Abstract
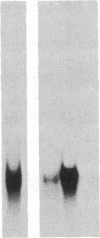
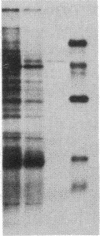
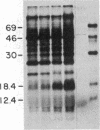
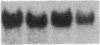
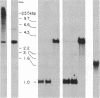
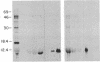
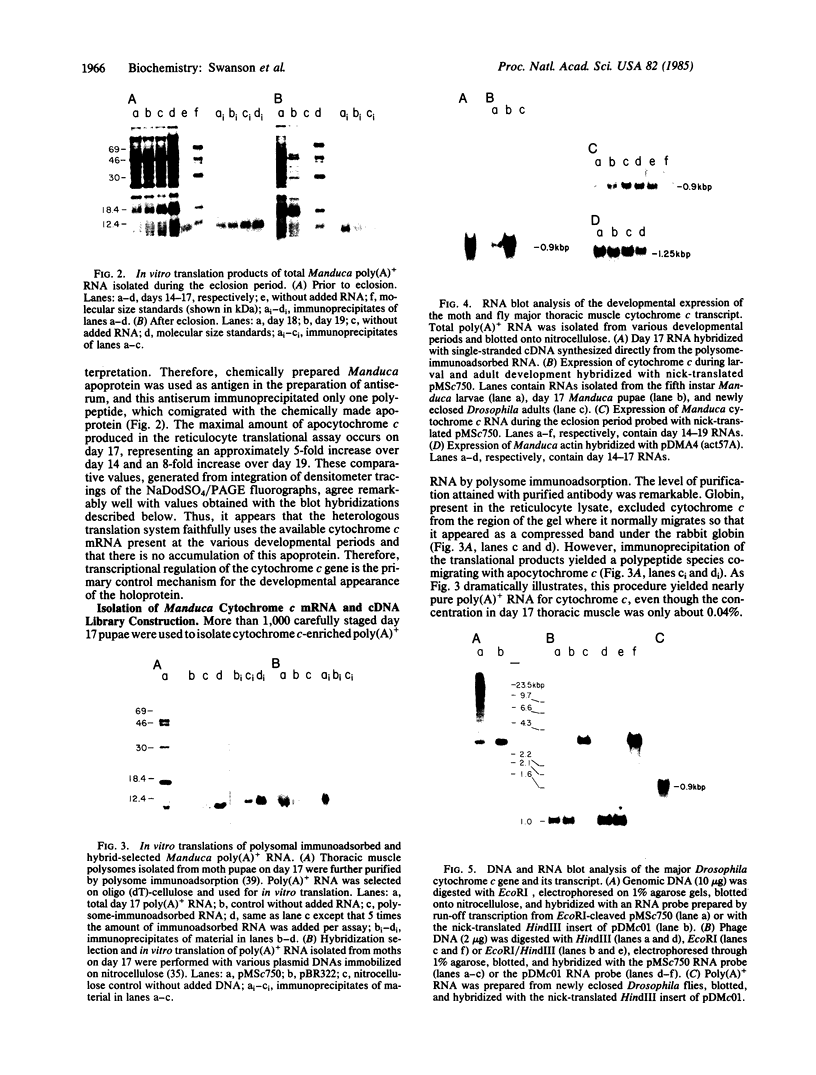
To investigate tissue-specific developmental regulation of mitochondrial biogenesis, we studied the expression of the Manduca sexta (tobacco hornworm moth) thoracic muscle cytochrome c gene during adult eclosion and used this information to obtain a cDNA clone for this gene, which in turn was used to isolate the corresponding Drosophila melanogaster gene. Over the 3 days prior to adult Manduca emergence, mitochondrial inner membranes become progressively more electron dense and lamellar, and, while there is no accumulation of apocytochrome c, the amount of the holoprotein increases 40-fold per insect thorax. As determined by in vitro translation and blot hybridization analysis, the major thoracic muscle cytochrome c gene is primarily regulated at the transcriptional level, with cytochrome c mRNA increasing from less than 0.01% to 0.04% of total poly(A)+ RNA and declining to an undetectable level by day 2 after eclosion. Furthermore, the ratio of cytochrome c to the other cytochromes remains the same at all times, indicating that these components of the respiratory chain follow coordinated developmental programs. By using polysome immunoadsorption, a poly(A)+ RNA population of greater than or equal to 95% cytochrome c mRNA was isolated from thoracic muscle tissue and was used to construct a cDNA library, which was screened by hybrid selection/translation. We report the sequence of one of those clones, pMSc750, and its use to isolate the major thoracic muscle cytochrome c gene of Drosophila.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Boss M. A., Dreyfuss G., Baltimore D. Localization of the Abelson murine leukemia virus protein in a detergent-insoluble subcellular matrix: architecture of the protein. J Virol. 1981 Nov;40(2):472–481. doi: 10.1128/jvi.40.2.472-481.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brautigan D. L., Ferguson-Miller S., Margoliash E. Mitochondrial cytochrome c: preparation and activity of native and chemically modified cytochromes c. Methods Enzymol. 1978;53:128–164. doi: 10.1016/s0076-6879(78)53021-8. [DOI] [PubMed] [Google Scholar]
- Candia Carnevali M. D., Reger J. F. Slow-acting flight muscles of Saturniid moths. J Ultrastruct Res. 1982 Jun;79(3):241–249. doi: 10.1016/s0022-5320(82)90001-6. [DOI] [PubMed] [Google Scholar]
- Chan S. K., Margoliash E. Biosynthesis of cytochrome c in developing pupae of Samia cynthia. J Biol Chem. 1966 May 25;241(10):2252–2255. [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Dretzen G., Bellard M., Sassone-Corsi P., Chambon P. A reliable method for the recovery of DNA fragments from agarose and acrylamide gels. Anal Biochem. 1981 Apr;112(2):295–298. doi: 10.1016/0003-2697(81)90296-7. [DOI] [PubMed] [Google Scholar]
- Faye G., Leung D. W., Tatchell K., Hall B. D., Smith M. Deletion mapping of sequences essential for in vivo transcription of the iso-1-cytochrome c gene. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2258–2262. doi: 10.1073/pnas.78.4.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyrberg E. A., Kindle K. L., Davidson N., Kindle K. L. The actin genes of Drosophila: a dispersed multigene family. Cell. 1980 Feb;19(2):365–378. doi: 10.1016/0092-8674(80)90511-5. [DOI] [PubMed] [Google Scholar]
- Grunstein M., Hogness D. S. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3961–3965. doi: 10.1073/pnas.72.10.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L., Lalonde B., Gifford P., Alani E. Distinctly regulated tandem upstream activation sites mediate catabolite repression of the CYC1 gene of S. cerevisiae. Cell. 1984 Feb;36(2):503–511. doi: 10.1016/0092-8674(84)90243-5. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Jemmerson R., Margoliash E. Topographic antigenic determinants on cytochrome c. Immunoadsorbent separation of the rabbit antibody populations directed against horse cytochrome. J Biol Chem. 1979 Dec 25;254(24):12706–12716. [PubMed] [Google Scholar]
- Kessler S. W. Cell membrane antigen isolation with the staphylococcal protein A-antibody adsorbent. J Immunol. 1976 Nov;117(5 Pt 1):1482–1490. [PubMed] [Google Scholar]
- Kraus J. P., Rosenberg L. E. Purification of low-abundance messenger RNAs from rat liver by polysome immunoadsorption. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4015–4019. doi: 10.1073/pnas.79.13.4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Limbach K. J., Wu R. Characterization of two Drosophila melanogaster cytochrome c genes and their transcripts. Nucleic Acids Res. 1985 Jan 25;13(2):631–644. doi: 10.1093/nar/13.2.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry C. V., Weiss J. L., Walthall D. A., Zitomer R. S. Modulator sequences mediate oxygen regulation of CYC1 and a neighboring gene in yeast. Proc Natl Acad Sci U S A. 1983 Jan;80(1):151–155. doi: 10.1073/pnas.80.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Hardison R. C., Lacy E., Lauer J., O'Connell C., Quon D., Sim G. K., Efstratiadis A. The isolation of structural genes from libraries of eucaryotic DNA. Cell. 1978 Oct;15(2):687–701. doi: 10.1016/0092-8674(78)90036-3. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., Kleid D. G. Nucleotide sequence of the rightward operator of phage lambda. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1184–1188. doi: 10.1073/pnas.72.3.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura S., Arpin M., Hannum C., Margoliash E., Sabatini D. D., Morimoto T. In vitro synthesis and posttranslational uptake of cytochrome c into isolated mitochondria: role of a specific addressing signal in the apocytochrome. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4368–4372. doi: 10.1073/pnas.78.7.4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Montgomery D. L., Hall B. D., Gillam S., Smith M. Identification and isolation of the yeast cytochrome c gene. Cell. 1978 Jul;14(3):673–680. doi: 10.1016/0092-8674(78)90250-7. [DOI] [PubMed] [Google Scholar]
- Nedergaard J., Cannon B. Overview--preparation and properties of mitochondria from different sources. Methods Enzymol. 1979;55:3–28. doi: 10.1016/0076-6879(79)55003-4. [DOI] [PubMed] [Google Scholar]
- Pedersen P. L., Schnaitman C. A. The oligomycin-sensitive adenosine diphosphate-adenosine triphosphate exchange in an inner membrane matrix fraction of rat liver mitochondria. J Biol Chem. 1969 Sep 25;244(18):5065–5074. [PubMed] [Google Scholar]
- Pelham H. R., Jackson R. J. An efficient mRNA-dependent translation system from reticulocyte lysates. Eur J Biochem. 1976 Aug 1;67(1):247–256. doi: 10.1111/j.1432-1033.1976.tb10656.x. [DOI] [PubMed] [Google Scholar]
- Ricciardi R. P., Miller J. S., Roberts B. E. Purification and mapping of specific mRNAs by hybridization-selection and cell-free translation. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4927–4931. doi: 10.1073/pnas.76.10.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb J. A. Maintenance of imaginal discs of Drosophila melanogaster in chemically defined media. J Cell Biol. 1969 Jun;41(3):876–885. doi: 10.1083/jcb.41.3.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla R. C., Agne K. M., Wu R. Cytochrome c gene-related sequences in mammalian genomes. Proc Natl Acad Sci U S A. 1982 Feb;79(3):739–743. doi: 10.1073/pnas.79.3.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., Stewart J. W., Margoliash E., Parker J., Campbell W. The structural gene for yeast cytochrome C. Proc Natl Acad Sci U S A. 1966 Jun;55(6):1498–1504. doi: 10.1073/pnas.55.6.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. R., Calvo J. M. Nucleotide sequence of the E coli gene coding for dihydrofolate reductase. Nucleic Acids Res. 1980 May 24;8(10):2255–2274. doi: 10.1093/nar/8.10.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. O., Birnstiel M. L. A simple method for DNA restriction site mapping. Nucleic Acids Res. 1976 Sep;3(9):2387–2398. doi: 10.1093/nar/3.9.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste W. H. Molecular proportion of the fixed cytochrome components of the respiratory chain of Keilin-Hartree particles and beef heart mitochondria. Biochim Biophys Acta. 1966 Jan 11;113(1):175–178. doi: 10.1016/s0926-6593(66)80132-7. [DOI] [PubMed] [Google Scholar]
- WILLIAMS J. N., Jr A METHOD FOR THE SIMULTANEOUS QUANTITATIVE ESTIMATION OF CYTOCHROMES A, B, C1, AND C IN MITOCHONDRIA. Arch Biochem Biophys. 1964 Sep;107:537–543. doi: 10.1016/0003-9861(64)90313-3. [DOI] [PubMed] [Google Scholar]
- Zimmermann R., Hennig B., Neupert W. Different transport pathways of individual precursor proteins in mitochondria. Eur J Biochem. 1981 Jun 1;116(3):455–460. doi: 10.1111/j.1432-1033.1981.tb05357.x. [DOI] [PubMed] [Google Scholar]
- Zimmermann R., Paluch U., Neupert W. Cell-free synthesis of cytochrome c. FEBS Lett. 1979 Dec 1;108(1):141–146. doi: 10.1016/0014-5793(79)81196-5. [DOI] [PubMed] [Google Scholar]
- Zinn K., DiMaio D., Maniatis T. Identification of two distinct regulatory regions adjacent to the human beta-interferon gene. Cell. 1983 Oct;34(3):865–879. doi: 10.1016/0092-8674(83)90544-5. [DOI] [PubMed] [Google Scholar]