Abstract
BACKGROUND
Obesity, metabolic syndrome, and type 2 diabetes are major public health challenges. Recently, interest has surged regarding the possible role of the intestinal microbiota as potential novel contributors to the increased prevalence of these 3 disorders.
CONTENT
Recent advances in microbial DNA sequencing technologies have resulted in the widespread application of whole-genome sequencing technologies for metagenomic DNA analysis of complex ecosystems such as the human gut. Current evidence suggests that the gut microbiota affect nutrient acquisition, energy harvest, and a myriad of host metabolic pathways.
CONCLUSION
Advances in the Human Microbiome Project and human metagenomics research will lead the way toward a greater understanding of the importance and role of the gut microbiome in metabolic disorders such as obesity, metabolic syndrome, and diabetes.
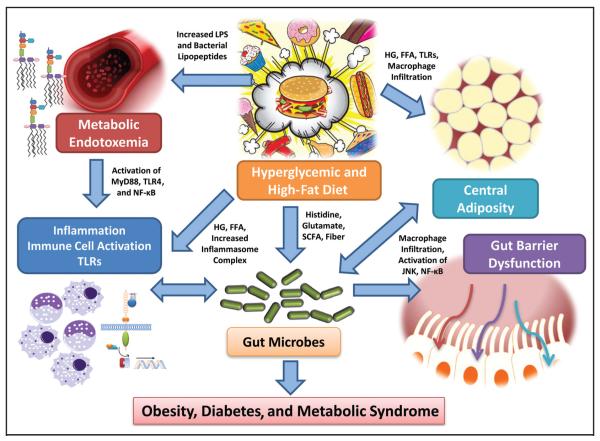
Obesity, metabolic syndrome, and type 2 diabetes are major public health challenges, affecting approximately 26 million children and adults in the US. More than 8% of the US population has diabetes, of which 17.9 million people have the metabolic syndrome (1). During the past 20 years, obesity has dramatically increased in prevalence in the US. More than 1 in 3 US adults (36%) are obese, and approximately 12.5 million (17%) of children and adolescents (age 2–19 years) are obese (2). In the US in 2010 (2), all of the states had a prevalence of obesity of over 20%. The heterogeneity of these disorders has been demonstrated through both anthropometric and genetic studies. These metabolic disorders are believed to be caused by a combination of genetic susceptibilities and lifestyle changes. Recently, interest has surged in the possible role of the intestinal microbiome as a potential contributor to the rapidly increased prevalence of obesity (3–5). This review focuses on recent advances in the understanding of the gut microbiome and techniques to assess the microbiome and its relationship to human body metabolism, obesity, metabolic syndrome, and type 2 diabetes (Fig. 1).
Fig. 1. Hyperglycemia (HG) and increased free fatty acids (FFA), which are hallmarks of obesity, metabolic syndrome, and diabetes, combined with a high-fat, high–glycemic load diet, could result in increased activation of the inflammasome complex as well as increase the activation of macrophages via increased TLR activation and nuclear factor κB (NF-κB) activation.
Increased metabolic endotoxemia may occur and activate the TLR4 pathway via the adapter protein, MyD88, leading to immune cell activation and inflammation. Also, macrophages could infiltrate the adipose tissue and activate mitogen-activated protein kinases, such as c-Jun aminoterminal kinase (JNK) and NF-κB, resulting in increased cross-talk and adipose-tissue–derived adipokines. A hyperglycemic and high fat diet could also result in changes to the gut microbiome by altering the content of histidine, glutamate, SCFAs, and other factors and promote gut-barrier dysfunction and conditions prevalent in obesity, metabolic syndrome, and diabetes by altering the host response. All of these metabolic alterations that result in increased systemic inflammation, macrophage activity, and TLR activation contribute to the increased cardiometabolic burden in obesity, diabetes, and metabolic syndrome.
The Human Gut Microbiome: The Toolkit behind the Science
The widespread application of 16S rRNA gene sequencing for detection of bacterial pathogens and microbial ecology has provided a robust technical platform for the evaluation of the bacterial composition of the human microbiome. Sequencing of 2 primary targets within bacterial 16S rRNA genes yielded valuable compositional data pertaining to the human fecal microbiome of 242 healthy adults (6, 7). In the Human Microbiome Project, 18 different body sites were sampled and sequenced. Stool specimens were the single specimen type used to study the intestinal microbiome. Previously published studies demonstrated the variation in composition of the gut microbiome among locations within the gastrointestinal tract in different mammalian species. For example, 16S rRNA gene sequencing has been deployed to study the maturation of murine cecal microbiota, and these studies demonstrated the existence of a large number of yet-unidentified bacteria that inhabit the mammalian intestine (6). Such sequencing strategies, which are culture independent, are essential for determining bacterial composition of the microbiome and its relative stability and diversity over time. Thus, it is essential to develop robust experimental models of the human microbiome to delineate important mechanistic processes in the development of human disease states.
Advances in sequencing technologies have resulted in the widespread application of whole-genome (WG)3 sequencing technologies for metagenomic DNA analysis of complex ecosystems such as the human intestine (7). WG sequencing strategies provide microbial compositional as well as functional information. WG data can be used to infer bacterial composition, and these data yield information similar to that generated by 16S rRNA gene sequencing. The genome sequences of highly abundant species are well represented in a set of random shotgun reads, whereas less abundant species are represented by fewer sequences generated in a next-generation sequencing run. This relative richness permits the comprehensive measurement of the compositional responses of an ecosystem to dietary changes, drug therapy, epigenetic alterations, and environmental perturbations. Alternatively, most genes (usually approximately 2000 genes per bacterium) in the microbiome are sequenced so that metabolic and other functional pathways can be evaluated in each individual’s metagenome. Functional WG data provide opportunities to find out which metabolic pathways are affected and how the microbiome may contribute mechanistically to health and disease states. This technology creates the formidable challenge of managing vast data sets. Advances in next-generation DNA sequencing yielded 576.7 Gb of microbial DNA sequence data, which were generated with an Illumina™ genome analyzer (Illumina) from total DNA from the stool samples of 124 European adults (8). The relationship between the commensal microbiota that comprise the gut microbiota and those that are in the intestinal barrier is complex and differs spatially throughout different areas of the gastrointestinal tract. Fecal metagenomics measures ecosystem changes in stool or the distal intestine, but it does not compare the microbiomes in different regions of the intestine. It is also important to note that metagenomic analysis of fecal samples does not include all important molecular interactions within the gastrointestinal tract. Turnbaugh et al. have proposed the idea of a core set of functions within the microbiome, and the tools of proteomics and metabolomics may be required for more in-depth functional analyses (7, 9). From a systems perspective, metagenomic analyses may provide further details on specific intraindividual changes and thus have major implications for personalized medicine strategies.
Metatranscriptomics, metaproteomics, and metabonomics will be useful to explore the functional aspects of the gut microbiome from the top down. Realtime analysis of the intestinal microbiome is a useful tool in the development of personalized approaches to targeted therapies. Metabonomics can be described as the study of metabolic responses to chemicals, the environment, and diseases and involves the computational analysis of spectral metabolic data that provide information on temporal changes to specific metabolites. In addition, metabonomics provides global metabolic profiling of an individual in real time. It is possible, with such approaches, to elucidate complex pathways and networks that are altered in specific disease states. The combination of metabolic profiling and metagenomic studies of gut microbiota permits the study of host and microbial metabolism in great detail. Such analysis of functional components of the microbiome that affect metabolism and human health is referred to as functional metagenomics.
Metagenomics and the science of the human microbiome have arrived at the forefront of biology primarily because of major technical and conceptual developments. The major technical development was the deployment in many centers of next-generation DNA sequencing technologies with greatly enhanced capabilities for sequencing collections of microbial genomes in the metagenome. Technological advances have created new opportunities for the pursuit of large-scale sequencing projects that were difficult to imagine a decade ago. The key conceptual development was the emerging paradigm of the essential nature of complex microbial communities and their importance to mammalian biology and human health and disease. The Human Microbiome Project was approved in May 2007 as 1 of 2 major components (in addition to the human epigenomics program) of NIH RoadMap version 1.5 (now known as the Common Fund). Recently, 2 seminal reports from the Human Microbiome Project consortium (10, 11) described investigations in which a population of 242 healthy adults were sampled at 15 or 18 body sites up to 3 times, 5177 microbial taxonomic profiles were generated from 16S rRNA genes, and more than 3.5 T bases of metagenomic sequences were generated. In addition, in parallel, the Human Microbiome Project consortium has sequenced approximately 800 human-associated reference genomes. This resource will provide a framework for future studies of disease states and a reference collection of healthy human microbiome data. The data set will enable future investigations into the epidemiology and ecology of the human microbiome in various disease states, and treatment strategies will evolve from these studies. Using compositional and functional approaches, the relationships between pathological variations in the gut microbiome and several disease states have been delineated.
Urine metabolomics provides an opportunity for studies of the microbiome’s impact on whole-body metabolism. The advantages of using urinary samples include relatively large sample volumes and the convenience of noninvasive collection. In addition, urine samples can be used for the investigation of the chronology of metabolic changes and thus are a valuable tool for investigations related to the pathogenesis or progression of disease and for screening and diagnosis as well as prognostic evaluation. The methods commonly used for metabolic profiling of urine include procedures such as nuclear magnetic resonance (NMR) spectroscopy, LC-MS, GC-MS, and gas chromatography TOF mass spectrometry (GC-TOFMS). In a recent seminal report, the Nicholson group described a method for urine collection and storage that emphasizes the importance of midstream urine collection and the addition of urease before the freezing of urine samples. This method will eventually be used for metabolic profiling. Before analyses by GC-MS–based techniques, urease activity is terminated with ethanol or methanol and then derivatized by subjecting the sample to oximation followed by trimethylsilyl derivatization (12). Because of the various sample preparation steps, it is important to use biological QC samples and check the validity of the data that are obtained from GC-MS–based techniques that use principal component analysis. GC-MS–based metabolomic studies include several steps such as baseline correction, noise reduction, deconvolution, peak area calculation, and retention time alignment, and these steps help to generate consistent data. Several different commercially available software programs can assist in such correction strategies before data analyses. Also, in urinary metabolic profiling, it is important to normalize data (on the basis of volume, creatinine content, and other variables) to obtain meaningful information and minimize the influence of dilution. Using this protocol, investigators were able to undertake high-throughput metabolic profiling of approximately 400–600 metabolites in 120 urine samples per week. High-throughput analyses by NMR spectroscopy or MS are metabolic-profiling strategies that are widely used to provide global metabolic overviews of human metabolism (13–16). Coupled with computational multivariate analysis, these methods provide a deeper understanding of disease states and can lead to biomarker discovery. This approach facilitates the quantification of environmental influences on the host genome and human health. As part of large-scale clinical studies, this analytical strategy has been successfully applied to disease states such as hypertension (17), ischemic heart disease (18), diabetes (19), and obesity (20).
Metabonomics can be quite challenging because the chemical space associated with the endogenous metabolites is highly diversified, and thus complete metabolic information for any sample is hard to decipher completely. Common analytical technologies used in metabolomics and metabonomics include NMR spectroscopy, LC-MS, and GC-MS, as well as GC-TOFMS. These different analytical techniques have their own strengths and weaknesses and are usually used in an integrated fashion such that each of these analytical platforms can provide complementary data; the selection of particular analytical techniques depends on the study questions that are being posed. NMR has the advantage of being rapid, nondestructive to samples, and applicable to intact biomaterials rich in chemical structural information. NMR requires minimal sample preparation and can be used to investigate a mixture of or several different metabolites in a single sample. However, MS-based strategies have the advantages of increased sensitivity, accuracy, precision, and reproducibility compared to NMR. Furthermore, the coupling of GC to TOFMS offers several additional advantages such as reduced analysis time and greater accuracy with respect to peak deconvolution.
The Gut Microbiome: Beyond Composition to Function and Metabolism
The gut microbial community includes approximately 1014 bacteria that normally reside in the gastrointestinal tract, reaching a microbial cell number that greatly exceeds the number of human cells of the body. The collective genome of these microorganisms (the microbiome) contains millions of genes (a rapidly expanding number) compared to roughly 20 000–25 000 genes in the human genome. This microbial “factory” contributes to a broad range of biochemical and metabolic functions that the human body could not otherwise perform (21). Although diet-induced changes in gut microbiota occur within a short time frame (1–3–4 days after a diet switch), the changes are readily reversible (22, 23). In animal models, the ratio of the most prominent intestinal bacterial phyla, the Bacteroidetes and Firmicutes, is altered in response to dietary changes (22, 23). Disruption of the energy equilibrium leads to weight gain. Mouse model studies have demonstrated the relationship between energy equilibrium, diet, and the composition of the gut microbiome. Transplantation of the gut microbiota from obese donors resulted in increased adiposity in recipients compared to a similar transfer from lean donors.
Recent evidence suggests that the gut microbiota affect nutrient acquisition, energy harvest, and a myriad of host metabolic pathways (24). Recent findings raise the possibility that the gut microbiota has an important role in regulating weight and may be partly responsible for the development of obesity. Initial evidence of the relationship between obesity and gut microbial composition was reported 3 decades ago, when surgically induced weight loss through gastric bypass surgery and weight gain through lesions of the ventromedial hypothalamic nucleus were found to be associated with changes in gut microbial ecology (25, 26). These earlier studies used culture-dependent methods, which detect a minority of microbes harbored in the gut. In recent years, the ability to obtain a thorough picture of gut microbial communities has improved by the introduction of molecular, culture-independent techniques based on ribosomal 16S rRNA gene sequencing. Jumpertz et al. (27) performed an inpatient energy balance study in 12 lean and 9 obese individuals as they consumed 2 calorically distinct diets for brief periods of time, and these investigators simultaneously monitored the gut microbiota by performing pyrosequencing studies of bacterial 16S rRNA genes present in feces and by measuring ingested and stool calories by bomb calorimetry. This study showed that altered nutrient load (i.e., high calories vs low calories) induced rapid changes in the bacterial composition of the human gut microbiota, and these changes correlated well with stool energy loss in lean individuals. Increased proportions of Firmicutes and corresponding reductions in Bacteroidetes taxa were associated with an increased energy harvest of approximately 150 kcal. These data point to a strong link between gut microbiome composition and nutrient absorption in humans, and such studies need to be confirmed with larger numbers of study participants.
The gut microbiome is very important in maintaining both gastrointestinal and immune function as well as being crucial for the digestion of nutrients, and this notion has been confirmed by studies of germ-free mice (28–30). Important metabolic functions of the gut microbiome include the catabolism of dietary toxins and carcinogens, synthesis of micronutrients, fermentation of indigestible food substances, and assisting in the absorption of electrolytes and minerals. In addition, the production of short-chain fatty acids (SCFAs) by the gut microbiome affects growth and differentiation of enterocytes and colonocytes. Differences in the metabolic activities of the gut microbiome may contribute to variation in caloric extraction from ingested dietary substances, storage of calories in adipose tissue, and energy availability for microbial proliferation. Such differences in the gut microbiome are also responsible for the variation in the ability of an individual’s capacity to harvest energy, which may explain aspects of obesity. Differences in gut microbial composition and its metabolic efficiency may be responsible for the predisposition of an individual to metabolic disorders such as obesity and diabetes (31).
The gut microbiome can affect whole-body metabolism and alter physiological parameters in multiple body compartments (32). In one study (33), gnotobiotic mice had increased quantities of phosphocholine and glycine in the liver and increased quantities of bile acids in the intestine. The gut microbiome also influences kidney homeostasis by modulating quantities of key cell-volume regulators such as betaine and choline (33). A more recent study showed specific differences in the patterns of bile acids present and reduced overall bile acid diversity in germ-free vs conventional rats (34). Compared to conventional rats, germ-free rats have increased concentrations of conjugated bile acids that can accumulate in the liver and the heart.
The Gut Microbiome and Carbohydrate Metabolism
Carbohydrates are an important nutritional component for mammals and the mammalian microbiome, including the gut microbiota. Mammals absorb simple sugars, including galactose and glucose, in the proximal jejunum via specific sugar transporters. Mammalian enzymes hydrolyze disaccharides (sucrose, lactose, maltose) and starches to constituent monosaccharides, but have limited abilities to hydrolyze other polysaccharides. As a consequence, every day a bulk of undigested plant polysaccharides (cellulose, xylan, and pectin) and partially digested starch reaches microbial communities in the distal gut. By hosting metabolically active microbiota capable of hydrolyzing complex carbohydrates, mammals avoid the need to evolve complex enzymes that are required to break down the variety of polysaccharides in the diet. Microbes, by contrast, contain many genes encoding a variety of carbohydrate-active enzymes (CAZymes) in the human microbiome (35). Microbial CAzymes that constitute the mammalian host repertoire include glycoside hydrolases, carbohydrate esterases, glycosyl transferases, and polysaccharide lyases (35). Microbes gain access to abundant readily fermentable carbon sources that would otherwise be wasted by the host and may use these complex carbohydrate substrates to sustain viable, functionally robust microbial communities and generate bioactive signals that affect mammalian metabolism.
Intestinal bacterial taxa differ with respect to their abilities to utilize dietary and host-derived carbohydrates (e.g., mucus components) (23, 36). Bacteriodetes (23, 36) also have been demonstrated to easily assimilate dietary carbohydrates, because members of this bacterial phylum possess several carbohydrate utilization pathways. However, in situations of dietary carbohydrate starvation, gut bacteria catabolize mucins in the gastrointestinal tract as a carbohydrate source, thereby potentially compromising the mucus layer adjacent to the epithelium. In addition to Bacteroides, strains of the genus Bifidobacterium contain genes encoding glycan-foraging enzymes that enable these gut bacteria to acquire nutrients from host-derived glycans (37). Besides their capacity to hydrolyze starch, gut microbes have developed the ability to degrade numerous plant and host-derived glycoconjugates (glycans) and glycosaminoglycans including cellulose, chondroitin sulfate, hyaluronic acid, mucins, and heparin. Microbial catabolic enzymes such as endoglycosidases may act on dietary substrates to release complex N-glycans from human milk and other dairy sources (38). Fluctuations in diet may have functional consequences for bacteria and the host so that the “cannibalization” of indigenous mammalian carbohydrates may result in augmentation of beneficial features, prevention of diseases, or predisposition to different disease states. For example, bifidobacteria grown on human milk oligosaccharides stabilize tight junction formation in the epithelium and promote the secretion of the antiinflammatory cytokine, interleukin-10 (39). The biogeography of the microbiome may be relevant because specific genes/pathways such as simple carbohydrate transport phosphotransferase systems are more prominent in the small intestine than in the colon (40). Probing into pathways which are affected by alterations in the gut microbiome (e.g., carbohydrate storage and utilization) will yield new knowledge on the role of human-associated microbes in the development of several metabolic disorders in humans.
The Gut Microbiome and Fatty Acid Metabolism
Intestinal bacteria including probiotics produce a diverse array of fatty acids that may have health-promoting effects. Intestinal bifidobacteria produce conjugated linoleic acid (CLA), and CLA appears to modulate the fatty acid composition in the liver and adipose tissue in murine models (41). In addition to conjugated and free fatty acids, intestinal bacteria generate SCFAs (i.e., acetate, butyrate, propionate) by fermenting dietary carbohydrates (fiber) that humans cannot digest themselves. A recent study showed that germ-free mice are devoid of SCFAs, indicating the importance of the gut microbiota for SCFA production in the intestine (42). Acetate is the dominant SCFA type in humans, and this SCFA appears to play an intriguing role in the modulation of 5′AMP-activated protein kinase activity and macrophage infiltration in adipose tissue (43). SCFAs such as propionate can be used for de novo glucose or lipid synthesis and serve as an energy source for the host.
SCFAs may function as microbe-derived signals that influence carbohydrate metabolism and gut physiology by stimulating mammalian peptide secretion and serving as energy sources for gut epithelial cells. SCFAs can stimulate glucagon-like peptide 1 (GLP-1) secretion via the G-protein–coupled receptor FFAR2 (free fatty acid receptor 2) in the colonic mucosa (44). By stimulating GLP-1 secretion, bacterial SCFAs provide signals that suppress glucagon secretion, induce glucose-dependent insulin secretion, and promote glucose homeostasis. An enteroendocrinological pathway is proposed in which SCFAs stimulate the secretion of peptide YY, a hormone that is released by ileal and colonic epithelial cells in response to feeding and seems to suppress the appetite (45). High-fat diets supplemented with butyrate prevented and reversed insulin resistance in dietary-obese mice. At the same time, butyrate-producing bacteria and fecal butyrate concentrations decline with diets containing reduced amounts of specific carbohydrates (46). The SCFA propionate modulates energy homeostasis by promoting GPR41 (G protein-coupled receptor 41)-mediated activation of sympathetic neurons, in contrast to ketone bodies (47). The ability to modulate sympathetic outflow provides another mechanism linking the gut microbiome to the enteric nervous system, energy expenditure, and metabolic homeostasis.
Roux-en-Y gastric bypass (RYGB) surgery is a major bariatric intervention to treat morbid obesity. Before surgery, increased quantities of Bacteroidetes were observed, but reductions in Bacteroidetes and enhanced quantities of Proteobacteria were detected following surgery (48). These microbial population shifts likely change the metabolite profiles and relative preponderance of different fatty acids, including SCFAs. These results are supported by a recent animal study. Nonobese rats with RYGB had decreased amounts of Firmicutes and Bacteroidetes and significantly increased amounts (52-fold higher concentrations) of Proteobacteria compared with sham-operated rats (49). Obesity is a proinflammatory state. It was shown that the abundance of butyrate-producing Faecalibacterium prausnitzii species is negatively associated with biomarkers of inflammation before and after RYGB, indicating that this bacterial species may contribute to maintaining a healthy gut (48). Thus, surgical interventions in the gastrointestinal tract may have profound effects on gut microbial composition, SCFA production, and the mammalian immune system.
Interestingly, a recent study (50) has demonstrated that subtherapeutic administration of antibiotics alters the population structure of the gut microbiome as well as its metabolic capabilities. In this study, investigators administered subtherapeutic doses of antibiotics to young mice, resulting in increased adiposity in young mice and increased levels of the incretin GIP-1. In addition, these investigators observed substantial taxonomic changes in the microbiome (increased Lachnospiraceae and Firmicutes and decreased Bacteroidetes), changes in key genes involved in the metabolism of carbohydrates to SCFAs (increased levels of acetate, propionate, and butyrate), increases in colonic SCFA levels, and alterations in the regulation of hepatic metabolism of lipids and cholesterol. Thus, modulation of murine metabolic homeostasis can be achieved by altering the gut microbiota through antibiotic manipulation.
The Gut Microbiome and Amino Acid Metabolism
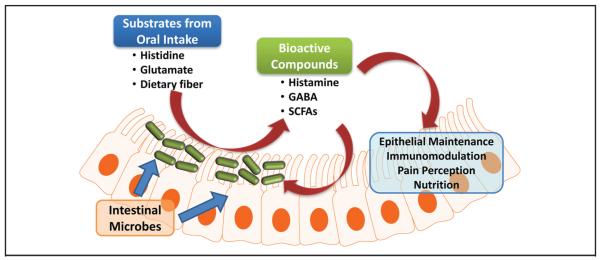
Beneficial microbes such as bifidobacteria and lactobacilli produce biologically active compounds derived from amino acids, including a variety of biogenic amines. Dietary components include proteins and peptides that may be hydrolyzed to amino acids by luminal proteinases and peptidases. Amino acids derived from dietary protein sources may serve as substrates for luminal bioconversion by the gut microbiome. Diverse microbial enzymes may contribute to mammalian amino acid metabolism by generating bioactive metabolites in the intestine. One such class of enzymes, amino acid decarboxylases, is widely prevalent in gut microbes, and these microbial enzymes, when combined with amino acid transport systems, link dietary compounds with microbial metabolism and signaling with the gut mucosa (Fig. 2).
Fig. 2. Intestinal microbes may play an important role in host-microbiota interactions via luminal conversion.
Nutrients consumed by the host may be converted by intestinal microbes into several bioactive compounds that could affect the health and disease states of the host and the intestinal microbiota. SCFAs = short-chain fatty acids. Reproduced with permission from Hemarajata et al. (78).
Combinations of metabolomics strategies, including MS, HPLC, and NMR, are leading to discoveries of metabolites and small compounds derived from the human microbiome. With the use of hydrophilic interaction liquid chromatography–HPLC (HILIC-HPLC), bioactive molecules derived from gut microbes were isolated as antiinflammatory, tumor necrosis factor (TNF)-inhibitory compounds and HILIC-HPLC fractions were analyzed by MS and NMR. One such microbial signal and biogenic amine, histamine, was identified and quantified in TNF-inhibitory HILICHPLC fractions derived from Lactobacillus reuteri found in breast milk and the gut (51). Histamine is produced from L-histidine via histidine decarboxylase, which is present in some fermentative bacteria including probiotic lactobacilli. One constituent of the gut microbiome, L. reuteri, is able to convert a dietary component, L-histidine, into an immunoregulatory signal, histamine, which suppresses proinflammatory TNF production via histamine type 2 receptors in the intestinal epithelium. Other examples of microbe-facilitated amino acid metabolism include the generation of γ-amino butyric acid (GABA) from glutamate via glutamate decarboxylase (52) and the production of putrescine from ornithine. The identification of these bacterial bioactive metabolites and their corresponding mechanisms of action with respect to immunomodulation may lead to improved antiinflammatory strategies for chronic immune-mediated diseases. Such antiinflammatory amino acid metabolites may ameliorate pathologic processes in obesity and diabetes.
The Gut Microbiome and Body Metabolism: Obesity and Inflammation
The incidence of overweight and obesity has reached epidemic proportions. Data reported by the CDC and the National Health and Nutrition Examination Survey indicated that, in 2008, an estimated 1.5 billion adults were overweight, and more than 200 million men and almost 300 million women were obese by these criteria. Worldwide obesity has more than doubled in the last 2 decades. Obesity is associated with a cluster of metabolic and systemic disorders such as insulin resistance, type 2 diabetes, fatty liver disease, atherosclerosis, and hypertension. The major cause of obesity is a positive energetic balance resulting from an increased energy intake from the diet and a decreased energy output associated with low physical activity. In addition to alterations in diet and physical activity resulting in obesity, genetic differences contribute to obesity and cause differences in energy storage and expenditure. Furthermore, growing evidence suggests that the gut microbiota represents an important factor contributing to the host response to nutrients. A landmark study by Turnbaugh et al. (53) was one of the first studies to show how the gene content in the gut microbiota contributes to obesity. The microbiomes obtained from the distal gut of genetically obese leptin-deficient mice (ob/ob) and their lean littermates (ob/+ and +/+) were compared. In this study, investigators reported that the microbiota in the ob/ob mice contained genes encoding enzymes that hydrolyze indigestible dietary polysaccharides. Increased amounts of fermentation end products (such as acetate and butyrate) and decreased calories were found in the feces of obese mice. These data suggest that the gut microbiota in this mouse model promoted the extraction of additional calories from the diet.
The composition of the gut microbiome seems to be important in regulating body weight (54). To demonstrate this point, investigators conducted experiments in which they transplanted the gut microbiota of either ob/ob mice or lean mice to lean gnotobiotic mice. After 2 weeks, mice that received microbiota from the ob/ob mice were able to extract more calories from food and also showed a significantly greater fat gain than mice that received the microbiota from lean mice. Thus, differences in caloric extraction of ingested food substances may be largely a result of the composition of the gut microbiota. These data support a pivotal role for the gut microbiome in the pathogenesis of obesity and obesity-related disorders.
Manipulation of the gut microbiota may be an important therapeutic strategy to regulate energy balance in individuals who are obese, diabetic, or have a diagnosis of metabolic syndrome. In genetically obese, ob/ob mice and their lean counterparts fed the same polysaccharide-rich diet, Ley et al. analyzed bacterial 16S rRNA gene sequences from the cecal microbiota and reported that ob/ob mice had 50% fewer Bacteroidetes and correspondingly more Firmicutes than their lean littermates and this difference was unrelated to differences in food consumption.
Backhed et al. (55) confirmed these findings and found that young, conventionally reared mice had a 40% higher body fat content and 47% higher gonadal fat content than germ-free mice, although their food consumption was less than their germ-free counterparts. When the distal gut microbiota from young, conventionally-reared mice were transplanted into the gnotobiotic mice, a 60% increase in body fat within 2 weeks was observed, without any increase in food consumption or energy expenditure. This increase in body fat was accompanied by increased insulin resistance, adipocyte hypertrophy, and increased concentrations of circulating leptin and glucose. Mechanistic studies demonstrated that the microbiota promoted absorption of monosaccharides from the gut and induced hepatic lipogenesis in the host. These responses were largely mediated via upregulation of 2 signaling proteins, ChREBP (carbohydrate response element-binding protein) and liver SREBP-1 (sterol response element-binding protein type-1). In addition, with genetically modified fasting-induced adipocyte factor (Fiaf)-knockout mice, gut microbes were also shown to suppress intestinal Fiaf (56).
Several studies have highlighted the pivotal role of inflammation in the metabolic processes leading to the metabolic syndrome, obesity, and diabetes. Cani et al. (57–59) postulated another mechanism linking the intestinal microbiota to the development of obesity. The authors hypothesized that bacterial lipopolysaccharide (LPS) derived from gram-negative bacteria residing in the gut microbiota may be the trigger for increased inflammation observed in high-fat diet–induced metabolic syndrome. In a series of experiments in mice fed a high-fat diet, the investigators showed evidence of pronounced endotoxemia, associated with reductions in both gram-negative (Bacteroides-related bacteria) and gram-positive bacteria (Eubacterium rectale—Clostridium coccoides group and bifidobacteria), and an increased ratio of gram-negative to gram-positive bacteria. The authors of this report suggested that chronic metabolic endotoxemia induces obesity, insulin resistance, and diabetes.
In human experiments, Ley et al. (60) and Ravussin et al. (61) serially monitored the fecal gut microbiota in 12 obese individuals who participated in a weight-loss program for a year by following either a fat-restricted or carbohydrate-restricted low-calorie diet. Similar to experiments in mice, in humans a relative abundance of microbiota that belonged to the Bacteroidetes and Firmicutes phyla was found, and the microbiota showed remarkable intraindividual stability over time. Before the initiation of the low-calorie diet, a relative abundance of Firmicutes and decreased amounts of Bacteroidetes were detected in the obese participants compared with the nonobese controls. After weight loss, increased amounts of Bacteroidetes (3% to 15%) and a decreased abundance of Firmicutes were observed, and these changes correlated with the percentage of weight loss and not with changes in dietary caloric content. These human studies confirmed animal data suggesting that alterations in gut microbial composition are associated with obesity. Cause and effect relationships between obesity and changes in the gut microbiota remain unclear. Kalliomäki et al., in a prospective study of children from birth to age 7 years (62), collected fecal specimens at 6 and 12 months of age. This report documented an abundance of Bifidobacterium taxa and decreased proportions of Staphylococcus aureus in children who were whose weight was within reference intervals at age 7 years than in those who were overweight or obese. Although they did not examine factors such as diet and physical activity, these data suggest that differences in the composition of the gut microbiota precede overweight and obesity status. Antibiotics have also been shown to pervasively affect gut microbial composition. A 5-day course of orally administered ciprofloxacin decreased substantially the diversity of the fecal microbial community (63). In this study, although most of the microbial community revived within 4 weeks after administration of ciprofloxacin, some other genera failed to reappear even after treatment with the antibiotic for 6 months (63).
The Gut Microbiome and Metabolism: Diabetes and the Metabolic Syndrome
Toll like receptors (TLRs) are pattern recognition receptors that are important in mediating inflammation and immunity. Increased amounts of TLRs are present on cell surfaces in patients with obesity, diabetes, and metabolic syndrome (63). Recently, investigators have explored the role of the gut microbiome in regulating TLR-mediated insulin resistance. Mice deficient in the microbial pattern-recognition receptor TLR5 displayed hyperphagia, became obese, and developed features of the metabolic syndrome, including hypertension, hypercholesterolemia, and insulin resistance secondary to dysregulation of interleukin-1β signaling (43). When gut microbiomes from these mice were transplanted into germ-free mice with an intact toll-like receptor 5 (TLR5) gene, recipient mice developed similar features of the metabolic syndrome, which suggests that the intestinal microbiome was the key determinant of this disease phenotype. In another study, TLR2-deficient mice developed obesity, insulin resistance, and glucose intolerance, and the gut microbiomes of TLR2-deficient mice had a greater abundance of Firmicutes and fewer Actinobacteria of the genus Bifidobacterium (64). Administration of an antibiotic cocktail eliminated many of the Firmicutes and resulted in improved insulin activity and glucose tolerance. In addition to improved insulin activity and glucose tolerance, because lower levels of Bifidobacterium spp. contribute to increased gut permeability, this change in the gut microbiome may result in a leaky gut and yield increased concentrations of endotoxins such as LPS in the circulation. The immune system recognizes LPS as a microbial pattern triggering TLR signaling and causing inflammation. Both obesity and inflammation tend to cause diabetes, so the loss of TLR2 in these mice leads to changes in their gut bacteria, which result in a greater risk of diabetes mellitus. Indeed, increased amounts of TLR2 have been observed on monocytes of patients with metabolic syndrome, type 1 and type 2 diabetes compared with matched controls (65–72). TLR2 deficiency in diabetic mice results in decreased development of complications of diabetes such as diabetic nephropathy (68). Possibly, the gut microbiome plays a crucial role in regulating diabetic vasculopathies, and this area will be an important area of future investigation.
With regard to the role of the microbiome in metabolic syndrome and associated abnormalities, studies in germ-free mice demonstrate that they are protected from obesity, insulin resistance, dyslipidemia, and fatty liver disease/nonalcoholic steatohepatitis when fed a high-fat Western diet (69). In contrast, following the colonization with microbiota from conventionally raised mice, the body fat content in the originally germ-free mice increased up to 60% in 14 days. This was associated with increased insulin resistance, although the food intake was reduced. The metabolic syndrome affects 1 in 3 US adults and leads to an increased propensity of diabetes and cardiovascular disease (70). In a single human study in patients with the metabolic syndrome, Zupancic et al. (71) studied Amish men and women with varying body mass indices. In 310 study participants, gut microbiota were characterized by deep pyrosequencing of bar-coded PCR amplicons from the V1–V3 region of the 16S rRNA gene. They were able to identify 3 communities of interacting bacteria in the gut microbiota, analogous to previously identified gut enterotypes. Network analysis identified 22 bacterial species and 4 operational taxonomic units that were either positively or inversely correlated with metabolic syndrome traits, suggesting that certain members of the gut microbiota can contribute to the metabolic syndrome. It is important that future studies focus on delineation of specific components of the gut microbiome that contribute to visceral obesity, dysglycemia, dyslipidemia, hypertension, and insulin resistance associated with this population (71). Nonalcoholic fatty liver disease is the hepatic manifestation of metabolic syndrome and the leading cause of chronic liver disease in the Western world. Using different mouse models of inflammasome deficiency, such as mice deficient in Asc (apoptosis-associated speck-like protein containing a caspase recruitment domain), NLRP3 (nucleotide-binding domain, leucine rich family, pyrin containing 3), caspase, or interleukin 18, the authors showed significant alterations in gut microbiota as evidenced by increased members of Bacteriodetes and decreased members of Firmicutes in these mouse models. More severe hepatic steatosis and inflammation were found, as evidenced by increased TLRs (mainly TLR 4 and 9) and secretion of hepatic TNF-α. Importantly, the authors speculated that increased hepatic steatosis was due to intestinal bacterial products acting as agonists for TLR4 and 9 and entering the liver via the portal circulation (72). Furthermore, these pathologic changes result in exacerbation of hepatic steatosis and obesity (72). Thus, altered interactions between the gut microbiota and the host, produced by defective inflammasome sensing, may govern the rate of progression of multiple abnormalities associated with metabolic syndrome.
Intestinal microbiomes have also been studied in relation to insulin resistance in patients with type 2 diabetes. Larsen et al. reported that there was a significant reduction in the relative abundance of Firmicutes and Clostridia in adults with type 2 diabetes when they used the technique of deep tag-encoded sequencing (73). Additionally, the ratios of Bacteroidetes to Firmicutes and Bacteroides–Prevotella to C. coccoides–Eubacterium rectale groups were correlated with increased fasting glucose levels in these patients. In this study, Larsen et al. showed that in addition to the decreased abundance of Firmicutes, the Betaproteobacteria levels were significantly increased in diabetic patients compared to non-diabetic controls and their abundance significantly correlated with their plasma glucose concentrations (r = 0.46, P < 0.05) (73). Such findings are intriguing and prompt questions regarding how microbial composition and corresponding metabolites may influence whole-body metabolism in humans and contribute to insulin resistance and diabetes.
Interestingly, the gut microbiome also regulates type 1 diabetes. Type 1 diabetes is an autoimmune disease, which is due to the specific destruction of the endocrine insulin-secreting pancreatic β cells resulting in an insulinopenic state. Type 1 diabetes also predisposes to microvascular and macrovascular complications. Data have emerged on the critical role of the gastrointestinal microbiota in the protection or the triggering of type 1 diabetes (74). In 2 models of diabetes, the NOD (nonobese diabetic) mouse and the biobreeding (BB) rat, the incidence of spontaneous type 1 diabetes mellitus can be affected by the microbial environment in the animal-housing facility or by exposure to microbial stimuli (75). Furthermore, the recognition of bacterial determinants from intestinal microbiota may trigger type 1 diabetes. TLRs are innate pattern-recognition receptors involved in host defense and maintenance of tissue integrity. TLR signaling is mediated through the adapter protein MyD88, and the deletion of MyD88 protects from atherosclerosis. Mice lacking MyD88 were protected against insulitis (74), and this phenomenon depends on commensal microbes because germ-free MyD88 knockout mice develop robust diabetes.
In BB rats (a model of type 1 diabetes), Lactobacillus species present in feces (L. johnsonii and L. reuteri) were negatively correlated with type 1 diabetes development (76), possibly via modulation of the intestinal mucosal protein and oxidative stress response leading to lower quantities of proinflammatory cytokines such as interferon γ. Thus, therapeutic modulation aimed at altering the gut microbiome may be beneficial in retarding the development of diabetes.
Alterations in the gut microbiota contribute to the development of autoimmune disorders such as type 1 diabetes. Fecal samples were obtained from 4 pairs of matched participants in a case control study (77). More than 30 billion nucleotide bases of Illumina shotgun metagenomic data were analyzed, and the findings revealed significantly increased proportions of pathways and modules involved in carbohydrate metabolism and stress responses in cases compared to controls. Other differences included the relative quantities of genes involved in adhesion, motility, and sulfur metabolism, which were more abundant in cases, whereas genes with roles in DNA and protein metabolism, amino acid synthesis, and aerobic respiration were more abundant in controls. The 16S rRNA data were also mined for indications of changes in microbial composition. At the phylum level, numbers of Actinobacteria, Bacteroidetes, and Proteobacteria were significantly increased in cases, whereas Firmicutes, Fusobacteria, Tenericutes, and Verrucomicrobia were higher in controls (P < 0.001). At the genus level, the abundance of Bacteroides was much greater in cases, whereas Prevotella was much more abundant in controls. Furthermore, the total number of bacteria that produce lactic acid and butyrate was greater in controls than in cases. Thus, these data suggest that such lactateand butyrate-producing bacteria may be beneficial and maintain a healthy gut and that dysregulation of these bacteria can lead to reduction in optimal mucin synthesis, as identified in individuals with autoimmune diseases, and contribute to development of type 1 diabetes. These exciting findings need to be confirmed in larger study populations.
In conclusion, randomized clinical studies should help define the features of the gut microbiome that contribute to obesity and diabetes epidemics in defined populations. In addition, mechanistic studies of the human microbiome will be instructive and have therapeutic implications. Furthermore, advances in technology, such as 16S rRNA sequencing, WG metagenomics, and metabolomics in metabolic diseases will enable scientists to mine large data sets for disease-contributing features. Large databases and bioinformatics resources such as those derived from the Human Microbiome Project will lead the way toward a greater understanding of the importance and role of the gut microbiome in metabolic disorders such as obesity, metabolic syndrome, and diabetes, and these studies may provide therapeutic strategies to reduce the aggregate cardiometabolic burden in human populations.
Acknowledgments
Research Funding: NIH P30 DK56338-06A2; J. Versalovic, NIH UH3 DK083990, NIH R01 AT004326-01A1, and NIH R01 DK065075-01.
Footnotes
Author Contributions: All authors confirmed they have contributed to the intellectual content of this paper and have met the following 3 requirements: (a) significant contributions to the conception and design, acquisition of data, or analysis and interpretation of data; (b) drafting or revising the article for intellectual content; and (c) final approval of the published article.
Authors’ Disclosures or Potential Conflicts of Interest: Upon manuscript submission, all authors completed the author disclosure form. Disclosures and/or potential conflicts of interest:
Employment or Leadership: None declared.
Consultant or Advisory Role: None declared.
Stock Ownership: None declared.
Honoraria: None declared.
Expert Testimony: None declared.
Patents: None declared.
Nonstandard abbreviations: WG, whole genome; NMR, nuclear magnetic resonance; GC-TOFMS, gas chromatography TOF mass spectrometry; SCFA, short-chain fatty acid; CAZymes, carbohydrate-active enzymes; CLA, conjugated linoleic acid; GLP-1, glucagon-like peptide 1; RYGB, Roux-en-Y gastric bypass; HILIC-HPLC, hydrophilic interaction liquid chromatography–HPLC; TNF, tumor necrosis factor; GABA, γ-amino butyric acid; Fiaf, fasting-induced adipocyte factor; LPS, lipopolysaccharide; TLR, Toll-like receptor; BB, biobreeding.
References
- 1.Cowie CC, Rust KF, Ford ES, Eberhardt MS, Byrd-Holt DD, Li C, et al. Full accounting of diabetes and pre-diabetes in the U.S. population in 1988-1994 and 2005-2006. Diabetes Care. 2009;32:287–94. doi: 10.2337/dc08-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.D’Adamo E, Santoro N, Caprio S. Metabolic syndrome in pediatrics: old concepts revised, new concepts discussed. Pediatr Clin North Am. 2011;58:1241–55. xi. doi: 10.1016/j.pcl.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 3.Backhed F. Host responses to the human microbiome. Nutr Rev. 2012;70(Suppl 1):S147. doi: 10.1111/j.1753-4887.2012.00496.x. [DOI] [PubMed] [Google Scholar]
- 4.Tehrani AB, Nezami BG, Gewirtz A, Srinivasan S. Obesity and its associated disease: a role for microbiota? Neurogastroenterol Motil. 2012;24:305–11. doi: 10.1111/j.1365-2982.2012.01895.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harris K, Kassis A, Major G, Chou CJ. Is the gut microbiota a new factor contributing to obesity and its metabolic disorders? J Obes. 2012;2012:879151. doi: 10.1155/2012/879151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kibe R, Sakamoto M, Hayashi H, Yokota H, Benno Y. Maturation of the murine cecal microbiota as revealed by terminal restriction fragment length polymorphism and 16S rRNA gene clone libraries. FEMS Microbiol Lett. 2004;235:139–46. doi: 10.1016/j.femsle.2004.04.023. [DOI] [PubMed] [Google Scholar]
- 7.Turnbaugh PJ, Gordon JI. The core gut microbiome, energy balance and obesity. J Physiol. 2009;587:4153–8. doi: 10.1113/jphysiol.2009.174136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A. 2010;108(Suppl 1):4516–22. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–4. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–14. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.The Human Microbiome Project Consortium A framework for human microbiome research. Nature. 2012;486:215–21. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan EC, Pasikanti KK, Nicholson JK. Global urinary metabolic profiling procedures using gas chromatography-mass spectrometry. Nat Protoc. 2011;6:1483–99. doi: 10.1038/nprot.2011.375. [DOI] [PubMed] [Google Scholar]
- 13.Verberkmoes NC, Russell AL, Shah M, Godzik A, Rosenquist M, Halfvarson J, et al. Shotgun metaproteomics of the human distal gut microbiota. ISME J. 2009;3:179–89. doi: 10.1038/ismej.2008.108. [DOI] [PubMed] [Google Scholar]
- 14.Nicholson JK, Lindon JC, Holmes E. ‘Metabonomics’: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999;29:1181–9. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- 15.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A. 2009;106:3698–703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nicholson JK, Connelly J, Lindon JC, Holmes E. Metabonomics: a platform for studying drug toxicity and gene function. Nat Rev Drug Discov. 2002;1:153–61. doi: 10.1038/nrd728. [DOI] [PubMed] [Google Scholar]
- 17.Holmes E, Loo RL, Stamler J, Bictash M, Yap IK, Chan Q, et al. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature. 2008;453:396–400. doi: 10.1038/nature06882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brindle JT, Antti H, Holmes E, Tranter G, Nicholson JK, Bethell HW, et al. Rapid and noninvasive diagnosis of the presence and severity of coronary heart disease using 1H-NMR-based metabonomics. Nat Med. 2002;8:1439–44. doi: 10.1038/nm1202-802. [DOI] [PubMed] [Google Scholar]
- 19.Dumas ME, Wilder SP, Bihoreau MT, Barton RH, Fearnside JF, Argoud K, et al. Direct quantitative trait locus mapping of mammalian metabolic phenotypes in diabetic and normoglycemic rat models. Nat Genet. 2007;39:666–72. doi: 10.1038/ng2026. [DOI] [PubMed] [Google Scholar]
- 20.Stella C, Beckwith-Hall B, Cloarec O, Holmes E, Lindon JC, Powell J, et al. Susceptibility of human metabolic phenotypes to dietary modulation. J Proteome Res. 2006;5:2780–8. doi: 10.1021/pr060265y. [DOI] [PubMed] [Google Scholar]
- 21.Neish AS. Microbes in gastrointestinal health and disease. Gastroenterology. 2009;136:65–80. doi: 10.1053/j.gastro.2008.10.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science. 2001;292:1115–8. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 23.Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN, Sonnenburg JL. Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell. 2010;141:1241–52. doi: 10.1016/j.cell.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–7. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 25.Clement K. Bariatric surgery, adipose tissue and gut microbiota. Int J Obes. 2011;35(Suppl 3):S7–15. doi: 10.1038/ijo.2011.141. [DOI] [PubMed] [Google Scholar]
- 26.Calvani R, Miccheli A, Capuani G, Tomassini Miccheli A, Puccetti C, Delfini M, et al. Gut microbiome-derived metabolites characterize a peculiar obese urinary metabotype. Int J Obes. 2010;34:1095–8. doi: 10.1038/ijo.2010.44. [DOI] [PubMed] [Google Scholar]
- 27.Jumpertz R, Le DS, Turnbaugh PJ, Trinidad C, Bogardus C, Gordon JI, Krakoff J. Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. Am J Clin Nutr. 2011;94:58–65. doi: 10.3945/ajcn.110.010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gravitz L. Microbiome: the critters within. Nature. 2012;485:S12–3. doi: 10.1038/485s12a. [DOI] [PubMed] [Google Scholar]
- 29.Tilg H, Kaser A. Gut microbiome, obesity, and metabolic dysfunction. J Clin Invest. 2011;121:2126–32. doi: 10.1172/JCI58109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Faith JJ, McNulty NP, Rey FE, Gordon JI. Predicting a human gut microbiota’s response to diet in gnotobiotic mice. Science. 2011;333:101–4. doi: 10.1126/science.1206025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kallus SJ, Brandt LJ. The intestinal microbiota and obesity. J Clin Gastroenterol. 2011;46:16–24. doi: 10.1097/MCG.0b013e31823711fd. [DOI] [PubMed] [Google Scholar]
- 32.Zhao L, Shen J. Whole-body systems approaches for gut microbiota-targeted, preventive healthcare. J Biotechnol. 2010;149:183–90. doi: 10.1016/j.jbiotec.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 33.Claus SP, Tsang TM, Wang Y, Cloarec O, Skordi E, Martin FP, et al. Systemic multicompartmental effects of the gut microbiome on mouse metabolic phenotypes. Mol Syst Biol. 2008;4:219. doi: 10.1038/msb.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swann JR, Want EJ, Geier FM, Spagou K, Wilson ID, Sidaway JE, et al. Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. Proc Natl Acad Sci U S A. 2010;108(Suppl 1):4523–30. doi: 10.1073/pnas.1006734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cantarel BL, Lombard V, Henrissat B. Complex carbohydrate utilization by the healthy human microbiome. PLoS One. 2012;7:e28742. doi: 10.1371/journal.pone.0028742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–9. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 37.Turroni F, Bottacini F, Foroni E, Mulder I, Kim JH, Zomer A, et al. Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc Natl Acad Sci U S A. 2010;107:19514–9. doi: 10.1073/pnas.1011100107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garrido D, Nwosu C, Ruiz-Moyano S, Aldredge D, German JB, Lebrilla CB, Mills DA. Endo-beta-N-acetylglucosaminidases from infant-gut associated bifidobacteria release complex N-glycans from human milk glycoproteins. Mol Cell Proteomics. 2012;11:775–85. doi: 10.1074/mcp.M112.018119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chichlowski M, De Lartigue G, German JB, Raybould HE, Mills DA. Bifidobacteria isolated from infants and cultured on human milk oligosaccharides affect intestinal epithelial function. J Pediatr Gastroenterol Nutr. 2012;55:321–7. doi: 10.1097/MPG.0b013e31824fb899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zoetendal EG, Raes J, van den Bogert B, Arumugam M, Booijink CC, Troost FJ, et al. The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. ISME J. 2012;6:1415–26. doi: 10.1038/ismej.2011.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O’Shea EF, Cotter PD, Stanton C, Ross RP, Hill C. Production of bioactive substances by intestinal bacteria as a basis for explaining probiotic mechanisms: bacteriocins and conjugated linoleic acid. Int J Food Microbiol. 2012;152:189–205. doi: 10.1016/j.ijfoodmicro.2011.05.025. [DOI] [PubMed] [Google Scholar]
- 42.Martin FP, Wang Y, Sprenger N, Yap IK, Lundstedt T, Lek P, et al. Probiotic modulation of symbiotic gut microbial-host metabolic interactions in a humanized microbiome mouse model. Mol Syst Biol. 2008;4:157. doi: 10.1038/msb4100190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carvalho BM, Guadagnini D, Tsukumo DM, Schenka AA, Latuf-Filho P, Vassallo J, et al. Modulation of gut microbiota by antibiotics improves insulin signalling in high-fat fed mice. Diabetologia. 2012;55:2823–34. doi: 10.1007/s00125-012-2648-4. [DOI] [PubMed] [Google Scholar]
- 44.Tolhurst G, Heffron H, Lam YS, Parker HE, Habib AM, Diakogiannaki E, et al. Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein-coupled receptor FFAR2. Diabetes. 2012;61:364–71. doi: 10.2337/db11-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou J, Hegsted M, McCutcheon KL, Keenan MJ, Xi X, Raggio AM, Martin RJ. Peptide YY and proglucagon mRNA expression patterns and regulation in the gut. Obesity (Silver Spring) 2006;14:683–9. doi: 10.1038/oby.2006.77. [DOI] [PubMed] [Google Scholar]
- 46.Gao Z, Yin J, Zhang J, Ward RE, Martin RJ, Lefevre M, et al. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes. 2009;58:1509–17. doi: 10.2337/db08-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kimura I, Inoue D, Maeda T, Hara T, Ichimura A, Miyauchi S, et al. Short-chain fatty acids and ketones directly regulate sympathetic nervous system via G protein-coupled receptor 41 (GPR41) Proc Natl Acad Sci U S A. 2011;108:8030–5. doi: 10.1073/pnas.1016088108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Furet JP, Kong LC, Tap J, Poitou C, Basdevant A, Bouillot JL, et al. Differential adaptation of human gut microbiota to bariatric surgery-induced weight loss: links with metabolic and low-grade inflammation markers. Diabetes. 2010;59:3049–57. doi: 10.2337/db10-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li JV, Ashrafian H, Bueter M, Kinross J, Sands C, le Roux CW, et al. Metabolic surgery profoundly influences gut microbial-host metabolic cross-talk. Gut. 2011;60:1214–23. doi: 10.1136/gut.2010.234708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cho I, Yamanishi S, Cox L, Methe BA, Zavadil J, Li K, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012;30:621–6. doi: 10.1038/nature11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thomas CM, Hong T, van Pijkeren JP, Hemarajata P, Trinh DV, Hu W, et al. Histamine derived from probiotic lactobacillus reuteri suppresses TNF via modulation of PKA and ERK signaling. PLoS One. 2012;7:e31951. doi: 10.1371/journal.pone.0031951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim JY, Chung EJ, Kim JH, Jung KY, Lee WY. Response to steroid treatment in anti-glutamic acid decarboxylase antibody-associated cerebellar ataxia, stiff person syndrome and polyendocrinopathy. Mov Disord. 2006;21:2263–4. doi: 10.1002/mds.21041. [DOI] [PubMed] [Google Scholar]
- 53.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–31. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 54.Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102:11070–5. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Backhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci U S A. 2007;104:979–84. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Greiner T, Backhed F. Effects of the gut microbiota on obesity and glucose homeostasis. Trends Endocrinol Metab. 2011;22:117–23. doi: 10.1016/j.tem.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 57.Delzenne NM, Neyrinck AM, Backhed F, Cani PD. Targeting gut microbiota in obesity: effects of prebiotics and probiotics. Nat Rev Endocrinol. 2011;7:639–46. doi: 10.1038/nrendo.2011.126. [DOI] [PubMed] [Google Scholar]
- 58.Cani PD, Delzenne NM. Involvement of the gut microbiota in the development of low grade inflammation associated with obesity: focus on this neglected partner. Acta Gastroenterol Belg. 2010;73:267–9. [PubMed] [Google Scholar]
- 59.Cani PD, Delzenne NM. Interplay between obesity and associated metabolic disorders: new insights into the gut microbiota. Curr Opin Pharmacol. 2009;9:737–43. doi: 10.1016/j.coph.2009.06.016. [DOI] [PubMed] [Google Scholar]
- 60.Ley RE. Obesity and the human microbiome. Curr Opin Gastroenterol. 2009;26:5–11. doi: 10.1097/MOG.0b013e328333d751. [DOI] [PubMed] [Google Scholar]
- 61.Ravussin Y, Koren O, Spor A, LeDuc C, Gutman R, Stombaugh J, et al. Responses of gut microbiota to diet composition and weight loss in lean and obese mice. Obesity. 2011;20:738–47. doi: 10.1038/oby.2011.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kalliomaki M, Collado MC, Salminen S, Isolauri E. Early differences in fecal microbiota composition in children may predict overweight. Am J Clin Nutr. 2008;87:534–8. doi: 10.1093/ajcn/87.3.534. [DOI] [PubMed] [Google Scholar]
- 63.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A. 2010;108(Suppl 1):4554–61. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Caricilli AM, Picardi PK, de Abreu LL, Ueno M, Prada PO, Ropelle ER, et al. Gut microbiota is a key modulator of insulin resistance in TLR 2 knockout mice. PLoS Biol. 2011;9:e1001212. doi: 10.1371/journal.pbio.1001212. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 65.Dasu MR, Devaraj S, Park S, Jialal I. Increased toll-like receptor (TLR) activation and TLR ligands in recently diagnosed type 2 diabetic subjects. Diabetes Care. 2010;33:861–8. doi: 10.2337/dc09-1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jialal I, Huet BA, Kaur H, Chien A, Devaraj S. Increased toll-like receptor activity in patients with metabolic syndrome. Diabetes Care. 2012;35:900–4. doi: 10.2337/dc11-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Creely SJ, McTernan PG, Kusminski CM, Fisher fM, Da Silva NF, Khanolkar M, et al. Lipopolysaccharide activates an innate immune system response in human adipose tissue in obesity and type 2 diabetes. Am J Physiol Endocrinol Metab. 2007;292:E740–7. doi: 10.1152/ajpendo.00302.2006. [DOI] [PubMed] [Google Scholar]
- 68.Devaraj S, Tobias P, Kasinath BS, Ramsamooj R, Afify A, Jialal I. Knockout of toll-like receptor-2 attenuates both the proinflammatory state of diabetes and incipient diabetic nephropathy. Arterioscler Thromb Vasc Biol. 2011;31:1796–804. doi: 10.1161/ATVBAHA.111.228924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Frazier TH, DiBaise JK, McClain CJ. Gut microbiota, intestinal permeability, obesity-induced inflammation, and liver injury. JPEN J Parenter Enteral Nutr. 2011;35:14S20S. doi: 10.1177/0148607111413772. [DOI] [PubMed] [Google Scholar]
- 70.Alberti KG, Eckel RH, Grundy SM, Zimmet PZ, Cleeman JI, Donato KA, et al. Harmonizing the metabolic syndrome: a joint interim statement of the International Diabetes Federation Task Force on Epidemiology and Prevention; National Heart, Lung, and Blood Institute; American Heart Association; World Heart Federation; International Atherosclerosis Society; and International Association for the Study of Obesity. Circulation. 2009;120:1640–5. doi: 10.1161/CIRCULATIONAHA.109.192644. [DOI] [PubMed] [Google Scholar]
- 71.Zupancic ML, Cantarel BL, Liu Z, Drabek EF, Ryan KA, Cirimotich S, et al. Analysis of the gut microbiota in the old order Amish and its relation to the metabolic syndrome. PLoS One. 2012;7:e43052. doi: 10.1371/journal.pone.0043052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Henao-Mejia J, Elinav E, Jin C, Hao L, Mehal WZ, Strowig T, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179–85. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Larsen N, Vogensen FK, van den Berg FW, Nielsen DS, Andreasen AS, Pedersen BK, et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One. 2010;5:e9085. doi: 10.1371/journal.pone.0009085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, et al. Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature. 2008;455:1109–13. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pozzilli P, Signore A, Williams AJ, Beales PE. NOD mouse colonies around the world–recent facts and figures. Immunol Today. 1993;14:193–6. doi: 10.1016/0167-5699(93)90160-M. [DOI] [PubMed] [Google Scholar]
- 76.Valladares R, Sankar D, Li N, Williams E, Lai KK, Abdelgeliel AS, et al. Lactobacillus johnsonii N6.2 mitigates the development of type 1 diabetes in BB-DP rats. PLoS One. 2010;5:e10507. doi: 10.1371/journal.pone.0010507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brown CT, Davis-Richardson AG, Giongo A, Gano KA, Crabb DB, Mukherjee N, et al. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS One. 2011;6:e25792. doi: 10.1371/journal.pone.0025792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hemarajata P, Versalovic J. Effects of probiotics on gut microbiota: mechanisms of intestinal immunomodulation and neuromodulation. Therap Adv Gastroenterol. 2013;6:39–51. doi: 10.1177/1756283X12459294. [DOI] [PMC free article] [PubMed] [Google Scholar]