red-dATP for d) then was added to the corresponding spot to allow the correct incorporation of a specific fluorescent nucleotide complementary to the corresponding base on the template. As shown in Scheme 2, four distinct fluorescence signals with different colors were generated by using the ScanArray scanner, matching the fluorescence emission produced by the fluorescent nucleotide incorporated on that particular spot. These results confirmed that the fluorescent nucleotides were incorporated accurately in a base-specific manner into the self-primed DNA after primer extension, and the synthesized DNA chip was thermally stable at high temperature. This result established the feasibility of carrying out DNA-extension reaction on the chip constructed by 1,3-dipolar azide-alkyne cycloaddition coupling chemistry. The nonspecific binding of DNA and free nucleotides on the chip contributed to <10% of the fluorescence signal (Supporting Materials and Methods).
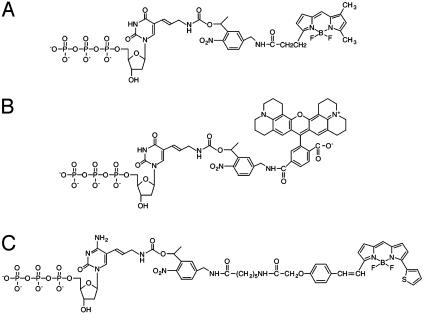
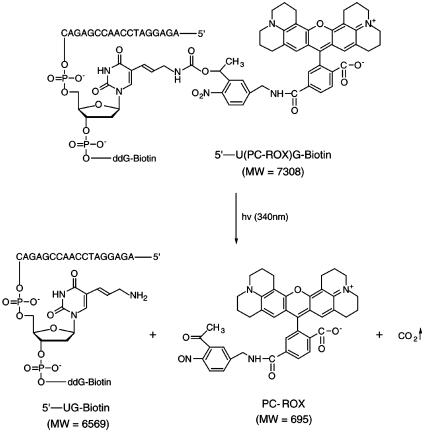
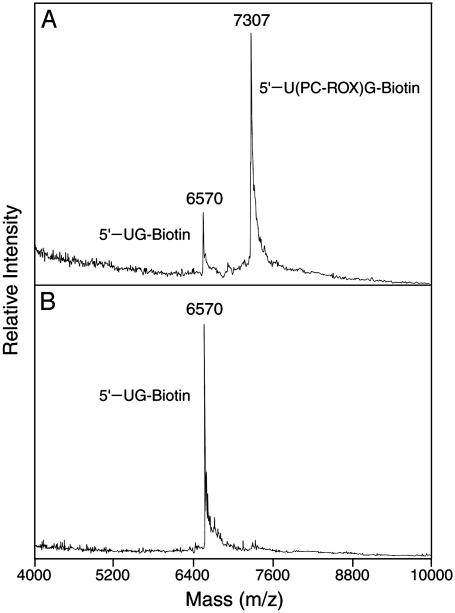
To demonstrate the feasibility of carrying out DSS on the chip, three fluorescent deoxynucleotide analogues (dUTP-PC-Bodipy-FL-510, dCTP-PC-Bodipy-650, and dUTP-PC-ROX) (Fig. 1) were synthesized and used to perform a sequencing reaction on the solid surface. The nucleotide analogues were synthesized by a similar synthetic method as reported before (15). These nucleotide analogues have different fluorescent dyes attached to the 5 position of the base (U/C) through a PC 2-nitrobenzyl linker. Previously we demonstrated that nucleotide analogue dUTP-PC-Bodipy-FL-510 can be incorporated faithfully by DNA polymerase into a growing DNA strand in a solution-phase polymerase reaction and that its incorporation does not inhibit the addition of the subsequent nucleotide. We also have shown that near-UV irradiation leads to the efficient release of the fluorophore, ensuring that the previous fluorescent signal does not leave any residue that otherwise could interfere with the detection of the subsequent nucleotide (15). Here we characterized the other two nucleotide analogues dCTP-PC-Bodipy-650 and dUTP-PC-ROX in a similar manner and have included the data for dUTP-PC-ROX as an example. We performed a DNA-extension reaction using dUTP-PC-ROX, ddGTP-biotin, and a synthetic template (100 bp) corresponding to a portion of exon 7 of the human p53 gene to generate DNA-extension product 5′-U(PC-ROX)G-biotin (Fig. 2). The purified product, 5′-U(PC-ROX)G-biotin, was analyzed by MALDI-TOF MS as shown in Fig. 3A, in which a strong peak corresponding to 5′-U(PC-ROX)G-biotin (m/z: calcd, 7,308; found, 7,307) is observed. A small peak at m/z 6,570 that corresponds to the photocleaved fragment 5′-UG-biotin (m/z: calcd, 6,569) was observed also. This peak is produced due to the photocleavage induced by the nitrogen laser pulse (337 nm) used for ionization in MALDI-TOF MS analysis. These results indicated that dUTP-PC-ROX was incorporated efficiently into the growing DNA strand by a DNA polymerase and that its incorporation does not hinder the addition of the subsequent nucleotide.
Fig. 1.
Structures of dUTP-PC-Bodipy-FL-510 (A), dUTP-PC-ROX (B), and dCTP-PC-Bodipy-650 (C).
Fig. 2.
DNA product 5′-U(PC-ROX)G-biotin, formed by incorporating a dUTP-PC-ROX into a primer in a polymerase reaction and its photocleavage, producing DNA fragment 5′-UG-biotin and PC-ROX. MW, molecular weight.
Fig. 3.
MALDI-TOF MS spectra of the DNA-extension product 5′-U(PC-ROX)G-biotin (m/z: calcd, 7,308; found 7,307) obtained by using dUTP-PC-ROX and its photocleavage product 5′-UG-biotin (m/z: calcd, 6,569; found, 6,570). (A) Without irradiation. (B) After 3 min of irradiation of the 5′-U(PC-ROX)G-biotin (λirr ≈ 340 nm).
Complete photocleavage of the fluorophore, ROX, from DNA-extension product 5′-U(PC-ROX)G-biotin is essential for the successful application of dUTP-PC-ROX in the DSS approach. We investigated the photocleavage efficiency of the DNA-extension product by MALDI-TOF MS. Three minutes of UV irradiation at 340 nm of the solution containing the extension product, 5′-U(PC-ROX)G-biotin, eliminated the corresponding peak (m/z: 7,307) in the MALDI-TOF mass spectrum but significantly increased the signal of the photocleaved fragment, 5′-UG-biotin (m/z: 6,570), as shown in Fig. 3B. These data confirm that the fluorescent dye is released completely from the DNA template in solution within 3 min of UV light irradiation at 340 nm.
The procedure for DSS on the chip is illustrated in Fig. 4 Upper, and the corresponding fluorescence image for each step is shown in Fig. 4 Lower. The self-primed DNA was first extended by using DNA polymerase and dUTP-PC-Bodipy-FL-510, complementary to the A on the template. After vigorous washing, the extension of the primer was confirmed by observing a blue signal (the emission of Bodipy-FL-510) in the scanner with 488-nm excitation, which is produced only if dUTP-PC-Bodipy-FL-510 is incorporated successfully into the DNA. After washing and detection, near-UV irradiation was applied to cleave the fluorophore from the DNA, which (in principle) should remove the blue signal efficiently. The scanned fluorescence images obtained from our experiments are shown in Fig. 4 Lower. After the first incorporation by dUTP-PC-Bodipy-FL-510, a blue signal was detected (Fig. 4, step 1). This blue signal from Bodipy-FL-510 was almost completely removed after 10 min of near-UV irradiation, producing an image with fluorescence levels close to background (Fig. 4, step 2). The integrated fluorescence intensity on the spot, obtained by using the scanner software, indicated that >97% of the original fluorescence signal was removed by photocleavage. To illustrate the continuity of this reaction on the solid surface, we next used dCTP-PC-Bodipy-650, complementary to the “G” on the template, to produce a red signal (emission from Bodipy-650) at the same position on the chip in the scanner with 633-nm excitation (Fig. 4, step 3), which again was almost entirely removed after photocleavage (Fig. 4, step 4). We repeated this process three more times, first using dUTP-PC-ROX, which gave an orange signal (emission from ROX with 594-nm excitation) after incorporation (Fig. 4, step 5), then dGTP with dUTP-PC-Bodipy-FL-510 (Fig. 4, step 7), and finally dATP with dUTP-PC-ROX (Fig. 4, step 9). The fluorescence intensity obtained after five cycles of incorporation indicated that the incorporation efficiency was ≈90%. In this manner, we identified a 7-nt sequence in the DNA template by alternate incorporation, detection, and photocleavage using the three PC nucleotide analogues.
Fig. 4.
Schematic representation of DSS on a chip using three PC fluorescent nucleotides (Upper) and the scanned fluorescence images for each step of DSS on a chip (Lower). 1, Incorporation of dUTP-PC-Bodipy-FL-510; 2, photocleavage of PC-Bodipy-FL-510; 3, incorporation of dCTP-PC-Bodipy-650; 4, photocleavage of PC-Bodipy-650; 5, incorporation of dUTP-PC-ROX; 6, photocleavage of PC-ROX; 7, incorporation of dGTP and dUTP-PC-Bodipy-FL-510; 8, photocleavage of PC-Bodipy-FL-510; 9, incorporation of dATP and dUTP-PC-ROX; 10, photocleavage of PC-ROX.
In conclusion, we successfully constructed a DNA chip using the 1,3-dipolar azide-alkyne cycloaddition reaction, offering a chemistry to covalently and chemoselectively immobilize DNA on the solid surface. Furthermore, we demonstrated the feasibility of performing the DNA polymerase reaction continuously on the chip using PC fluorescent nucleotides. Primer-extension and photocleavage reactions were performed successfully on the chip, confirming that DSS can be carried out on the solid surface. Because the photocleavage is performed in each cycle of nucleotide addition, any residual fluorescent dye that is left over from the previous cycle will be photocleaved further, ensuring a constant low background. The covalent and chemoselective 1,3-dipolar cycloaddition coupling chemistry for immobilizing DNA on a surface and the library of PC fluorescent nucleotides will also facilitate the development of single-molecule DNA sequencing and digital gene-expression analysis approaches. With the current efficiency of nucleotide incorporation and photocleavage, we expect to be able to sequence at least 25 bases per spot on the chip, which is sufficient for single-nucleotide polymorphism detection and gene-expression measurement. By reducing the photocleavage time and improving the read length and incorporation efficiency, this approach potentially can be developed into a high-throughput DNA-analysis system for whole-genome sequencing, single-nucleotide polymorphism detection, and applications in pharmacogenetics.
Supplementary Material
Acknowledgments
This work is supported by National Science Foundation Sensing and Imaging Initiative Grant 0097793, a Packard Fellowship for Science and Engineering, and Center of Excellence in Genomic Science Grant P50 HG002806 from the National Institutes of Health.
Abbreviations: DSS, DNA sequencing by synthesis; PC, photocleavable; Bodipy, 4,4-difluoro-4-bora-3α,4α-diaza-s-indacene; FAM, 5-carboxyfluorescein; HRMS, high-resolution MS; MALDI-TOF, matrix-assisted laser desorption ionization–time-of-flight; FAB, fast atom bombardment; NHS, N-hydroxysuccinimide; dH2O, deionized water; ROX, 6-carboxy-X-rhodamine.
References
- 1.Collins, F. S., Green, E. D., Guttmacher, A. E. & Guyer, M. S. (2003) Nature 422, 835–847. [DOI] [PubMed] [Google Scholar]
- 2.Thomas, J. W., Touchman, J. W., Blakesley, R. W., Bouffard, G. G., Beckstrom-Sternberg, S. M., Margulies, E. H., Blanchette, M., Siepel, A. C., Thomas, P. J. & McDowell, J. C., et al. (2003) Nature 424, 788–793. [DOI] [PubMed] [Google Scholar]
- 3.Pennisi, E. (2000) Science 288, 1146–1147. [DOI] [PubMed] [Google Scholar]
- 4.Smith, L. M., Sanders, J. Z., Kaiser, R. J., Hughes, P., Dodd, C., Connell, C. R., Heiner, C., Kent, S. B. H. & Hood, L. E. (1987) Nature 321, 674–679. [DOI] [PubMed] [Google Scholar]
- 5.Ju, J., Ruan, C., Fuller, C. W., Glazer, A. N. & Mathies, R. A. (1995) Proc. Natl. Acad. Sci. USA 92, 4347–4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ronaghi, M., Uhlen, M. & Nyren, P. (1998) Science 281, 363–365. [DOI] [PubMed] [Google Scholar]
- 7.Fu, D. J., Tang, K., Braun, A., Reuter, D., Darnhofer-Demar, B., Little, D. P., O'Donnell, M. J., Cantor, C. R. & Koster, H. (1998) Nat. Biotechnol. 16, 381–384. [DOI] [PubMed] [Google Scholar]
- 8.Roskey, M. T., Juhasz, P., Smirnov, I. P., Takach, E. J., Martin, S. A. & Haff, L. A. (1996) Proc. Natl. Acad. Sci. USA 93, 4724–4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edwards, J. R., Itagaki, Y. & Ju, J. (2001) Nucleic Acids Res. 29, e104. Available at http://nar.oupjournals.org/cgi/content/full/29/21/e104. [DOI] [PMC free article] [PubMed]
- 10.Drmanac, S., Kita, D., Labat, I., Hauser, B., Schmidt, C., Burczak, J. D. & Drmanac, R. (1998) Nat. Biotechnol. 16, 54–58. [DOI] [PubMed] [Google Scholar]
- 11.Kasianowicz, J. J., Brandin, E., Branton, D. & Deamer, D. W. (1996) Proc. Natl. Acad. Sci. USA 93, 13770–13773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braslavsky, I., Hebert, B., Kartalov, E. & Quake, S. R. (2003) Proc. Natl. Acad. Sci. USA 100, 3960–3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitra, R. D., Shendure, J., Olejnik, J., Olejnik, E. K. & Church, G. M. (2003) Anal. Biochem. 320, 55–65. [DOI] [PubMed] [Google Scholar]
- 14.Ju, J., Li, Z., Edwards, J. & Itagaki, Y. (2003) U.S. Patent 6,664,079.
- 15.Li, Z., Bai, X., Ruparel, H., Kim, S., Turro, N. J. & Ju, J. (2003) Proc. Natl. Acad. Sci. USA 100, 414–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pleasants, J. C., Guo, W. & Rabenstein, D. L. (1989) J. Am. Chem. Soc. 111, 6553–6558. [Google Scholar]
- 17.Huyghues-Despointes, B. M. P. & Nelson, J. W. (1992) Biochemistry 31, 1476–1483. [DOI] [PubMed] [Google Scholar]
- 18.Shena, M., Shalon, D., Davis, R. W. & Brown, P. O. (1995) Science 270, 467–470. [DOI] [PubMed] [Google Scholar]
- 19.Wang, D. G., Fan, J., Siao, C., Berno, A., Young, P., Sapolsky, R., Ghandour, G., Perkins, N., Winchester, E. & Spencer, J., et al. (1998) Science 280, 1077–1082. [DOI] [PubMed] [Google Scholar]
- 20.Debouck, C. & Goodfellow, P. N. (1999) Nat. Genet. 1, 48–50. [DOI] [PubMed] [Google Scholar]
- 21.Beier, M. & Hoheisel, J. D. (1999) Nucleic Acids Res. 27, 1970–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adessi, C., Matton, G., Ayala, G., Turcatti, G., Mermod, J., Mayer, P. & Kawashima, E. (2000) Nucleic Acids Res. 28, e87. Available at http://nar.oupjournals.org/cgi/content/full/28/20/e87. [DOI] [PMC free article] [PubMed]
- 23.Lindroos, K., Liljedahl, U., Raitio, M. & Syvänen, A. (2001) Nucleic Acids Res. 29, e69. Available at http://nar.oupjournals.org/cgi/content/full/29/13/e69. [DOI] [PMC free article] [PubMed]
- 24.Seo, T. S., Li, Z., Ruparel, H. & Ju, J. (2003) J. Org. Chem. 68, 609–612. [DOI] [PubMed] [Google Scholar]
- 25.Rostovtsev, V. V., Green, J. G., Fokin, V. V. & Sharpless, K. B. (2002) Angew. Chem. Int. Ed. Engl. 41, 2596–2599. [DOI] [PubMed] [Google Scholar]
- 26.Tornφe, C. W., Christensen, C. & Meldal, M. (2002) J. Org. Chem. 67, 3057–3064. [DOI] [PubMed] [Google Scholar]
- 27.Wang, Q., Chan, T. R., Hilgraf, R., Fokin, V. V., Sharpless, K. B. & Finn, M. G. (2003) J. Am. Chem. Soc. 125, 3192–3193. [DOI] [PubMed] [Google Scholar]
- 28.Link, A. J. & Tirrell, D. A. (2003) J. Am. Chem. Soc. 125, 11164–11165. [DOI] [PubMed] [Google Scholar]
- 29.Speers, A. E., Adam, G. C. & Cravatt, B. F. (2003) J. Am. Chem. Soc. 125, 4686–4687. [DOI] [PubMed] [Google Scholar]
- 30.Fazio, F., Bryan, M. C., Blixt, O., Paulson, J. C. & Wong, C. (2002) J. Am. Chem. Soc. 124, 14397–14402. [DOI] [PubMed] [Google Scholar]
- 31.Chrisey, L. A., Lee, G. U. & O'Ferrall, C. E. (1996) Nucleic Acids Res. 24, 3031–3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Antao, V. P., Lai, S. Y. & Tinoco, I., Jr. (1991) Nucleic Acids Res. 19, 5901–5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.