Abstract
In most eukaryotic cells, the C-terminal amino acid of α-tubulin is aromatic (Tyr in mammals and Phe in Saccharomyces cerevisiae) and is preceded by two glutamate residues. In mammals, the C-terminal Tyr of α-tubulin is subject to cyclic removal from the peptide chain by a carboxypeptidase and readdition to the chain by a tubulin–Tyr ligase. There is evidence that tubulin–Tyr ligase suppression and the resulting accumulation of detyrosinated (Glu) tubulin favor tumor growth, both in animal models and in human cancers. However, the molecular basis for this apparent stimulatory effect of Glu tubulin accumulation on tumor progression is unknown. Here we have developed S. cerevisiae strains expressing only Glu tubulin and used them as a model to assess the consequences of Glu tubulin accumulation in cells. We find that Glu tubulin strains show defects in nuclear oscillations. These defects are linked to a markedly decreased association of the yeast ortholog of CLIP170, Bik1p, with microtubule plus-ends. These results indicate that the accumulation of Glu tubulin in cells affects microtubule tip complexes that are important for microtubule interactions with the cell cortex.
Microtubules are fibrous structures in the cytoplasm of eukaryotic cells that play a vital role in cell organization, motility, and division. Microtubule functions involve cell-cycle-dependent changes in polymer organization and dynamics that rely to a large extent on the biochemical properties of the microtubule building block, the α/β-tubulin heterodimer. Tubulin self-assembles to form different structures (including dynamic microtubules), interacts with a large variety of other proteins, binds a number of drugs or nucleotides with high specificity, and has GTPase enzymatic activity (1). Additionally, tubulin is subject to special posttranslational modifications, including a detyrosination–tyrosination cycle in which the C-terminal Tyr residue of α-tubulin is cyclically removed from the peptide chain by a carboxypeptidase (tubulin carboxypeptidase) and readded to the chain by tubulin–Tyr ligase (TTL). The detyrosination–tyrosination cycle is present in many cell types and phyla, but its functional meaning has been elusive. However, there is recent evidence that the detyrosination–tyrosination cycle is important for the control of cancer cell proliferation. TTL is systematically suppressed during tumor progression in animal models, with resulting accumulation of detyrosinated (Glu) tubulin (2). Additionally, TTL is frequently lost during tumor growth in human cancers. In human breast cancers, TTL loss and the resulting Glu tubulin accumulation are associated with increased tumor aggressiveness (3).
Here, we have used the budding yeast Saccharomyces cerevisiae as a model system to examine the cellular consequences of Glu tubulin accumulation. In budding yeast, α-tubulin is encoded by two genes, TUB1 and TUB3 (4). The TUB1 gene is essential for growth of normal haploid strain, whereas TUB3 is not (4). The α-tubulin C-termini encoded by both genes display a Glu-Glu-Phe sequence, analogous to the Glu-Glu-Tyr sequence found in mammals. To test the role of the C-terminal aromatic residue of α-tubulin, we have constructed yeast strains expressing either wild-type tubulin (Phe tubulin) or mutated α-tubulin lacking the C-terminal amino acid (Glu tubulin). We find drastic perturbations of nuclear oscillations in Glu tubulin clones, apparently related to perturbation of the composition of microtubule plus-end complexes. We propose that similar perturbations in cancer cells may interfere with the cell physiology and favor tumor progression.
Materials and Methods
Yeast Strains and Media. The S. cerevisiae strains and plasmid used are listed in Table 1. The TUB1 and tub1-Glu strains were obtained by using the “plasmid shuff le” technique (5). pRB539Glu was obtained from pRB539 by integration of a stop codon in place of the C-terminal Phe codon in the TUB1 coding sequence.
Table 1. Yeast strains and plasmids.
| Genotype | Source | |
|---|---|---|
| Strains | ||
| TUB1 | MATa/α ade4/ADE4, his3-200/his3-200, leu2-3_112/leu2-3_112, ura3-52/ura3-52, LYS2/lys2-801, tub1::HIS3/tub1::HIS3, tub3::TRP1/tub3::TRP1 pRB539 | Ref. 5 |
| tub1-Glu | TUB1 except pRB539Glu | This study |
| iTUB1 | MATa/α ade4/ADE4, his3-200/his3-200, leu2-3_112/leu2-3_112, ura3-52/ura3-52, LYS2/lys2-801, tub1::HIS3/tub1::HIS3, promtub1::TUB1-LEU2/promtub1::TUB1-LEU2, tub3::TRP1/tub3::TRP1 | This study |
| itub1-Glu | TUB1 except promtub1::tub1-Glu-LEU2/promtub1::tub1-Glu-LEU2 | This study |
| W303 | MATa/MATα, ura3-52/ura3-52; trp1Δ2/trp1Δ2; leu2-3_112/leu2-3_112; his3-11/his3-11; ade2-1/ade2-1; can1-100/can1-100 | Euroscarf |
| Plasmids | ||
| pRB539 | TUB1 LEU2 CEN | Ref. 5 |
| pRB539Glu | tub1-Glu LEU2 CEN | This study |
| pB1225 | BIM1-GFP URA3 CEN | Ref. 6 |
| pB681 | BIK1-GFP URA3 2μ | Ref. 7 |
Integrated strains (iTUB1 and itub1-Glu) were constructed by cloning pBR539 and pRB539Glu (SalI and AatII) in a pUC18 plasmid (SalI and AatII). pUC18-TUB1 and pUC18-tub1-Glu were cut with NsiI and used for integration in the TUB1 locus by using the plasmid shuffle technique (5).
Growing Conditions and Antibodies. Cells were grown at 30°C to midlogarithmic growth phase. For the temperature shift experiment, cells were either left at 30°C or shifted to 10°C for 4 days. Growth was monitored by cell counting. Benomyl was supplied by Aldrich.
For Western blotting, we used α-tubulin mAb YOL1/34 (Sera-Lab, Crawley Down, Sussex, U.K.). Intact α-tubulin (Phe tubulin) was detected with mAb YL1/2 (6), Glu tubulin was detected with polyclonal antibody NS8, which was obtained by injecting rabbits with the peptide STAEEEE linked to keyhole limpet hemocyanin (eight injections of 100 μg; Neosystem, Strasbourg, France). The antibody was affinity-purified on immobilized peptide before use. Carboxypeptidase A comes from Sigma–Aldrich and is used for 10 min at 30°C at a concentration of 2 μg/ml.
Video Microscopy and Quantification. For time-lapse video microscopy examination, cells were mixed with a suspension containing 6 μm of latex beads (10% vol/vol; Polysciences) laid on coverslips and fenced with mineral oil. The coverslips were mounted in a rose chamber covered by another coverslip. Time-lapse Z sequences were collected either on a Leica (Deerfield, IL) or Zeiss microscope controlled by metamorph software (Universal Imaging, Media, PA). The acquisition time was 200–500 ms. Typically, 18–20 sequential z-axis images were collected in 0.3-μm steps every 30 s. Spindle-tracking and fluorescent-counting was performed automatically by metamorph software in maximal intensity projections computed from the original three-dimensional datasets.
Results
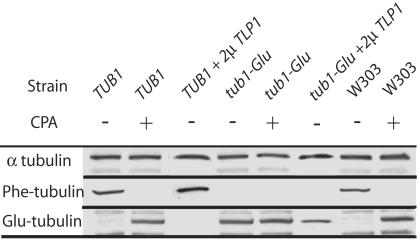
Analysis of the α-Tubulin C Terminus in Wild-Type and Mutant S. cerevisiae Strains. We tested whether the TUB1 and tub1-Glu clones faithfully expressed the tubulin variant encoded by the plasmid gene or whether tubulin composition was posttranslationally modified by T complex protein or TTL activity in these clones. To perform this test, the presence of Phe tubulin and Glu tubulin was assessed in protein preparations from both strains by using Western blotting (Fig. 1). As an additional control, a wild-type W303 strain was also analyzed. When the extracts were probed with Phe tubulin antibody (Fig. 1); cell extracts from TUB1 and W303 strains gave a strong signal. By contrast, extracts from the tub1-Glu strain showed no signal. When the same extracts were probed with Glu tubulin antibody (Fig. 1), extracts from the tub1-Glu strain gave a strong signal, whereas extracts from TUB1 and W303 clones gave no signal. When TUB1 or W303 extracts were exposed to carboxypeptidase A to cleave the C-terminal Phe-residue (7) before immunoblotting, the Phe tubulin signals were abolished, whereas strong signals were observed with Glu tubulin antibody (Fig. 1, row CPA). These results indicate that TUB1 and tub1-glu clones express only Phe tubulin and Glu tubulin, respectively, and suggest that there is neither T complex protein nor TTL activity in S. cerevisiae in our experimental conditions. However, a putative TTL-like protein (TLP), YBR094W/TLP1, has been identified in S. cerevisiae (8). To test whether this TTL-related protein had TTL-like activity, we cloned the YBR094W/TLP1 gene in a high-copy number plasmid (2μ TLP1 construct). The overexpression of the corresponding protein was checked by Western blotting (data not shown). Such overexpression did not induce detectable amounts of Phe tubulin in tub1-Glu strains (Fig. 1, lane tub1-Glu + 2μ TLP1). Thus, despite structural homology with the mammalian TTL, the YBR094W/TLP1 protein is apparently devoid of tubulin–Phe ligase activity.
Fig. 1.
Analysis of the α-tubulin C terminus in control and mutant strains. Western blotting of whole-cell extracts from TUB1 and tub1-Glu, expressing or not the Tlp1p protein (2μ TLP1) and W303 strains. Extracts were exposed or not to carboxypeptidase A (CPA) before immunoblotting. Primary antibodies were directed against total α-tubulin (YOL34), Phe tubulin (YL1/2), or Glu tubulin (NS8) as indicated.
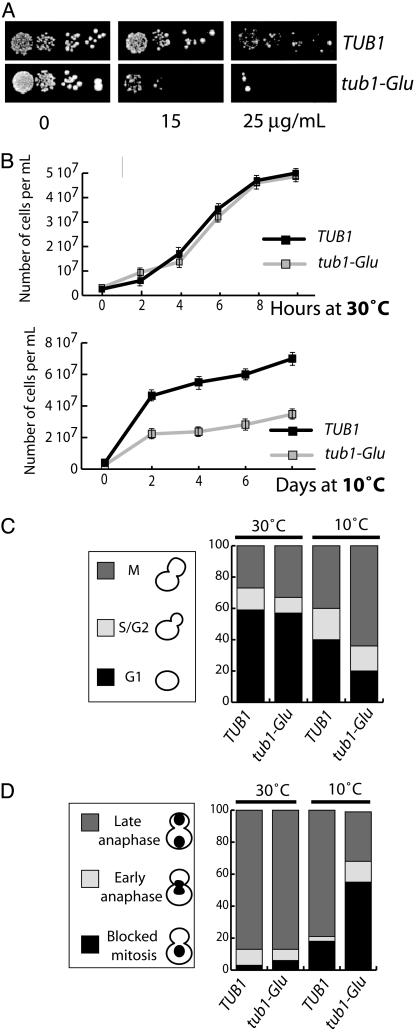
tub1-Glu Strain Shows Benomyl Supersensitivity and Growth Defect. Supersensitivity to the microtubule depolymerizing drug benomyl is a common occurrence in both α-tubulin and β-tubulin mutants (5). Sensitivity of the TUB1 and tub1-Glu cells to benomyl was tested by spotting fixed numbers of cells on rich plates containing different concentrations of the drug (15–25 μg/ml) (Fig. 2A) and examining cell growth. In the presence of benomyl, cell growth impairment was markedly enhanced in tub1-Glu compared with TUB1 cells, indicating supersensitivity to benomyl in cells expressing Glu tubulin. Supersensitivity to benomyl can be indicative of an α-tubulin cold-sensitive mutation (4). Therefore, we examined cell growth in tub1-Glu and TUB1 strains at both warm and cold temperatures (Fig. 2B). The two strains showed similar growth kinetics at 30°C. In contrast, cell growth was substantially impaired in the tub1-glu strain at 10°C (Fig. 2B). The same cells also showed impaired viability at 10°C compared with controls (data not shown). These results indicate cold-sensitive impairment of cell growth and viability in tub1-Glu strain.
Fig. 2.
Benomyl supersensitivity and mitotic block in tub1-Glu strains. (A) Benomyl sensitivity. Yeast cells of the indicated genotype were spotted on solid rich media containing the indicated benomyl concentration. (B) Cell growth. At each time point a minimum of 100 cells were counted. Four independent experiments were run. Growth rates were similar at 30°C in both strains. At 10°C, the tub1-Glu strain showed impaired growth. (C) Mitotic block in tub1-Glu cells. The percent of unbudded cells (G1), cells containing small buds (S), and cells with large buds (M) were scored in TUB1 and tub1-Glu strain (n = 150) at both warm and cold temperatures as indicated. (D) Inhibition of nuclear division and the position of DNA in control and mutant cells. Cells grown at 30°C and 10°C were stained with Hoescht, and the nucleus number and position were determined by fluorescence microscopy (n = 150).
Delay in Anaphase Onset in tub1-Glu Strains. We used both direct cell observation and flow cytometry analysis to test whether the impaired cell growth observed in tub1-Glu strains at 10°C was related to defects at a particular stage of the cell cycle. Direct observation showed an accumulation of cells with a large bud in tub1-glu clones compared with controls (Fig. 2C) and an accumulation of cells with replicated DNA in flow cytometry analysis (data not shown), suggesting a mitotic block in tub1-Glu cells.
We tested whether the block occurred before or after nuclear division. At permissive temperature, ≈90% of the cells with large buds had two nuclei (one in the mother cell and one in the bud) in TUB1 and tub1-Glu cells (Fig. 2D). In contrast, at restrictive temperature, the two clones differed strikingly, with 80% of the cells with two nuclei in control cells, compared with only 30% in the tub1-Glu cells. The remaining tub1-Glu cells contained a single nucleus located in the mother cell. These results indicate a delay in anaphase onset in a large proportion of tub1-Glu cells at restrictive temperature.
Inhibition of Nuclear Oscillations in Glu Tubulin Strains. Blockage in mitosis before nuclear division has been observed in situations in which the nucleus never enters the bud in anaphase because of abnormal nucleus positioning or motion (9–11). These data prompted us to examine nuclear behavior in cells expressing either Phe tubulin or Glu tubulin. In the experiments reported in the following sections, the spindle and the microtubule plus-ends were labeled by expressing either GFP-Bim1p or Bik1p fusion protein in yeast cells (12, 13). Nuclear oscillation and microtubule end-labeling were examined with similar results in strains with plasmid (TUB1 and tub1-Glu) and integrated (iTUB1 and itub1-Glu) tubulin genes.
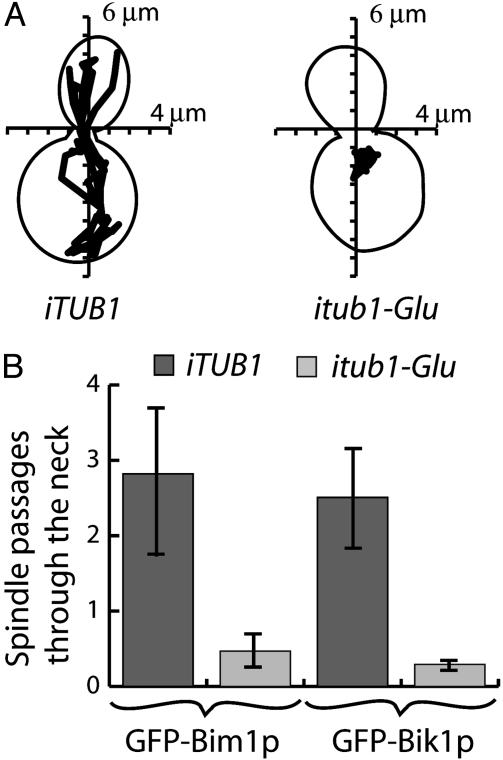
In pilot experiments using cells expressing GFP-Bim1p before and during mitosis, we found no obvious difference between Phe tubulin and Glu tubulin cells up to anaphase (data not shown). However, major differences were evident between Phe tubulin and Glu tubulin clones with respect to nuclear oscillations and spindle penetration into the bud. Although large nuclear oscillations commonly occurred in Phe tubulin clones (Fig. 3A), nuclei remained generally immotile and close to the neck in Glu tubulin clones (Fig. 3A). As a consequence, penetration of spindle in the bud was a rare event in Glu tubulin cells (Fig. 3B). Similar results were observed in cells expressing GFP-Bik1p. We verified that spindle motion was quantitatively similar in cells expressing either GFP-Bim1p or GFP-Bik1p, suggesting that differences between Phe tubulin and Glu tubulin cells did not depend on the microtubule end-marker used for analysis (Table 2).
Fig. 3.
Inhibition of nuclear oscillations in Glu tubulin strains. (A) Tracking of spindle motions over a 30-min time duration was performed, and a representative example is shown in both control and mutated strains. (B) Quantitative analysis of spindle passage through the neck in control or mutated strains expressing GFP-Bim1p (number of films: control, n = 5; mutated, n = 11) or GFP-Bik1p (number of films: control, n = 16; mutated, n = 21) as indicated. The ordinate represent the absolute number of spindle passages through the neck during the 30 min of the observation.
Table 2. Quantitative analysis of spindle motion.
| Strain | Maximum speed, μm/min | Minimum speed, μm/min | Average speed, μm/min | Maximum amplitude, μm | Minimum amplitude, μm | Average amplitude, μm |
|---|---|---|---|---|---|---|
| iTUB1 GFP-Bim 1p | 0.8 ± 0.16 | 0.014 ± 0.004 | 0.220 ± 0.05 | 3.2 ± 0.76 | 0.14 ± 0.06 | 1.34 ± 0.2 |
| iTUB1 GFP-Bik 1p | 0.7 ± 0.05 | 0.014 ± 0.003 | 0.211 ± 0.001 | 2.9 ± 0.27 | 0.24 ± 0.05 | 1.45 ± 0.2 |
| itub1-Glu GFP-Bim 1p | 0.4 ± 0.05 | 0.006 ± 0.001 | 0.095 ± 0.009 | 1.3 ± 0.27 | 0.06 ± 0.01 | 0.63 ± 0.17 |
| itub1-Glu GFP-Bik 1p | 0.4 ± 0.06 | 0.006 ± 0.001 | 0.111 ± 0.001 | 1.9 ± 0.40 | 0.07 ± 0.01 | 0.56 ± 0.2 |
Spindle position for all filmed cells was determined at 30-s intervals by using metamorph software. The speed and amplitude of spindle motions were derived from positional data by using the same software. The spindle speed was calculated between two successive time points as the ratio of displacement/elapsed time. Amplitudes were measured at each time point as the distance of the spindle center from its original position at t = 0. Data are shown ± SEM.
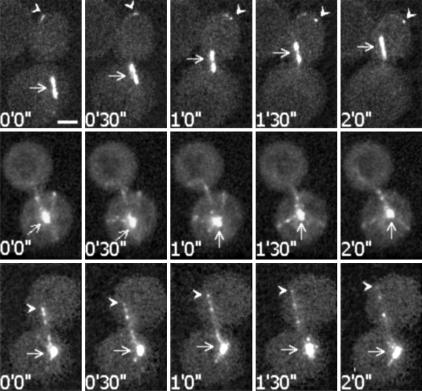
Microtubule Behavior in Anaphase Phe Tubulin and Glu Tubulin Cells. Nuclear oscillations involve the guidance of microtubules toward the bud cortex, followed by the capture of clustered microtubule plus-ends at the cortex, with subsequent microtubule sliding and spindle pulling. Such events were observed in Phe tubulin clones expressing GFP-Bim1p (Fig. 4 Top) (14). By contrast, in GFP-Bim1p expressing Glu tubulin cells, microtubule orientation often remained apparently random during the period of observation (Fig. 4 Middle). The spindle seemed subject to noncoordinated forces resulting in no significant movement (Fig. 4 Middle). In some cells, a preferential orientation to the bud was observed, but microtubule plus-ends did not display clear clustering as in Phe tubulin cells (Fig. 4 Bottom). Although microtubule plus-ends could reach the bud cortex, microtubule sliding and spindle migration in the bud through the neck was a rare event.
Fig. 4.
Video microscopy analysis of spindle and microtubule behavior in Phe tubulin or Glu tubulin strains expressing GFP-Bim1p. (Top) Phe tubulin GFP-Bim1p strain during a nuclear oscillation event. Arrows indicate the spindle. Arrowheads indicate microtubules plus-ends. Microtubule ends were apparently captured at the cell cortex with subsequent microtubule sliding and spindle pulling. (Middle) One aspect observed in Glu tubulin GFP-Bim1p strains. Arrows indicate the spindle. In Glu tubulin strains, microtubules often seemed to lack directional cues and showed little net spindle motion. (Bottom) Another aspect observed in Glu tubulin GFP-Bim1p strains. Arrows indicate the spindle. Arrowheads indicate microtubules plus-ends. In some cases, microtubules had apparent directional cues. However, microtubule ends were not clustered, and microtubule capture and sliding at the cell cortex were rarely observed. Time points corresponding to image capture are indicated. (Bar, 2 μm.)
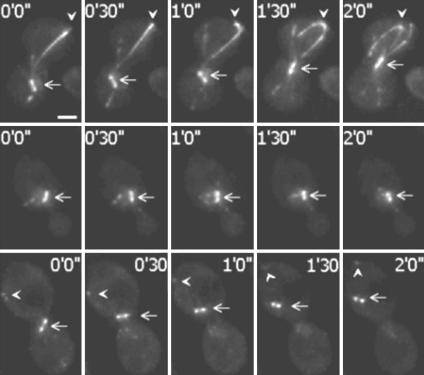
Qualitatively similar observations were made with GFP-Bik1p-expressing cells (Fig. 5). However, in addition to reduced spindle motion, major differences in the distribution of GFP-Bik1p were observed between Phe tubulin and Glu tubulin cells. In Phe tubulin cells, microtubule ends were intensely labeled with GFP-Bik1p, which features bundled microtubule ends and long-trailing comets (Fig. 5 Top). In contrast, even in the rare cells showing significant spindle motion, the labeling of microtubule ends with GFP-Bik1p was barely detectable in Glu tubulin cells, and comets were absent or evanescent (Fig. 5 Middle and Bottom).
Fig. 5.
Video microscopy analysis of spindle and microtubule behavior in Phe tubulin and Glu tubulin strains expressing GFP-Bik1p. (Top) Phe tubulin GFP-Bik1p strain during a nuclear oscillation. Arrows indicate spindle. Arrowheads indicate microtubules plus-ends. With GFP-Bik1p, long comets were observed on the bundled microtubules. Such comets remained conspicuous during microtubule capture and sliding at the cell cortex. (Middle) One aspect observed in Glu microtubule GFP-Bik1p strains. Arrows indicate the spindle. Microtubules apparently lack directional cues. Microtubule plus-ends are poorly labeled with GFP-Bik1p, and no comet is visible. (Bottom) Glu tubulin GFP-Bik1p strain during a nuclear oscillation. Arrows indicate spindle with a translocation event. Arrowheads indicate microtubules plus-ends. Nuclear oscillations were rarely observed in Glu tubulin strains (Fig. 3B). During such oscillations, the GFP-Bik1p labeling remained conspicuously different from controls, with barely apparent and evanescent GFP-Bik1p comets and poorly labeled microtubule ends. Times when images are captured are indicated. (Bar, 2 μm.)
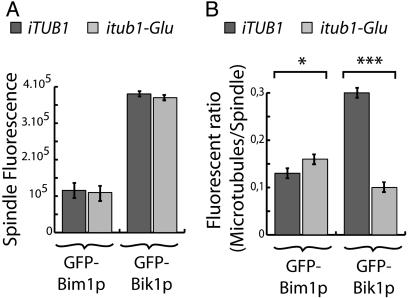
In quantitative analysis, spindle labeling was similar in Glu tubulin and Phe tubulin cells, whether GFP-Bim1p or GFP-Bik1p was used as a marker (Fig. 6A). The ratio of GFP-Bim1p fluorescence in the microtubule ends to GFP-Bim1p in the spindle was slightly higher in Glu tubulin cells compared with Phe tubulin cells. A similar analysis with GFP-Bik1p showed a dramatic 3-fold decrease of the microtubule ends/spindle fluorescence ratio in Glu tubulin cells compared with Phe tubulin cells (Fig. 6B). Taken together, these results indicate major and specific defects in the interaction of Bik1p with microtubule ends in Glu tubulin cells.
Fig. 6.
Quantitative analysis of GFP-Bim1p and GFP-Bik1p labeling in Phe tubulin and Glu tubulin strains. (A) Spindle fluorescence measured in arbitrary units by using metamorph software. Spindle labeling was averaged by using the first 10 images of each filmed cell. Labeling was stronger with GFP-Bik1p compared with GFP-Bim1p in both Phe strains and Glu strains. No statistically significant differences were observed between the two types of strains. (B) Ratio of microtubule/spindle fluorescence labeling in Phe tubulin and Glu tubulin strains. With GFP-Bim1p, the ration of microtubule/spindle fluorescence was slightly (but significantly) higher in Glu tubulin cells compared with Phe tubulin cells. In contrast, with GFP-Bik1p, the same ratio was strongly reduced in Glu tubulin cells compared with Phe tubulin cells, indicating impaired association of GFP-Bik1p with microtubule plus-ends in the Glu tubulin cells. Results were analyses with ANOVA. *, P < 0.05; ***, P < 0.001.
Discussion
Absence of Cyclic Modification of the C-Terminal α-Tubulin Amino Acid in S. cerevisiae. The tyrosination cycle is highly conserved among eukaryotes and has hitherto been found in most cells where it has been searched for, with the exception of the fission yeast, Schizosaccharomyces pombe (15). In S. cerevisiae, the C-terminal amino acid of α-tubulin is a Phe, not a Tyr, but the two amino acids are similar, and both can be added to the tubulin molecule by the mammalian TTL (7). Additionally, S. cerevisiae contains a TTL-related protein that has been identified based on sequence similarities and structural homology with the mammalian TTL (8). The existence of a phenylalanination–dephenylalanination cycle in S. cerevisiae was therefore a definite possibility. However, in this study, we found that tubulin remains fully Phe in TUB1 cells and fully Glu in tub1-Glu cells, in the presence or absence of overexpressed TTL-related protein. This finding strongly indicates that there is no turnover of the α-tubulin C-terminal Phe in budding yeast. The absence of TTL activity allowed a direct assay of the role of the C-terminal Phe of α-tubulin, by comparing strains containing either only Phe tubulin or only Glu tubulin.
Nuclear Oscillation Defects in Glu Tubulin Strains. In our study, expression of Glu tubulin instead of Phe tubulin in S. cerevisiae cells resulted in a severe impairment of nuclear motion through the neck into the bud, probably explaining the delay in anaphase onset observed at restrictive temperature in Glu tubulin cells. Other cold-sensitive tubulin mutations, including tub2–401, induce a defect in anaphase onset (16, 17). In tub2–401 clones the mitotic block affects all cells uniformly. In contrast, a mitotic block is observed in only a proportion of tub1-Glu cells, may be because nuclear motion through the neck is impaired but not completely impossible in such cells.
Nuclear movements in yeast involve nuclear migration toward the bud, alignment and retention of the spindle at the neck, and nuclear oscillations through the aperture of the budded cell at anaphase onset. Motor proteins play a key role in this process by means of the regulation of microtubule dynamics and generation of pushing or pulling forces along astral microtubules (dynein, ref. 18; Kip2p, ref. 19; Kip3p, ref. 20; and Kar3p, ref. 21).
Several spindle polarity determinants, including septin ring, Kar9p, Bni1p, and Kip3p, provide positional cues for directing the nucleus to the neck (18, 22–24). A cytoplasmic dynein-dependent pathway is responsible for microtubule pulling at the cell cortex during the pronounced spindle oscillations at the neck of a budded cell and contributes to the forces required to pull the nucleus through the aperture between mother cell and bud (18, 25, 26). In tub1-Glu and itub1-Glu cells, nuclear positioning to the neck did not show obvious alterations, whereas microtubule pulling, nuclear oscillations, and nucleus migration into the bud were drastically reduced. These reduced spindle movements were associated with a dramatic decrease of Bik1p association with microtubule ends. Such a reduction could have several origins, including (i) a reduced rate of microtubule polymerization (27) or a decrease in microtubule number (27) in Glu tubulin cells, (ii) detection bias linked to differences in microtubule bundling between Phe tubulin and Glu tubulin cells, or (iii) intrinsic anomalies in the Bik1p interaction with Glu microtubule ends. Differences in microtubule growth rates or number and detection bias would, however, similarly affect the Bim1p and Bik1p signals at microtubule ends. The comparable Bim1p signals observed between Phe microtubule ends and Glu microtubule ends are strong indications that the decrease of Bik1p fluorescence at microtubule ends in Glu tubulin strains corresponds to a bona fide depletion of Bik1p at Glu microtubule tips.
Bik1p is the yeast ortholog of CLIP170, a protein known to be a key factor for dynein-dependent microtubule capture and pulling at the cell cortex (28–30). Therefore, it seems logical that a perturbation of Bik1p association with microtubule ends perturbs nuclear oscillations, known to be dynein-dependent. In addition to apparent perturbation of microtubule pulling at the cortex, microtubules in Glu tubulin cells spend most of the time in disorganized arrays, lacking a preferential orientation. Such aster-like organization can be observed in Phe tubulin strains but is generally short-timed. CLIP170 is known to be required for polarization of microtubule array in epithelial cells (31). Bik1p may be similarly required for the directional control of microtubules in yeast.
Whereas Bik1p is depleted at microtubule ends in Glu tubulin cells, its association with spindle microtubules is apparently normal in the same cells, suggesting that the absence of the C-terminal Phe residue does not directly impair Bik1p interaction with microtubules. Apparently, the mammalian ortholog of Bik1p, CLIP170, has several modes of interaction with microtubules. In vitro, CLIP170 behaves as a classical microtubule-associated protein associating with the microtubule lattice (32, 33). It seems likely that Bik1p has similar microtubule-associated protein-like behavior and that this accounts for the strong staining of spindle microtubules with Bik1p. The microtubule end-tracking behavior of CLIP170 in vivo is not fully understood; it apparently requires recognition of a specific feature of the microtubule distal end and subsequent release from an older, more proximal part of the microtubule (27). Such a complex sequence of binding and dissociation events has not been observed in vitro as yet, indicating that it may require cell regulators. Microtubule tip complexes contain signaling proteins, such as G proteins or G protein exchange factors (31, 34). The composition of the tip complex may be regulated by a signaling cascade, affecting the intrinsic microtubule binding activity of tip components. Our results may reflect an involvement of the α-tubulin C terminus in such signaling cascades.
From Yeast to Cancer? In mammalian cells, CLIP170 is apparently crucial for microtubule interactions with the dynein–dynactin cortical complexes and for resulting correct spindle orientation (35). CLIP170 is also associated with kinetochores and is probably central for the dynein-dependent morphogenesis of the spindle (36, 37). If our observations in yeast apply to mammalian tubulin, several dynein-dependent functions may be affected by tubulin detyrosination in mammalian cells, and this would obviously increase the probability of genomic instability. This study identifies dynein complexes as possible molecular targets affected by the accumulation of Glu tubulin and opens new clues to elucidate the mechanisms through which the tyrosination cycle may interfere with the cell physiology and favor tumor growth.
Acknowledgments
We thank Dr. Frank Solomon for his generous gift of plasmids and strains, Dr. Jennifer Tirnauer for providing the GFP-Bim1p plasmid, Dr. David Pellman for providing the GFP-Bik1p plasmids, and Dr. Yasmina Saoudi for help in imaging. This work was supported in part by a grant (to D.J.) from la Ligue Nationale contre le Cancer.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: TTL, tubulin-Tyr ligase; TLP, TTL-like protein.
References
- 1.Valiron, O., Caudron, N. & Job, D. (2001) Cell Mol. Life Sci. 58, 2069–2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lafanechere, L., Courtay-Cahen, C., Kawakami, T., Jacrot, M., Rudiger, M., Wehland, J., Job, D. & Margolis, R. L. (1998) J. Cell Sci. 111, 171–181. [DOI] [PubMed] [Google Scholar]
- 3.Mialhe, A., Lafanechere, L., Treilleux, I., Peloux, N., Dumontet, C., Bremond, A., Panh, M. H., Payan, R., Wehland, J., Margolis, R. L. & Job, D. (2001) Cancer Res. 61, 5024–5027. [PubMed] [Google Scholar]
- 4.Schatz, P. J., Solomon, F. & Botstein, D. (1986) Mol. Cell. Biol. 6, 3722–3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schatz, P. J., Solomon, F. & Botstein, D. (1988) Genetics 120, 681–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wehland, J., Schroder, H. C. & Weber, K. (1984) EMBO J. 3, 1295–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.MacRae, T. H. (1997) Eur. J. Biochem. 244, 265–278. [DOI] [PubMed] [Google Scholar]
- 8.Dideberg, O. & Bertrand, J. (1998) Trends Biochem. Sci. 23, 57–58. [DOI] [PubMed] [Google Scholar]
- 9.Bardin, A. J., Visintin, R. & Amon, A. (2000) Cell 102, 21–31. [DOI] [PubMed] [Google Scholar]
- 10.Bloecher, A., Venturi, G. M. & Tatchell, K. (2000) Nat. Cell Biol. 2, 556–558. [DOI] [PubMed] [Google Scholar]
- 11.Pereira, G., Hofken, T., Grindlay, J., Manson, C. & Schiebel, E. (2000) Mol. Cell 6, 1–10. [PubMed] [Google Scholar]
- 12.Tirnauer, J. S., O'Toole, E., Berrueta, L., Bierer, B. E. & Pellman, D. (1999) J. Cell Biol. 145, 993–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin, H., de Carvalho, P., Kho, D., Tai, C. Y., Pierre, P., Fink, G. R. & Pellman, D. (2001) J. Cell Biol. 155, 1173–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adames, N. R. & Cooper, J. A. (2000) J. Cell Biol. 149, 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alfa, C. E. & Hyams, J. S. (1991) Cell Motil. Cytoskeleton 18, 86–93. [DOI] [PubMed] [Google Scholar]
- 16.Huffaker, T. C., Thomas, J. H. & Botstein, D. (1988) J. Cell Biol. 106, 1997–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sullivan, D. S. & Huffaker, T. C. (1992) J. Cell Biol. 119, 379–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yeh, E., Yang, C., Chin, E., Maddox, P., Salmon, E. D., Lew, D. J. & Bloom, K. (2000) Mol. Biol. Cell 11, 3949–3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cottingham, F. R. & Hoyt, M. A. (1997) J. Cell Biol. 138, 1041–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeZwaan, T. M., Ellingson, E., Pellman, D. & Roof, D. M. (1997) J. Cell Biol. 138, 1023–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cottingham, F. R., Gheber, L., Miller, D. L. & Hoyt, M. A. (1999) J. Cell Biol. 147, 335–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kusch, J., Meyer, A., Snyder, M. P. & Barral, Y. (2002) Genes Dev. 16, 1627–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, L., Klee, S. K., Evangelista, M., Boone, C. & Pellman, D. (1999) J. Cell Biol. 144, 947–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller, R. K., Matheos, D. & Rose, M. D. (1999) J. Cell Biol. 144, 963–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheeman, B., Carvalho, P., Sagot, I., Geiser, J., Kho, D., Hoyt, M. A. & Pellman, D. (2003) Curr. Biol. 13, 364–372. [DOI] [PubMed] [Google Scholar]
- 26.Bloom, K. (2001) Curr. Biol. 11, R326–R329. [DOI] [PubMed] [Google Scholar]
- 27.Galjart, N. & Perez, F. (2003) Curr. Opin. Cell Biol. 15, 48–53. [DOI] [PubMed] [Google Scholar]
- 28.Valetti, C., Wetzel, D. M., Schrader, M., Hasbani, M. J., Gill, S. R., Kreis, T. E. & Schroer, T. A. (1999) Mol. Biol. Cell 10, 4107–4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vaughan, K. T., Tynan, S. H., Faulkner, N. E., Echeverri, C. J. & Vallee, R. B. (1999) J. Cell Sci. 112, 1437–1447. [DOI] [PubMed] [Google Scholar]
- 30.Dujardin, D. L. & Vallee, R. B. (2002) Curr. Opin. Cell Biol. 14, 44–49. [DOI] [PubMed] [Google Scholar]
- 31.Fukata, M., Watanabe, T., Noritake, J., Nakagawa, M., Yamaga, M., Kuroda, S., Matsuura, Y., Iwamatsu, A., Perez, F. & Kaibuchi, K. (2002) Cell 109, 873–885. [DOI] [PubMed] [Google Scholar]
- 32.Rickard, J. E. & Kreis, T. E. (1991) J. Biol. Chem. 266, 17597–17605. [PubMed] [Google Scholar]
- 33.Pierre, P., Scheel, J., Rickard, J. E. & Kreis, T. E. (1992) Cell 70, 887–900. [DOI] [PubMed] [Google Scholar]
- 34.Gundersen, G. G. (2002) Nat. Rev. Mol. Cell Biol. 3, 296–304. [DOI] [PubMed] [Google Scholar]
- 35.Busson, S., Dujardin, D., Moreau, A., Dompierre, J. & De Mey, J. R. (1998) Curr. Biol. 8, 541–544. [DOI] [PubMed] [Google Scholar]
- 36.Pfarr, C. M., Coue, M., Grissom, P. M., Hays, T. S., Porter, M. E. & McIntosh, J. R. (1990) Nature 345, 263–265. [DOI] [PubMed] [Google Scholar]
- 37.Rusan, N. M., Tulu, U. S., Fagerstrom, C. & Wadsworth, P. (2002) J. Cell Biol. 158, 997–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]