Abstract
Measles virus (MV) constitutes a principal cause of worldwide mortality, accounting for almost 1 million deaths annually. Although a live-attenuated vaccine protects against MV, vaccination efficiency of young infants is low because of interference by maternal antibodies. Parental concerns about vaccination safety further contribute to waning herd immunity in developed countries, resulting in recent MV outbreaks. The development of novel antivirals that close the vaccination gap in infants and silence viral outbreaks is thus highly desirable. We previously identified a microdomain in the MV fusion protein (F protein) that is structurally conserved in the paramyxovirus family and constitutes a promising target site for rationally designed antivirals. Here we report the template-based development of a small-molecule MV inhibitor, providing proof-of-concept for our approach. This lead compound specifically inhibits fusion and spread of live MV and MV glycoprotein-induced membrane fusion. The inhibitor induces negligible cytotoxicity and does not interfere with receptor binding or F protein biosynthesis or transport but prevents F protein-induced lipid mixing. Mutations in the postulated target site alter viral sensitivity to inhibition. In silico docking of the compound in this microdomain suggests a binding model that is experimentally corroborated by a structure-activity analysis of the compound and the inhibition profile of mutated F proteins. A second-generation compound designed on the basis of the interaction model shows a 200-fold increase in antiviral activity, creating the basis for novel MV therapeutics. This template-based design approach for MV may be applicable to other clinically relevant members of the paramyxovirus family.
The paramyxovirus family of negative stranded enveloped RNA viruses contains highly contagious, clinically important pathogens such as measles virus (MV), respiratory syncytial virus, and human parainfluenza viruses (hPIV) (1, 2). Although a live-attenuated vaccine protects against MV infection (3), the virus remains a principal cause of worldwide mortality, accounting for almost one million deaths per year (4). This is partially because of inefficient immunization of young infants resulting from immaturity of their immune systems and interference by transplacentally acquired maternal antibodies (5, 6). Furthermore, immunity against the live vaccine is less robust than natural immunity, and protection is less durable (7). Half-lives of protective antibodies have been estimated at 25 years or less (8, 9) creating a basis for spontaneous outbreaks in an aging population. In addition, parental concerns over vaccination safety, particularly in the United Kingdom, have contributed to such low vaccination coverage that MV outbreaks have occurred (10). Considering the mortality associated with primary MV infections and with secondary microbial infections because of MV-induced immunosuppression (11, 12) and considering that the only drug approved for treatment of some paramyxovirus infections, ribavirin, shows limited efficacy against MV (13), the development of novel therapeutics that control local outbreaks and close the immunization gap in young infants is a priority.
MV infection results from fusion of either the viral envelope or an infected cell with the plasma membrane of an uninfected cell (14, 15). The fusion process is initiated by insertion of a hydrophobic stretch of the fusion protein (F protein) ectodomain, the fusion peptide, into the target cell membrane. Further conformational rearrangements in the F protein ectodomain ultimately result in merging of the two membranes (14). To date, a crystal structure of a paramyxovirus fusion protein trimer is only available for the Newcastle disease virus F protein (16). We have generated an homology model of MV F protein (MV-F) (17) based on the coordinates for Newcastle Disease Virus F protein. Through molecular characterization of primary MV isolates with different fusogenicities (18–20), we have identified a cavity in the F protein ectodomain that is essential for F protein functionality and, hence, viral entry (17). Given that the biochemical properties of the F protein cavity are essential for fusion activity (17), this microdomain constitutes a promising target site for novel antivirals.
Inhibition of enveloped viruses at the stage of viral entry provides a route for therapeutic intervention, as evidenced by the peptidic HIV entry inhibitor T-20 (21). Other inhibitory peptides have demonstrated considerable in vitro potency against retroviruses (22, 23) and paramyxoviruses (24–27). Several obstacles hinder the production of peptidic antivirals, however. Virus-derived peptides may be immunogenic in vivo; larger peptides are often highly cost intensive to manufacture; and peptides frequently possess poor absorption from the gastrointestinal tract, necessitating i.v. delivery.
We therefore explored the development of nonpeptidic small molecules to inhibit MV entry. Multiple routes of administration are conceivable for these drug-like molecules and highly cost-effective production strategies can be easily achieved. Support for our approach comes from the previous identification of several small molecules that interfere with respiratory syncytial virus entry (28). These compounds were identified in large-scale random library screens, however, and limited information about the mechanistic basis of their activity is available, hampering directed improvement of their antiviral activity. Our template-based design approach allows the rational optimization of lead compounds and enables promising candidates to be analyzed for their ability to dock into the target domains of MV or related members of the paramyxoviridae family.
Materials and Methods
Cell Culture, Transfection, and Production of MV Stocks. Vero and HeLa cells were maintained in DMEM containing 10% FBS. Lipofectamine 2000 was used for transient transfection; cells were analyzed 18–24 h after transfection. MV stocks were grown and titered as described (29). Simian virus 5 (SV5) and hPIV2 were propagated and titered on Vero, and vaccinia virus was propagated and titered on HeLa cells.
Recombinant MV. MV particles were recovered as described (30). Recombinant viruses were confirmed by RT-PCR and DNA sequencing of the modified genes.
Compound Synthesis. Synthesis of lead compounds is described in Supporting Materials and Methods, which is published as supporting information on the PNAS web site. Structures were confirmed and the purity was determined by NMR, MS, HPLC, and elemental analysis. All compounds were dissolved in DMSO. The highest DMSO concentration used was 0.2% (vol/vol), at which no DMSO-related effect on cell viability or the degree of membrane fusion could be detected. As controls, cells were treated with DMSO at the highest concentration used in each experiment.
Compound Screen, Cytotoxicity, and Antiviral Activity. Cells were infected with MV carrying GFP as an additional transcription unit (31) at a multiplicity of infection of 0.1 plaque-forming unit (pfu) per cell for 1 h in the presence of 600 μM compound. Cytopathicity, GFP-induced fluorescence, and compound-induced cytotoxicity were monitored at 8-h time intervals. To quantify cytotoxicity, cells were incubated with compound for 20 h, and their proliferation rate was measured over 3 h by using a nonradioactive proliferation assay (Promega). To determine virus yields, virions were mixed with compounds and cells infected with 0.1 pfu per cell. Thirty hours after infection, yields of cell-associated viral particles were determined by 50% tissue culture infective dose (TCID50) titration.
Virus Entry Assays. For entry experiments, cells or virus inocula were pretreated with 300 μM oxazole 1 (OX-1) for 1 h at 25°C as indicated, followed by transfer to target cells and incubation at 37°C for 60 min in the presence or absence of 300 μM OX-1. Alternative attachment at 4°C for 60 min followed by fusion at 37°C for 20 min had no significant influence on final virus yields. When indicated, adsorbed extracellular virions were inactivated by 2-min acid treatment (40 mM sodium citrate/10 mM KCl/135 mM NaCl, pH 3.0) at 25°C, then cells were extensively washed and incubated at 37°C for 30 h. Yields of cell-associated virus were determined by TCID50 titration.
Quantitative Cell Fusion. β-galactosidase reporter assays to quantify membrane fusion were carried out essentially as described (17). Each compound concentration was assessed in at least two independent assays. To quantify viral glycoprotein-induced cytopathicity, cells were cotransfected with plasmid DNA encoding viral glycoproteins and transferred in aliquots of 1.5 × 105 cells to wells containing inhibitor. Cell viability as an indicator for fusion inhibition was determined 30 h after transfection in four replicates for each concentration by using a nonradioactive proliferation assay (Promega).
Lipid Mixing. Cells were cotransfected with plasmids as indicated and overlaid 16 h after transfection with rhodamine 18 (R18)-labeled African Green monkey-derived erythrocytes for 1 h at 4°C. Compound was added as indicated, and cells were incubated at 37°C for 30 min. Transfer of red fluorescence indicating lipid mixing was documented at a magnification of 400×.
In Vitro Protein Transcription/Translation. Rabbit reticulocyte lysates were mixed with 0.5 μg of plasmid DNA encoding MV-F under the control of the T7 promoter (pT7-MV-F), 20 μCi (1 Ci = 37 GBq) [35S]methionine, and OX-1 or DMSO. Samples were incubated at 30°C for 90 min, mixed with urea buffer (200 mM Tris, pH 6.8/8 M urea/5% SDS/0.1 mM EDTA/0.03% bromphenol blue/1.5% DTT), and fractionated on 12% polyacrylamide gels. Dried gels were exposed to XAR films (Kodak).
Surface Expression and Immunostaining. Transfected cells were labeled with 0.5 mg/ml sulfosuccinimidyl-2-(biotinamido) ethyl-1,3-dithiopropionate and harvested as described (20). Equal amounts of protein lysates were absorbed to Sepharose-coupled streptavidin and subjected to immunoblotting with antibodies specific for MV-F (17). For Western analysis, cells were harvested 30 h after infection in lysis buffer (50 mM Tris, pH 8.0/62.5 mM EDTA/0.4% deoxycholate/1% Igepal), 2.5 μg of protein lysate was mixed with urea buffer and analyzed by immunoblotting with antibodies against MV nucleocapsid protein.
Virus Adsorption. Infected cells were labeled with [35S]methionine for 16 h, and the released virions were purified on a 20–60% sucrose gradient (32). The equivalent of 20,000 cpm was adsorbed to target cells for 1 h at 4°C in the presence or absence of OX-1, and bound activity was determined with a scintillation counter.
Computational Methods. sybyl6.9 (Tripos Discovery Software, St. Louis) Connolly surfaces for analogs of OX-1 and acetamide 4 (AM-4, see Supporting Materials and Methods) of MV-F 94V residues within 10 Å of the ligand were constructed. The surfaces were docked with ligands both manually and automatically by using the dock algorithm (25). Complexes were refined by low temperature molecular dynamics (MD) (20 K, TVN ensemble; 5 ps). Binding-site changes for F(V94G) and F(V94M) were examined by using MD with the backbone initially held fixed (20 K, TVN ensemble; 1 ps), then continued without restrictions in the presence of the inhibitor (4 ps). This procedure circumvented unfolding of the protein in the absence of solvation while allowing the generation of a viable working model. All other MV-F variants described were treated similarly, and deformations of the binding site were visualized with Connolly surfaces.
Results
The cavity we previously identified in MV-F is located at the interface between the F protein neck and head regions (Fig. 6, which is published as supporting information on the PNAS web site). Our experiments found its relatively nonpolar nature to be important for F protein fusion activity (17), implicating compounds with a polar head and a nonpolar tail as inhibitor candidates.
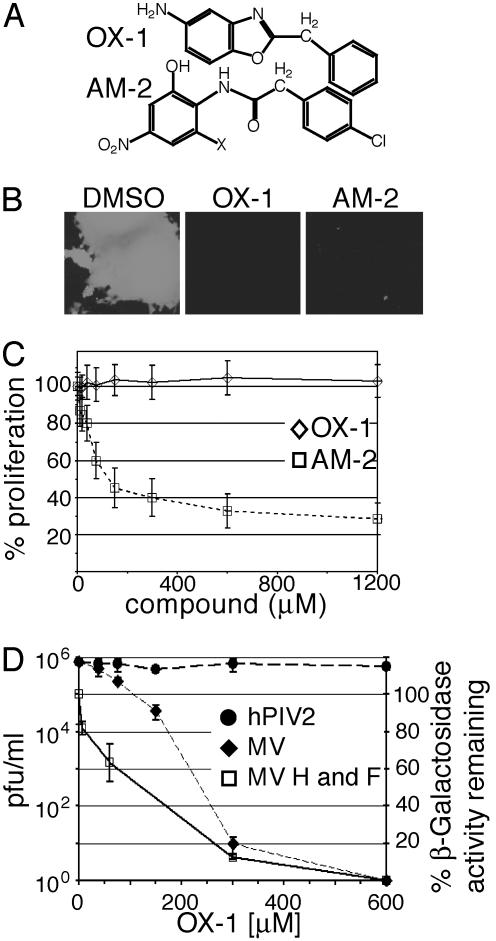
Identification of First-Generation Leads. Potential inhibitors are predicted to have molecular geometries of 7–12 Å in two orthogonal directions, sealing the cavity floor with a hydrophobic plug and reaching its top to engage in polar interactions with water or hydrophilic interactions at the cavity rim. To test this hypothesis, we analyzed antiviral activity of 32 representatives of different structural classes (Fig. 7, which is published as supporting information on the PNAS web site) that met these requirements. Activity was assessed against a recombinant MV strain carrying GFP as an additional transcription unit, which is genetically based on the Edmonston (MV-Edm) strain (30) and facilitates visualization of single infectious centers and multinucleated syncytia. Two compounds of different classes (Fig. 1A), OX-1 and AM-2, efficiently suppressed the formation of infectious centers (Fig. 1B). All other molecules tested lacked antiviral activity and/or were cytotoxic (data not shown).
Fig. 1.
Identification of a first-generation lead compound. (A) Structures of compounds OX-1 and AM-2. (B) Inhibitory activity of compounds OX-1 and AM-2. Cells were infected with MV-GFP in the presence of compounds or DMSO and photographed at a magnification of 400×. (C) Proliferation assay of cells incubated in the presence of compound. Values indicate the percentage of signal intensity as compared with cells treated with DMSO. (D) Cells infected with MV-Edm or hPIV2 were incubated in the presence of different OX-1 concentrations as indicated, and virus yields were determined by TCID50 titration (left axis). A β-galactosidase reporter assay was used for quantification of cell fusion mediated by transiently expressed MV glycoproteins in the presence of different OX-1 concentrations; the percentage of β-galactosidase activity as compared with cells treated with DMSO is given (right axis).
To assess cytotoxicity of OX-1 and AM-2, their effect on cell proliferation was determined. AM-2 revealed a 50% cytotoxic concentration (CC50) <50 μM (Fig. 1C), similar to its active dose (data not shown), suggesting that its antiviral effect might be attributable to cytotoxicity. In contrast, OX-1 displayed CC50 values ≫600 μM, indicating negligible cytotoxicity.
OX-1 Activity Against Live Virus and MV Glycoprotein-Mediated Membrane Fusion. To assess MV-specificity and quantify the inhibitory activity of OX-1, cells were infected with 0.1 pfu per cell each of MV-Edm or hPIV2, a paramyxovirus distantly related to MV, in the presence of different compound concentrations. The 50% inhibitory concentration (IC50) of OX-1 against live MV-Edm was ≈55 μM, whereas no inhibitory activity against hPIV2 was detected (Fig. 1D), thus indicating MV specificity of OX-1 inhibition.
Membrane fusion induced by transiently expressed MV glycoproteins should be impaired if OX-1 targets entry (rather than postentry) steps of the viral life cycle. A β-galactosidase reporter-based quantitative fusion assay (33) was used to test this hypothesis. OX-1 inhibitory activity determined in this assay (IC50 ≈ 100 μM) resembled the effect observed against live virus (Fig. 1D), suggesting a direct effect of the compound on the entry process.
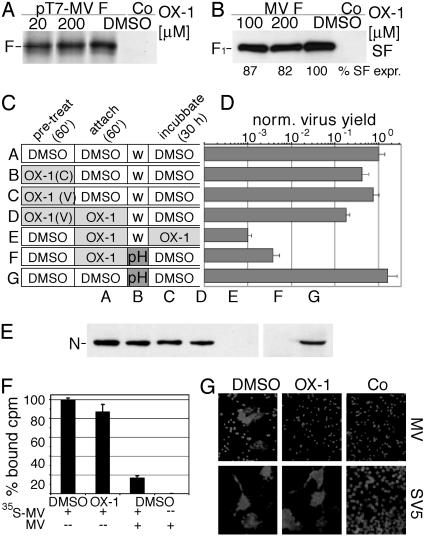
Protein Biosynthesis, Transport, and Virus Attachment Are Unaffected by OX-1. Supporting this conclusion, in vitro protein biosynthesis and evaluation of F protein surface steady-state levels demonstrated biochemically that F protein biosynthesis and intracellular transport are essentially unaffected at OX-1 concentrations corresponding to up to four IC50 (200 μM) (Fig. 2 A and B).
Fig. 2.
OX-1 prevents viral entry. (A) In vitro F protein transcription/translation in the presence of different OX-1 concentrations or DMSO. Control (Co) cells were treated with DMSO and transfected with a variant of plasmid pT7-MV-F harboring the F protein gene in reverse orientation to the T7 promoter. (B) Surface (SF) biotinylation of cleaved F1 protein displayed on transfected cells incubated in the presence of different OX-1 concentrations or DMSO. Biotinylated samples were detected by immunoblotting with antibodies directed against the F protein tail. Controls include DMSO-treated mock-transfected cells. (C) Scheme of pretreatment of cells [OX-1 (C)] or virus [OX-1 (V)], virus adsorption, and incubation in the presence of OX-1 or DMSO. Subsequent to adsorption, inocula were removed and cells either washed (w) or subjected to low-pH treatment (pH) followed by washing. (D) Virus yields on treatment as outlined in C were normalized for control experiments of cells treated with DMSO only or treated with DMSO and low pH. (E) Western analysis of MV nucleocapsid (N) protein synthesis in cells treated as outlined in C. (F) Adsorption of metabolically labeled virions to cells in the presence of compound. Control cells were treated with an excess of purified, nonlabeled virions (MV). Bound activity in cell lysates was determined by using a scintillation counter. (G) OX-1 prevents lipid mixing. Cells were cotransfected with plasmid DNA encoding MV-H and MV-F or SV5-hemagglutinin/neuraminidase and SV5-F, and overlaid with R18-labeled erythrocytes in the presence or absence of OX-1. Control cells were transfected with plasmid DNA encoding MV-H or SV5-hemagglutinin/neuraminidase, respectively.
OX-1 inhibition of entry could be due to interference with viral attachment to target cells mediated by the hemagglutinin (H) protein or to inhibition of F protein-mediated fusion. To assess these alternatives, inhibitor was added at different times during infection (Fig. 2C). Pretreating target cells or viral particles with OX-1 did not significantly affect virus yield (Fig. 2D, samples B and C). Likewise, removing compound and unbound particles after attachment in the presence of OX-1 caused only a minor reduction in virus yield, whereas attachment and continued incubation in the presence of compound substantially reduced virus yield (Fig. 2D, samples D and E) These observations indicate that inhibition is reversible and that receptor binding of HA is not impaired by the compound.
If OX-1 interferes with viral fusion, virions adsorbed to target cells in the presence of compound should remain sensitive to pH-3.0 treatment known to inactivate many enveloped viruses (34). Indeed, although low-pH treatment did not reduce virus yields in the absence of compound, low-pH treatment after adsorption to target cells in the presence of OX-1 resulted in a reduction in virus yield (Fig. 2D, samples F and G). The lower degree of inhibition compared to sample E is likely because of the escape of some particles from OX-1 inhibition or pH inactivation followed by secondary infections not prevented in the absence of inhibitor.
These findings were corroborated by immunoblot analysis of MV nucleocapsid protein in cell lysates (Fig. 2E) and by adsorption of purified, metabolically labeled virions to target cells at low temperatures in the presence of compound followed by removal of unbound virions and quantification of bound activity (Fig. 2F). In this assay, OX-1 induced no significant reduction of virion attachment.
OX-1 Prevents Lipid Mixing. Merging of the outer layers of the donor and target membranes results in lipid mixing, an early stage of the fusion process that can be assessed by examining the redistribution of lipid dyes, such as R18. OX-1 strongly suppressed R18 redistribution in MV-H- and MV-F-expressing cells (Fig. 2G), indicating that it prevents membrane merging. Dye redistribution in SV5-hemagglutinin/neuraminidase- and SV5-F-expressing control samples was unaffected by OX-1, confirming that the compound has no secondary inhibitory effect on R18 transfer.
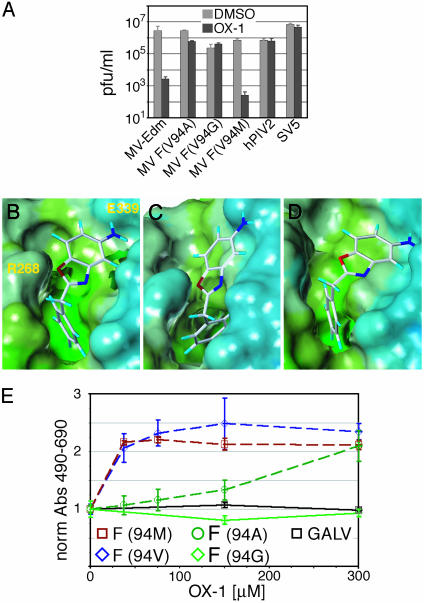
Mutations in the F Protein Cavity Induce Resistance to OX-1. We previously described MV recombinants MV-F(V94A), MV-F(V94G), and MV-F(V94M) (20) harboring point mutations in the F protein cavity domain. We observed greatly increased or complete resistance to inhibition by OX-1 of the V94A (IC50 = 124 μM) and V94G (IC50 = 600 μM) variants, respectively, whereas the V94M (IC50 = 20 μM) recombinant and the parental MV-Edm strain (IC50 = 55 μM) were efficiently inhibited (Fig. 3A). Importantly, most primary MV strains contain a methionine residue at this position. Yields of other members of the paramyxovirus family, such as hPIV2 (Figs. 1D and 3A) and SV5 (Fig. 3A) are not affected by OX-1. These observations thus underline that OX-1 does not interfere nonspecifically with MV replication; they furthermore indicate that the conformation of the F protein cavity is essential for OX-1 activity, suggesting this microdomain to be the binding site for the compound.
Fig. 3.
Point mutations and in silico docking support OX-1 binding to the F protein cavity. (A) OX-1 activity is target-specific. Cells were infected with MV recombinants harboring point mutations in the F protein cavity, parental MV, hPIV2, or SV5 in the presence of 200 μM OX-1 or DMSO, and virus yields were determined by TCID50 titration (MV and hPIV2) or plaque assay (SV5). (B) Model of the postulated inhibitor-binding site shown with a Connelly surface mapped with lipophilic properties. OX-1 was docked into the F protein cavity, and the system was subjected to low-temperature MD. (C) The conformational change to the MV-F binding site with the V94G mutation after MD. The binding site narrows, forcing the inhibitor out of the pocket as demonstrated by artificial docking of the compound. (D) Illustration of the F protein V94M mutant binding site after MD. (E) Quantitative inhibition assay of cells cotransfected with MV-H and MV-F-Edm or MV-F protein variants, or a gibbon ape leukemia virus (GALV)-derived glycoprotein for control, followed by incubation in the presence of compound. Values reflect inhibition of fusion and were normalized for fusion observed in the absence of compound.
Compound Docking. We performed in silico docking of OX-1 into the cavity followed by MD to refine the complex. A three-point interaction model (Fig. 3B) proved consistent with both the predicted size of the cavity and the distribution of polar and nonpolar centers (17). According to this model, the amino group of OX-1 engages in a hydrogen bond with E339 at the top of the site, the oxygen of the oxazole ring is anchored by R268, and the phenyl ring is buried in the hydrophobic base of the cavity. Automated docking of the compound by using the dock algorithm (35) suggested the same binding mode.
When introducing the V94A (data not shown) and V94G (Fig. 3C) mutations into the model and subjecting all residues within 10 Å to MD, the cavity is predicted to contract and fill the volume previously occupied by the valine side chain, thus forcing the ligand out of the cavity. Consequently, mutation of V94 to a residue of larger bulk, such as 94M, is predicted to expand the target area for OX-1 (Fig. 3D), allowing favorable docking.
The resistance profile of our mutant virions was corroborated by the inhibitory effect of OX-1 on fusion mediated by the corresponding F variants when transiently expressed. The V94M mutant showed a slightly increased sensitivity to inhibition, whereas the V94G variant was uninhibited and the V94A mutation resulted in an intermediate phenotype (Fig. 3E). Fusion induced by a plasmid-encoded fusogenic Gibbon Ape Leukemia virus envelope protein (36) was not inhibited, further confirming the specificity of OX-1 activity in the transient inhibition assay.
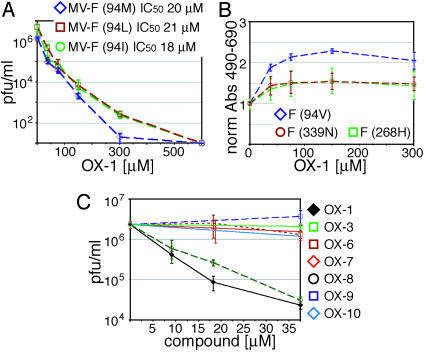
Variants of OX-1 and Cavity Modifications Support the Docking Model. To experimentally evaluate the quality of the docking model, we first generated recombinant virions carrying bulky 94L and 94I side chains. As predicted by the model, sensitivity of these recombinants to OX-1 was virtually identical to the 94M mutant (Fig. 4A) and ≈2-fold increased when compared with the parental MV-Edm.
Fig. 4.
Experimental evaluation of the binding model. (A) Cells were infected with different MV recombinants and incubated in the presence of a range of OX-1 concentrations as indicated, followed by TCID50 titration to determine virus yields. IC50 values are given. (B) Quantitative inhibition assay of cells cotransfected with MV-H and MV-F variants as indicated, followed by incubation in the presence of compound. Values reflect inhibition of fusion and were normalized for fusion observed in the absence of compound. (C) Cells infected with MV-Edm were incubated in the presence of a panel of OX-1 variants as indicated and subjected to TCID50 titration.
Second, when testing F protein variants harboring E339N and R268H mutations predicted to form less stable hydrogen bonds with OX-1, we observed a substantial reduction in inhibitory activity in the transient fusion assay (Fig. 4B). Less conserved changes at positions E339 and R268 or insertion of hydrophilic side chains into the base of the cavity ablate F protein fusion activity (data not shown) and could hence not be examined.
Third, a panel of structural variants of OX-1 carrying substituents that are predicted to interfere with docking to the cavity was generated. Exchange of the NH2 group in OX-1 for a NO2 group or a proton (OX-6 and OX-7, predicted to disrupt hydrogen bonding with E339) or addition of a NO2 or CH3 group to the OX-1 phenyl ring [OX-9 and OX-10 (Fig. 8, which is published as supporting information on the PNAS web site), postulated to prevent insertion of the ring into the base of the cavity] caused a substantial reduction in antiviral activity (Fig. 4C). In contrast, substitution of the oxygen in the OX-1 oxazole ring for nitrogen, OX-8 (Fig. 8), should not disrupt the hydrogen bond with R268. Consistent with this prediction, this variant showed virtually unchanged antiviral activity (Fig. 4C).
Lastly, the interaction model depends on the L-shaped conformation of OX-1 induced by the CH2 unit between its aromatic rings, given that the phenyl ring of a linear OX-1 variant would lie above the hydrophobic pocket and be unfavorably exposed to the aqueous phase. Indeed, antiviral activity of an essentially linear compound identical to OX-1 but lacking the CH2 moiety (OX-3) is greatly reduced (Fig. 4C). Thus, the effects of both cavity mutations on OX-1 susceptibility and compound variants on antiviral activity are supportive of the three-point interaction model.
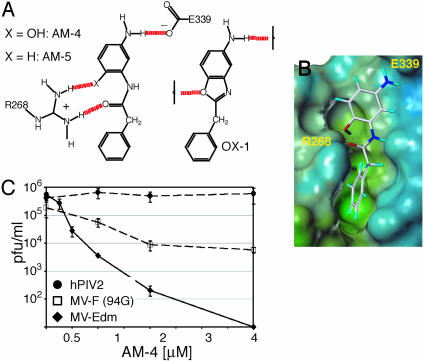
Design of Second-Generation Inhibitors Based on OX-1. Toward increasing the antiviral potency of OX-1, we aimed to optimize compound docking on the basis of the binding model. Following in silico screening of a variety of candidate structures, an acyclic variant of OX-1 incorporating an amide and a orthohydroxy group, AM-4 (Fig. 5A) appeared favorable based on its ability to form multiple hydrogen bonds with R268 (Fig. 5 A and B). When this compound was tested against MV, an ≈200-fold increase (IC50 = 260 nM) in antiviral activity was observed, whereas yields of hPIV2 were unaffected (Fig. 5C). The resistance profile of MV-94G (Fig. 5C) and MV-94A (data not shown) resembled that for OX-1, indicating high specificity of AM-4. Target specificity also excludes cytotoxic effects as a functional basis for AM-4 activity, although cell proliferation was reduced at elevated concentrations of the compound (data not shown). Based on a CC50 of 17 μM, a high therapeutic index (CC50/IC50) of 65 was calculated for AM-4. The importance of multiple hydrogen bonds to R268 is further supported by results obtained for an analog of AM-4 lacking the OH moiety. This compound, AM-5, (IC50 = 10.5 μM), although ≈5× more active than OX-1, is 40× less potent than AM-4.
Fig. 5.
Template-based development of second-generation MV inhibitors. (A) Structures of compounds AM-4 and AM-5. The scheme shows predicted hydrogen bonding for AM-4 in comparison with OX-1. The amide carbonyl and ortho-hydroxy groups of AM-4 engage in simultaneous hydrogen bonding with R268. (B) Connelly surface model of AM-4 docked into the F protein cavity, followed by low-temperature MD. (C) Antiviral activity of AM-4. Cells infected with MV-Edm, MV-F (V94G), or hPIV2 as indicated were incubated in the presence of different AM-4 concentrations, followed by TCID50 titration.
Discussion
In previous work, we identified a cavity in the MV-F protein that is essential for F protein functionality and, hence, constitutes an attractive target for novel antivirals (17). By implementing a template-design approach targeting this microdomain, we now report the development of a small molecule inhibitor with specific activity against MV. Characterization of this compound indicates that it interferes with F protein-mediated membrane fusion and, hence, viral entry as the molecular basis for inhibition. That the inhibitor induces negligible cytotoxicity, does not impair F protein synthesis and transport, and does not hinder virus attachment to target cells indicates prefusion and postentry steps not to contribute to its mechanism of antiviral activity. This finding is corroborated by the inhibition of membrane fusion induced by transiently expressed MV glycoproteins. Replication of the paramyxoviruses hPIV2 and SV5 and membrane fusion induced by a gibbon ape leukemia virus envelope protein are unaffected by OX-1, confirming target specificity of the molecule and further arguing against general interference with cellular functions or virus genome replication as the underlying mechanism of action.
A panel of MV recombinants with mutations in the F protein cavity shows altered sensitivity to OX-1-mediated inhibition, suggesting this microdomain to be the target site for the compound. Automated and manual docking of OX-1 into the F protein cavity indicates a three-point interaction model, which for all mutants predicts changes in sensitivity to OX-1 as determined experimentally. Previous studies (37–41) have used mutation-determined resistance to identify compound target sites in a similar manner. The docking model is further supported by our observation that structural alterations of OX-1 designed to impair docking into the target site reduce inhibitory activity. Additional support for its validity comes from the finding that second-generation molecules conceived on the basis of the model possess greatly increased antiviral activity.
The docking model provides a potential explanation for the MV specificity of OX-1 as compared with hPIV2 or SV5. Sequence alignment reveals differences in key amino acids that are predicted to anchor the inhibitor in MV-F, R268 (Q262 in hPIV2 and Q258 in SV5), and E339 (N333 in hPIV2 and N329 in SV5). Based on this linear comparison, the two charged residues in the postulated MV-F binding site are replaced by polar but neutral amino acids, each of which is shorter by two or one methylene carbon, respectively. These differences are compatible with those expected for altered ligand selectivity.
Considering the location of the F protein cavity at the intersection of the head and neck domains of the F protein trimer (17) at the mouths of radial channels that have been suggested to harbor the fusion peptide in the prefusion conformation (42), it may be involved in facilitating F protein structural rearrangements during exposure of the fusion peptide. By binding to the cavity, the compound could stabilize a metastable prefusion conformation of the F protein trimer and, hence, raise the energy barrier that must be overcome to initiate the conformational transitions ultimately resulting in membrane fusion. Alternatively, the compound may sterically interfere with an essential transitional conformation of the F protein trimer, thereby hindering progression of the molecule to a stable postfusion conformation.
We predict that rationally developed antivirals, in addition to their therapeutic potential, constitute valuable tools for the analysis of the mechanism of paramyxovirus entry. Thus, further characterization of the structural basis for fusion inhibition might contribute to our understanding of F protein-mediated membrane fusion and open additional routes for future improvements of compound activity. Considering that the overall structures of the F proteins are conserved in the paramyxovirus family, the identification of a target site for small molecule antivirals in the MV-F ectodomain suggests that this templatebased approach might be adaptable to other clinically relevant members of this virus family, such as respiratory syncytial virus, against which no vaccine is currently available.
Supplementary Material
Acknowledgments
We thank B. Moss for vaccinia virus vTF7–3; R. Vile for the GALV expression plasmid; R. Cattaneo for plasmids encoding MV-F and MV-H and antibodies directed against the F protein tail; M. Billeter for the MV rescue system; and D. Steinhauer and A. Hammond for critical reading of the manuscript. This work was supported by a Feodor Lynen Fellowship from the Alexander von Humboldt Foundation (to R.K.P.), National Institutes of Health Grant AI054337 (to R.W.C.), and Grant AI057157 to the Southeastern Regional Center of Excellence for Emerging Infections and Biodefense.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: MV, measles virus; F protein, fusion protein; MV-F, MV F protein; MV-Edm, Edmonston strain of MV; SV5, simian virus 5; CC50, 50% cytotoxic concentration; IC50, 50% inhibitory concentration; TCID50, 50% tissue culture infective dose; pfu, plaque-forming unit; R18, rhodamine 18; H, hemagglutinin; hPIV, human parainfluenza virus; MD, molecular dynamics; OX, oxazole; AM, acetamide.
References
- 1.Chua, K. B., Goh, K. J., Wong, K. T., Kamarulzaman, A., Tan, P. S., Ksiazek, T. G., Zaki, S. R., Paul, G., Lam, S. K. & Tan, C. T. (1999) Lancet 354, 1257–1259. [DOI] [PubMed] [Google Scholar]
- 2.Chua, K. B., Bellini, W. J., Rota, P. A., Harcourt, B. H., Tamin, A., Lam, S. K., Ksiazek, T. G., Rollin, P. E., Zaki, S. R., Shieh, W., et al. (2000) Science 288, 1432–1435. [DOI] [PubMed] [Google Scholar]
- 3.Hilleman, M. R. (2001) Vaccine 20, 651–665. [DOI] [PubMed] [Google Scholar]
- 4.World Health Organization. (2000) Health Systems: Improving Performance, The World Health Report 2000 (World Health Organization, Geneva).
- 5.Gans, H. A., Arvin, A. M., Galinus, J., Logan, L., DeHovitz, R. & Maldonado, Y. (1998) J. Am. Med. Assoc. 280, 527–532. [DOI] [PubMed] [Google Scholar]
- 6.Polack, F. P., Lee, S. H., Permar, S., Manyara, E., Nousari, H. G., Jeng, Y., Mustafa, F., Valsamakis, A., Adams, R. J., Robinson, H. L. & Griffin, D. E. (2000) Nat. Med. 6, 776–781. [DOI] [PubMed] [Google Scholar]
- 7.Putz, M. M., Bouche, F. B., de Swart, R. L. & Muller, C. P. (2003) Int. J. Parasitol. 33, 525–545. [DOI] [PubMed] [Google Scholar]
- 8.Mossong, J., Nokes, D. J., Edmunds, W. J., Cox, M. J., Ratnam, S. & Muller, C. P. (1999) Am. J. Epidemiol. 150, 1238–1249. [DOI] [PubMed] [Google Scholar]
- 9.Mossong, J., O'Callaghan, C. J. & Ratnam, S. (2000) Vaccine 19, 523–529. [DOI] [PubMed] [Google Scholar]
- 10.Jansen, V. A., Stollenwerk, N., Jensen, H. J., Ramsay, M. E., Edmunds, W. J. & Rhodes, C. J. (2003) Science 301, 804. [DOI] [PubMed] [Google Scholar]
- 11.Griffin, D. E. (2001) in Fields Virology, eds. Knipe, D. M. & Howley, P. M. (Raven, New York), Vol. 2, pp. 1401. [Google Scholar]
- 12.Patterson, J. B., Manchester, M. & Oldstone, M. B. (2001) Trends Mol. Med. 7, 85–88. [DOI] [PubMed] [Google Scholar]
- 13.Anderson, L. J., Parker, R. A. & Strikas, R. L. (1990) J. Infect. Dis. 161, 640–646. [DOI] [PubMed] [Google Scholar]
- 14.Lamb, R. A. (1993) Virology 197, 1–11. [DOI] [PubMed] [Google Scholar]
- 15.Lamb, R. A. & Kolakofsky, D. (1996) in Fundamental Virology, eds. Fields, B. N., Knipe, D. M. & Howley, P. M. (Lippencott–Raven, Philadelphia, PA), pp. 577–604.
- 16.Chen, L., Gorman, J. J., McKimm-Breschkin, J., Lawrence, L. J., Tulloch, P. A., Smith, B. J., Colman, P. M. & Lawrence, M. C. (2001) Structure (London) 9, 255–266. [DOI] [PubMed] [Google Scholar]
- 17.Plemper, R. K., Lakdawala, A. S., Gernert, K. M., Snyder, J. P. & Compans, R. W. (2003) Biochemistry 42, 6645–6655. [DOI] [PubMed] [Google Scholar]
- 18.Johnston, I. C., ter Meulen, V., Schneider-Schaulies, J. & Schneider-Schaulies, S. (1999) J. Virol. 73, 6903–6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Plemper, R. K., Hammond, A. L., Gerlier, D., Fielding, A. K. & Cattaneo, R. (2002) J. Virol. 76, 5051–5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plemper, R. K. & Compans, R. W. (2003) J. Virol. 77, 4181–4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Starr-Spires, L. D. & Collman, R. G. (2002) Clin. Lab. Med. 22, 681–701. [DOI] [PubMed] [Google Scholar]
- 22.Root, M. J., Kay, M. S. & Kim, P. S. (2001) Science 291, 884–888. [DOI] [PubMed] [Google Scholar]
- 23.Eckert, D. M. & Kim, P. S. (2001) Proc. Natl. Acad. Sci. USA 98, 11187–11192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lambert, D. M., Barney, S., Lambert, A. L., Guthrie, K., Medinas, R., Davis, D. E., Bucy, T., Erickson, J., Merutka, G. & Petteway, S. R., Jr. (1996) Proc. Natl. Acad. Sci. USA 93, 2186–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yao, Q. & Compans, R. W. (1996) Virology 223, 103–112. [DOI] [PubMed] [Google Scholar]
- 26.Rapaport, D., Ovadia, M. & Shai, Y. (1995) EMBO J. 14, 5524–5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Young, J. K., Hicks, R. P., Wright, G. E. & Morrison, T. G. (1997) Virology 238, 291–304. [DOI] [PubMed] [Google Scholar]
- 28.Meanwell, N. A. & Krystal, M. (2000) Drug Discovery Today 5, 241–252. [DOI] [PubMed] [Google Scholar]
- 29.Plemper, R. K., Hammond, A. L. & Cattaneo, R. (2001) J. Biol. Chem. 276, 44239–44246. [DOI] [PubMed] [Google Scholar]
- 30.Radecke, F., Spielhofer, P., Schneider, H., Kaelin, K., Huber, M., Dotsch, C., Christiansen, G. & Billeter, M. A. (1995) EMBO J. 14, 5773–5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ehrengruber, M. U., Hennou, S., Bueler, H., Naim, H. Y., Deglon, N. & Lundstrom, K. (2001) Mol. Cell Neurosci. 17, 855–871. [DOI] [PubMed] [Google Scholar]
- 32.Bellini, W. J., Trudgett, A. & McFarlin, D. E. (1979) J. Gen. Virol. 43, 633–639. [DOI] [PubMed] [Google Scholar]
- 33.Nussbaum, O., Broder, C. C., Moss, B., Stern, L. B., Rozenblatt, S. & Berger, E. A. (1995) J. Virol. 69, 3341–3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kizhatil, K. & Albritton, L. M. (1997) J. Virol. 71, 7145–7156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ewing, T. J., Makino, S., Skillman, A. G. & Kuntz, I. D. (2001) J. Comput. Aided Mol. Des. 15, 411–428. [DOI] [PubMed] [Google Scholar]
- 36.Diaz, R. M., Bateman, A., Emiliusen, L., Fielding, A., Trono, D., Russell, S. J. & Vile, R. G. (2000) Gene Ther. 7, 1656–1663. [DOI] [PubMed] [Google Scholar]
- 37.Kuritzkes, D. R. (2002) J. HIV Ther. 7, 87–91. [PubMed] [Google Scholar]
- 38.Sheldon, J. G. & Condra, J. H. (1999) Antiviral Ther. 4, 135–142. [PubMed] [Google Scholar]
- 39.Friedberg, E. C. & Fischhaber, P. L. (2003) Cell 113, 139–140. [DOI] [PubMed] [Google Scholar]
- 40.Poehlsgaard, J. & Douthwaite, S. (2003) Curr. Opin. Invest. Drugs 4, 140–148. [PubMed] [Google Scholar]
- 41.Giannakakou, P., Gussio, R., Nogales, E., Downing, K. H., Zaharevitz, D., Bollbuck, B., Poy, G., Sackett, D., Nicolaou, K. C. & Fojo, T. (2000) Proc. Natl. Acad. Sci. USA 97, 2904–2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Colman, P. M. & Lawrence, M. C. (2003) Nat. Rev. Mol. Cell Biol. 4, 309–319. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.