Abstract
Nucleus movement, positioning, and orientation is precisely specified and actively regulated within cells, and it plays a critical role in many cellular and developmental processes. Mutation of proteins that regulate the nucleus anchoring and movement lead to diverse pathologies, laminopathies in particular, suggesting that the nucleus correct positioning and movement is essential for proper cellular function. In motile cells that polarize toward the direction of migration, the nucleus undergoes controlled rotation promoting the alignment of the nucleus with the axis of migration. Such spatial organization of the cell appears to be optimal for the cell migration. Nuclear reorientation requires the cytoskeleton to be anchored to the nuclear envelope, which exerts pulling or pushing torque on the nucleus. Here we discuss the possible molecular mechanisms regulating the nuclear rotation and reorientation and the significance of this type of nuclear movement for cell migration.
Keywords: cell polarity, migration, nuclear reorientation, LINC, actin, microtubules, focal adhesions, FAK, dynein, myosin
Introduction
Nucleus contributes to the establishment of the polarized, asymmetrical profile of migrating cells. During migration, nucleus positions to the cell’s rear and promotes microtubule organizing center (MTOC) localization close to the cell center between the leading edge and the nucleus. MTOC positioning in front of the nucleus is a prerequisite for polarized microtubule growth from the MTOC toward the leading edge. Microtubules are selectively stabilized at the leading edge and they are thought to provide a unique track for directed vesicle trafficking toward the leading edge (Fig. 1A). The stereotypical localization of the nucleus to the cell’s rear and MTOC close to the cell center has been recognized as an indicator of the migratory polarity defining the axis of migration.1
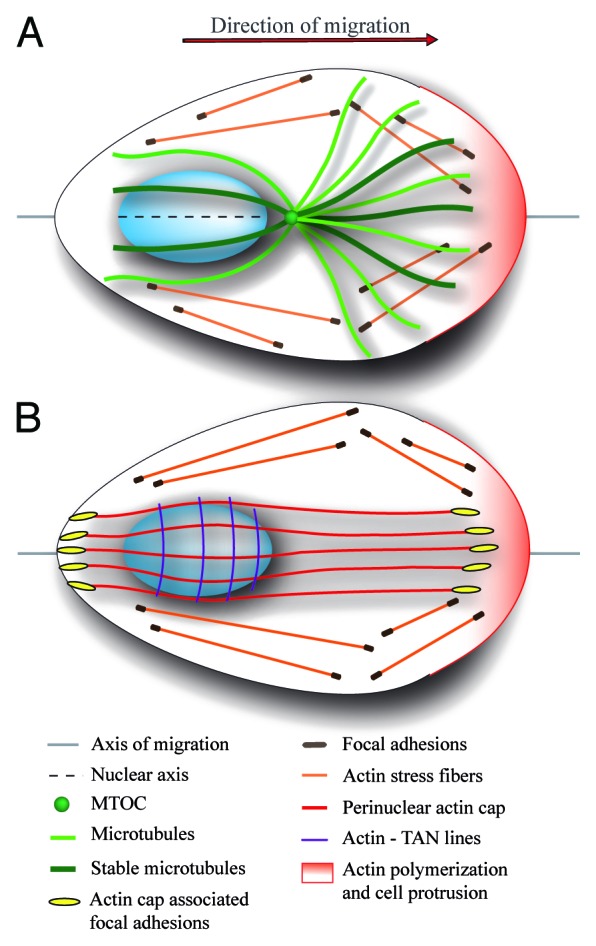
Figure 1. Schematic representation of an adherent migrating cell with front-rear polarity. Polarized cell displays conical shape with actin polymerization induced at the leading edge (pink) and limited at the cell rear. Cells are attached to the substrate through cell–matrix adhesion, such as focal contacts and adhesions, which connects extracellular matrix to cellular cytoskeleton. (A) In polarized cells, the oval-shaped nucleus localizes to the cell rear, MTOC in front of the nucleus close to the cell center, and microtubules are preferentially oriented toward the leading edge and they are stabilized at this location. The relative position of the nucleus and MTOC is an important marker of cell migration polarity defining the axis of migration. In addition, the longer nuclear axis is aligned with the axis of migration. (B) Specific types of actin filaments anchored to the dorsal side of the nucleus contribute to the establishment of asymmetric profile of the migrating cell. TAN lines are arranged perpendicular to axis of migration and drive the rearward movement of the nucleus during cell polarization. Perinuclear actin cap fibers span the nucleus and link the nucleus to subset of focal adhesions at cell periphery. Actin cap filaments are aligned with the axis of migration and longer nuclear axis and they probably stabilize nuclear orientation. It is not clear whether perinuclear actin cap and TAN lines co-exist in the same cell.
More recently, it was observed that during cell polarization the nucleus undergoes synchronous and temporally restricted rotational movement. This reorientation of the nucleus is characterized by the alignment of the longer nuclear axis with the direction of migration.2,3 Nuclear reorientation further promotes the establishment of bilateral symmetry characteristic for migrating fibroblasts (Fig. 1).
Nuclear reorientation is propelled by the cytoskeleton attached to the nucleus. Microtubules are the prime candidates for nuclear rotation as they have been shown to control nuclear movement in several cell types. However, recent identification of perinuclear actin cap4 and actin associated with TAN (transmembrane actin-associated nuclear) lines,5 two different actomyosin structures anchored to the nucleus, suggests that specific types of actin filaments may promote the establishment of migratory polarity and cellular locomotion (Fig. 1B). Here, we highlight the important features and mechanisms involved in the regulation of nuclear rotation and nuclear reorientation.
Nuclear rotation and reorientation in adherent cells
Nucleus movement and rotation was first observed 60 years ago in human nasal mucosa cells,6 and subsequently, in other cell types.7-9 These studies showed that nuclei migrated linearly through the cytoplasm or rotated around the axis clockwise or counterclockwise, or occasionally perpendicular to the substrate. Nuclear rotation was observed as a three-dimensional motion of chromatin domains associated with nucleoli, leading to the conclusion that the nuclear rotation is in fact karyoplasmic streaming.10 Nevertheless, other reports suggested that the whole cell nucleus rotates as the nucleoli maintained the rigid pattern during the rotation.11,12 Recent experiments using fluorescence labeling of discrete nuclear compartments have conclusively shown that the nuclear rotation involves the movement of an entire nucleus, including nuclear interior as well as the inner and outer nuclear membranes.13
Nuclear rotation is a nuclear movement around the nucleus axis perpendicular to the substratum. Nuclear rotation ranges from stochastically or oscillatory rotation of the nucleus back and forth within a few degrees only to more sustained and directional rotation that changes the nuclear orientation.2,3,13-17 Sustained nuclear rotation could be induced by diverse stimuli such as mechanical shear stress16 or cyclical stretches of the substrate.18 Nuclear rotation is also induced in two-dimensional migration models, where cells polarize and migrate into the wound made in a confluent monolayer of cells.2,3,17 In the wound healing model the nucleus appears to be relatively static in non-polarized cells present in the cell monolayer and in polarized cells migrating into the wound. Nuclear rotation, termed nuclear reorientation, occurs only temporally during the wound-induced cell polarization. We defined nuclear reorientation as controlled nuclear rotation allowing the nucleus to rotate in the “xy” plane until its longer axis is aligned with the axis of migration, i.e., perpendicular to the wound.3 Therefore, both rotation and reorientation are functionally similar, although the molecular players and the precise mechanism that control the rotation may vary according to the cell type.
Sustained nuclear rotation could be continuous, sometimes exceeding 360°.17-19 We suppose that constant nuclear rotation is a consequence of deregulated reorientation and lack of control over nuclear reorientation. To support this, continuous nucleus rotation can be experimentally induced by disruption of intermediate filaments19,20 and actomyosin contractility.17,21 Continuous nuclear rotation was also observed in cells deficient in lamin B113 or in wound-edge epithelial cells deficient in heterogeneous group of genes.22 These data suggest the existence of a complex molecular mechanism that regulates nuclear reorientation.
Nuclear reorientation and rotation requires the LINC complex
All three types of cytoskeleton, microtubules, actin filaments, and intermediate filaments, have been shown to associate with the components of nuclear envelope and to regulate nuclear movement including nuclear rotation. The key role in nuclear movement plays the nuclear lamina, particularly lamin A/C, which provides mechanical support and structural stability to the nucleus. Tight coupling of the nuclear lamina to the cytoskeleton allows the forces exerted by the cytoskeleton to move the nucleus (for a review. see refs. 23–26). Connection between the cell cytoskeleton and nuclear lamina is mediated by the LINC (Linker of Nucleoskeleton and Cytoskeleton) complex that passes both through the outer and inner nuclear membranes. The LINC complex is composed of SUN proteins that span the inner nuclear membrane and their N terminus interacts with nuclear lamina. The conserved SUN domain of SUN proteins localized within the perinuclear space interacts with the KASH domain of Syne/nesprin family of proteins that span the outer nuclear membrane. At the cytoplasmic side Syne/nesprin proteins interact with cytoskeletal components linking the nuclear lamina with the cell cytoskeleton. Actin filaments are the only components of the cell cytoskeleton known so far to interact directly with the LINC complex. The association is mediated by nesprin-1 and -2 and their calponin homology domains. Nesprin-1 and -2 also interact with microtubule motor proteins dynein and kinesin, which capture centrosomal microtubules, while nesprin-3 interacts with plectin, which links the LINC complex to intermediate filaments (for a review, see refs. 26 and 27). Kinesin also links microtubules to nesprin-4; nevertheless, the expression of nesprin-4 is restricted to epithelial secretory cells.28
The integrity of nuclear lamina and LINC complex is essential for the nuclear rotation, and deficiency of lamin A/C or disruption of the LINC complex by dominant negative versions of Sun or nesprin proteins impairs nuclear rotation and reorientation.2,3,18,29 The evidence also suggests the important role of the LINC complex and nucleus movement in the regulation of cell migration as disruption of nucleo-cytoskeletal link impairs cell migration.2,3,5,29-31 However, it should be noted that the nucleus–cytoskeleton association and nucleus movement could also be LINC-independent. For example, microtubules have been shown to interact with the nuclear pore complex components,32 nuclear envelope protein emerin,33 and nucleus rearward positioning could be LINC-independent in some cell types.3,34
Proposed mechanisms for nuclear reorientation
Nuclear rotation requires forces acting on the nucleus that are mediated by the cytoskeleton and cytoskeleton-associated motor proteins. The nuclear rotation in some cells is exclusively driven by microtubules and microtubule motor dynein, such as in wound edge NIH3T3 fibroblasts.17 In other cases, the inhibition of myosin motor proteins and actomyosin contractility revealed that actin is required for nuclear rotation18 or, conversely, serves as an anchoring system preventing nuclear rotation.17,19,21 Microtubules and actin could also have non-redundant functions because interfering with the function of either microtubules or actin cytoskeleton impedes nuclear rotation in lamin B1-deficient cells.13 In addition, intermediate filaments (IFs) are also implicated in nuclear rotation despite the fact that IFs have no motor protein.19,20 Thus, it appears that the requirements for the specific cytoskeletal structures vary depending on the cell type and experimental conditions used.
We propose three mechanisms based on microtubules and actin-associated motor proteins, although these mechanisms may cooperate in the nucleus reorientation. Dynein is accumulated at the leading lamellipodia of migrating cells and at the tips and alongside of microtubules. From these cortical and cytosolic anchoring sites dynein exerts pulling forces on microtubules that induce MTOC centration (for a review, see ref. 35). MTOC is connected to the nucleus e.g., by association of emerin with MTOC33 or by centrosomal microtubules captured by the LINC complex.26,27 In first model, cortical and lengthwise pulling forces move MTOC, which consequently induces nuclear rotation (Fig. 2A). Nevertheless, we observed that during the cell polarization MTOC moves toward the leading edge without significant nucleus movement (our unpublished data). Also in other studies the nucleus was shown to rotate independently of MTOC, indicating that the link between the nucleus and MTOC is weak or reversible16,17,19,36,37 and that the forces exerted by cortically and cytoplasmically anchored dynein are not sufficient to induce nuclear rotation.
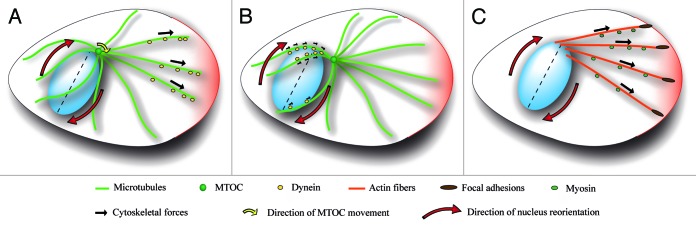
Figure 2. Schematic representation of hypothetical nuclear reorientation models. Forces exerted and transmitted by the cell cytoskeleton (black arrows) are transferred to the nucleus through LINC complex (not shown) to induce nucleus rotation and reorientation (red arrows). (A and B) Microtubules induce nuclear reorientation by forces exerted by microtubule-associated motor protein dynein. (A) Dynein pulls at the tips and alongside of microtubules to induce MTOC re-positioning close to the cell center (yellow arrow). Because MTOC associates with the nucleus, MTOC movement also induces nucleus reorientation (red arrows). (B) Dynein, through its interactions with nesprins, links microtubules to the nuclear envelope and pulls the nucleus as a huge cargo toward minus end of microtubules mediating nuclear reorientation. The asymmetric distribution of microtubules associated with nucleus is required to induce torque on the nucleus. (C) Actin cap fibers reorientate the nucleus. Actin cap fibers emanating from the focal adhesions at the leading edge associate with LINC complex at the nuclear envelope, predominantly at one pole of the nucleus. Nuclear reorientation is induced by actomyosin contractile forces between the leading edge and the nucleus.
Cortically and lengthwise exerted pulling forces can be supported by the microtubule motor proteins, particularly dyneins, which are associated with the nuclear envelope through the LINC complex.37 In this model (Fig. 2B), dynein pulls the nucleus as a huge cargo along the microtubules toward the minus end of microtubules emanating from MTOC.17,37 Transient asymmetric distribution of microtubules associated with the nucleus would create net torque on the nucleus, thus inducing its rotation. The nuclear rotation would be terminated when the asymmetry of the microtubular network is reversed.37
Recently identified perinuclear actin cap represents an additional player that may be involved in the regulation of nuclear reorientation. Actin cap is composed of contractile actomyosin filaments that interact with the LINC complex on the dorsal side of the nucleus and with focal adhesions at the cell periphery. Since actin cap fibers extend over the nucleus in a pole-to-pole manner and they are aligned with the longer nuclear axis and the axis of migration,4,38 it is possible that the actin cap serves as an anchoring structure stabilizing the nucleus in specific orientation. Consistently, we observed that the actin cap is disrupted directly above the nucleus during cell polarization and then reassembled when the cells are polarized, allowing the nucleus to rotate (our unpublished results). We also observed that during reassembly of the actin cap, actin fibers attach predominantly to one pole of the nucleus (our unpublished data). Since the actin cap is anchored at focal adhesions at the leading edge, it is tempting to speculate that actin cap fibers also induce nuclear rotation (Fig. 2C). The potential mechanism involves the attachment of actin cap fibers newly formed from the leading edge to the pole region of the nucleus. Consequently, the nucleus reorientation may be induced by actin-mediated forces (Fig. 2C).
Is nuclear reorientation regulated by the distant signaling at the leading edge?
Nuclear rotation and reorientation is likely a consequence of cytoskeleton rearrangement. Because cytoskeleton is to a large extent regulated by Rho family GTPases it is likely that Rho GTPase signaling also regulates nuclear rotation. Indeed, Cdc42 was shown to control nucleus rotation in cells exposed to shear stress.16
In our previous study3 we described that nucleus reorientation requires two signaling axes, LPA-mediated Rho activation and integrin/FAK/p190RhoGAP signaling leading to Rho inactivation (Fig. 3). Adhesion and LPA signaling are coordinated in order to induce nuclear reorientation. After wounding the cell monolayer, integrin engagement to ECM activates FAK and active FAK localizes with its downstream effector p190RhoGAP at the leading edge. Adhesion and integrin/FAK/p190RhoGAP signaling inhibits Rho transiently,39 suggesting that LPA-mediated Rho activation is inhibited at the leading edge. Thus, integration of adhesion and LPA stimuli at Rho constitutes a gradient-forming mechanism with the capability to differentially regulate Rho at the cell front and rear.
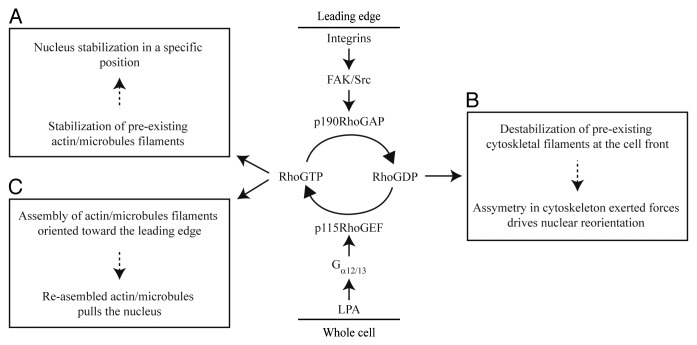
Figure 3. Signaling pathways involved in the regulation of nuclear reorientation. Two signaling pathways that converge at small GTPase Rho regulate cycling between active (GTP-bound) and inactive (GDP-bound) state of Rho allowing the cytoskeleton remodeling and subsequently nuclear reorientation. LPA signaling induces the activation of heterotrimeric G proteins, and consequently, RGS-RhoGEF, such as p115–RhoGEF, which directly activates Rho. Since LPA is a soluble mitogen it presumably activates Rho within cell uniformly. Active Rho regulates the stabilization of pre-existing cytoskeletal filaments that anchor the nucleus in immobile state (box A). Acute integrin engagement to ECM at the cell front activates FAK/Src signaling complex that recruits Rho inhibitor p190RhoGAP to the leading edge. Transient Rho inhibition at the leading edge leads to the destabilization of cytoskeletal filaments and consequent asymmetry in cytoskeletal forces may induce nucleus reorientation (box B). Since integrin-mediated Rho inactivation is transient, new cycle of Rho activation allows re-assembly and stabilization of new cytoskeletal filaments oriented toward the leading edge that may pull the nucleus and contribute to the nucleus reorientation (box C).
We suppose that signaling induced at the leading edge provides the molecular framework for the mechanism underlying nuclear reorientation. The actin fibers or microtubules and associated motor proteins remain anchored to the cell cortex and ECM structures on one side and to the nucleus on the other side.23,40-43 Therefore, through the cytoskeletal elements nucleus remains under isometric tension and it is relatively immobile in stationary, non-polarized cells (Fig. 3, box A). In response to migratory polarity cues like wounding, transient decrease of Rho activity at the cell front allows actin and microtubule disassembly promoting partial relaxation of the isometric tension. The relaxation of isometric tension could be sufficient for nuclear reorientation as it generates asymmetric distribution of cytoskeletal elements associated with nucleus to induce torque on the nucleus (Fig. 3, box B; see also Fig. 2B). In addition, it is possible that actin cap fibers that are attached to focal adhesions at the leading edge44 respond to Rho inactivation and their disassembly allows nucleus to rotate. Since integrin-mediated Rho inactivation is transient, Rho induces the re-assembly of cytoskeleton that may represent additional mechanism controlling nuclear rotation. De novo polymerized actin cap fibers and microtubules are oriented toward the leading edge (Fig. 3, box C). Actin or microtubule-mediated forces would then rotate the nucleus (Fig. 2A and C). Once the cells have polarized, formation of the actin-myosin fibers and stabilization of the microtubules restores isometric tension that put the break on nucleus rotation. Therefore, we speculate that signal-dependent changes in actin and microtubule dynamics constitute the molecular mechanisms that control nuclear rotation (Fig. 3).
Functional significance of nuclear orientation in migrating cells
There is growing body of evidence that the disruption of LINC complex impairs cell polarization and migration suggesting that the attachment of cytoskeleton to the nucleus plays an important role in cell motility.2,3,5,29-31 Nevertheless, the role of nuclear shape and nuclear orientation in cell migration has not been extensively studied. The nucleus frequently displays ovoid or elliptical shape with characteristic orientation in different cell types. For example, migrating fish keratocytes are fan-like shaped with broad lamellipodial protrusion at the cell front and elliptical nucleus oriented perpendicular to the axis of migration. On the contrary, in conically shaped fibroblasts the nucleus is oriented parallel to the axis of migration. This raises the question why cells orientate the nucleus and how nuclear orientation affects cellular functions, notably cell migration.
One obvious reason for specific nuclear orientation is that it may promote cell polarization, and thus, cell motility, as we proposed recently.3 It was found that disruption of the LINC complex specifically affects nucleus reorientation, MTOC and/or Golgi polarization, and cell migration.2,3,29 It indicates that the defect in MTOC/Golgi polarization and cell migration is a consequence of impaired nuclear reorientation.
Alternatively, specific nucleus orientation may help overcome the physical constraints facing the migrating cells. During keratocyte movement, the cell body is rolling together with the nucleus45 and the nuclear oval shape and its orientation perpendicular to the direction of migration may ease the nucleus rolling and cell locomotion. Nucleus rolling may also help to overcome the blockage of cytoplasmic granules accumulated in front of the nucleus, as described in C. elegans development.46 Such rolling is not probable in migrating fibroblasts because the longer nuclear axis is aligned with the direction of migration. Nevertheless, the oval-shaped nucleus may move forward like a “torpedo” through the cytoplasm to facilitate nucleus translocation as cells move. Similarly to fibroblasts migrating in 2D environment, cells migrating in 3D environment reorient the nucleus toward the direction of migration. It has been suggested that the nuclear reorientation helps the cells passage through the narrow pores in the collagen lattice.47
The characteristic orientation of the cell nucleus in motile cells allows us to speculate whether the nucleus itself is a polarized organelle. It has been described that the nuclear envelope is polarized as nesprin-4 accumulates asymmetrically at the pole of the nucleus distally to MTOC. Interestingly, overexpression of nesprin-4 leads to polarization of other nuclear components, including lamins and nuclear pore complex proteins.28 Given that chromatin interacts with lamins, the spatial positioning of chromatin within the nucleus could be regulated. Keeping the gene in specific location within the nucleus, and thus, within the cell, may promote specific transcripts to be delivered to the specific locations. To support this speculation, it has been demonstrated that migration-specific transcripts are preferentially delivered to the leading edge of migrating cells.48
Conclusion and Perspectives
A large body of recent work shows that precise nucleus location inside the cell is important for correct cellular functioning. In respect of motile cells, it is interesting that the cells possess the mechanism(s) that purposely move and orientate the nucleus in order to facilitate their motility. In particular, the rearward positioning of the nucleus and nuclear reorientation emerged to be important for the establishment of cell polarity and optimal for cellular migration. The regulatory mechanism(s) that move the nucleus could also be employed by cells migrating in 3D environment to facilitate their invasion, suggesting the possible role of the nucleus mechanics in the patho-physiological processes such as cancer cell invasion. Clearly, additional studies are required to understand the functional significance of the nucleus orientation in motile cells and to decipher the basic molecular mechanisms controlling this process.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by Czech Science Foundation grant 204/09/0614, EU FP7 Marie Curie IRG grant 231086 and by the institutional research concept RVO 61388971. TV was supported by J.E. Purkynje fellowship from the Academy of Sciences of the Czech Republic.
Glossary
Abbreviations:
- ECM
extracellular matrix
- FAK
focal adhesion kinase
- LINC
linker of nucleoskeleton and cytoskeleton
- KASH
Klarsicht, ANC-1, Syne Homology, LPA, lysophosphatidic acid
- MTOC
microtubule organizing center
- SUN
Sad1p, UNC-84
- TAN lines
transmembrane actin-associated nuclear lines
Footnotes
Previously published online: www.landesbioscience.com/journals/celladhesion/article/27761
References
- 1.Luxton GW, Gundersen GG. Orientation and function of the nuclear-centrosomal axis during cell migration. Curr Opin Cell Biol. 2011;23:579–88. doi: 10.1016/j.ceb.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Houben F, Willems CH, Declercq IL, Hochstenbach K, Kamps MA, Snoeckx LH, Ramaekers FC, Broers JL. Disturbed nuclear orientation and cellular migration in A-type lamin deficient cells. Biochim Biophys Acta. 2009;1793:312–24. doi: 10.1016/j.bbamcr.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Maninová M, Klímová Z, Parsons JT, Weber MJ, Iwanicki MP, Vomastek T. The reorientation of cell nucleus promotes the establishment of front-rear polarity in migrating fibroblasts. J Mol Biol. 2013;425:2039–55. doi: 10.1016/j.jmb.2013.02.034. [DOI] [PubMed] [Google Scholar]
- 4.Khatau SB, Hale CM, Stewart-Hutchinson PJ, Patel MS, Stewart CL, Searson PC, Hodzic D, Wirtz D. A perinuclear actin cap regulates nuclear shape. Proc Natl Acad Sci U S A. 2009;106:19017–22. doi: 10.1073/pnas.0908686106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luxton GW, Gomes ER, Folker ES, Vintinner E, Gundersen GG. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science. 2010;329:956–9. doi: 10.1126/science.1189072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pomerat CM. Rotating nuclei in tissue cultures of adult human nasal mucosa. Exp Cell Res. 1953;5:191–6. doi: 10.1016/0014-4827(53)90104-9. [DOI] [PubMed] [Google Scholar]
- 7.Leone V, Hsu TC, Pomerat CM. Cytological studies on HeLa, a strain of human cervical carcinoma. II. On rotatory movements of the nuclei. Z Zellforsch Mikrosk Anat. 1955;41:481–92. doi: 10.1007/BF00345357. [DOI] [PubMed] [Google Scholar]
- 8.Nakai J. Dissociated dorsal root ganglia in tissue culture. Am J Anat. 1956;99:81–129. doi: 10.1002/aja.1000990105. [DOI] [PubMed] [Google Scholar]
- 9.Capers CR. Multinucleation of skeletal muscle in vitro. J Biophys Biochem Cytol. 1960;7:559–66. doi: 10.1083/jcb.7.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Boni U, Mintz AH. Curvilinear, three-dimensional motion of chromatin domains and nucleoli in neuronal interphase nuclei. Science. 1986;234:863–6. doi: 10.1126/science.3775367. [DOI] [PubMed] [Google Scholar]
- 11.Paddock SW, Albrecht-Buehler G. Distribution of microfilament bundles during rotation of the nucleus in 3T3 cells treated with monensin. Exp Cell Res. 1986;163:525–38. doi: 10.1016/0014-4827(86)90083-2. [DOI] [PubMed] [Google Scholar]
- 12.Paddock SW, Albrecht-Buehler G. Rigidity of the nucleus during nuclear rotation in 3T3 cells. Exp Cell Res. 1988;175:409–13. doi: 10.1016/0014-4827(88)90205-4. [DOI] [PubMed] [Google Scholar]
- 13.Ji JY, Lee RT, Vergnes L, Fong LG, Stewart CL, Reue K, Young SG, Zhang Q, Shanahan CM, Lammerding J. Cell nuclei spin in the absence of lamin b1. J Biol Chem. 2007;282:20015–26. doi: 10.1074/jbc.M611094200. [DOI] [PubMed] [Google Scholar]
- 14.Bard F, Bourgeois CA, Costagliola D, Bouteille M. Rotation of the cell nucleus in living cells: a quantitative analysis. Biol Cell. 1985;54:135–42. doi: 10.1111/j.1768-322X.1985.tb00388.x. [DOI] [PubMed] [Google Scholar]
- 15.Park PC, De Boni U. Dynamics of nucleolar fusion in neuronal interphase nuclei in vitro: association with nuclear rotation. Exp Cell Res. 1991;197:213–21. doi: 10.1016/0014-4827(91)90425-T. [DOI] [PubMed] [Google Scholar]
- 16.Lee JS, Chang MI, Tseng Y, Wirtz D. Cdc42 mediates nucleus movement and MTOC polarization in Swiss 3T3 fibroblasts under mechanical shear stress. Mol Biol Cell. 2005;16:871–80. doi: 10.1091/mbc.E03-12-0910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levy JR, Holzbaur EL. Dynein drives nuclear rotation during forward progression of motile fibroblasts. J Cell Sci. 2008;121:3187–95. doi: 10.1242/jcs.033878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brosig M, Ferralli J, Gelman L, Chiquet M, Chiquet-Ehrismann R. Interfering with the connection between the nucleus and the cytoskeleton affects nuclear rotation, mechanotransduction and myogenesis. Int J Biochem Cell Biol. 2010;42:1717–28. doi: 10.1016/j.biocel.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 19.Gerashchenko MV, Chernoivanenko IS, Moldaver MV, Minin AA. Dynein is a motor for nuclear rotation while vimentin IFs is a “brake”. Cell Biol Int. 2009;33:1057–64. doi: 10.1016/j.cellbi.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 20.Hay M, De Boni U. Chromatin motion in neuronal interphase nuclei: changes induced by disruption of intermediate filaments. Cell Motil Cytoskeleton. 1991;18:63–75. doi: 10.1002/cm.970180107. [DOI] [PubMed] [Google Scholar]
- 21.Vicente-Manzanares M, Zareno J, Whitmore L, Choi CK, Horwitz AF. Regulation of protrusion, adhesion dynamics, and polarity by myosins IIA and IIB in migrating cells. J Cell Biol. 2007;176:573–80. doi: 10.1083/jcb.200612043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simpson KJ, Selfors LM, Bui J, Reynolds A, Leake D, Khvorova A, Brugge JS. Identification of genes that regulate epithelial cell migration using an siRNA screening approach. Nat Cell Biol. 2008;10:1027–38. doi: 10.1038/ncb1762. [DOI] [PubMed] [Google Scholar]
- 23.Dahl KN, Ribeiro AJ, Lammerding J. Nuclear shape, mechanics, and mechanotransduction. Circ Res. 2008;102:1307–18. doi: 10.1161/CIRCRESAHA.108.173989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Starr DA, Fridolfsson HN. Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu Rev Cell Dev Biol. 2010;26:421–44. doi: 10.1146/annurev-cellbio-100109-104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dupin I, Etienne-Manneville S. Nuclear positioning: mechanisms and functions. Int J Biochem Cell Biol. 2011;43:1698–707. doi: 10.1016/j.biocel.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 26.Gundersen GG, Worman HJ. Nuclear positioning. Cell. 2013;152:1376–89. doi: 10.1016/j.cell.2013.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Starr DA. A nuclear-envelope bridge positions nuclei and moves chromosomes. J Cell Sci. 2009;122:577–86. doi: 10.1242/jcs.037622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roux KJ, Crisp ML, Liu Q, Kim D, Kozlov S, Stewart CL, Burke B. Nesprin 4 is an outer nuclear membrane protein that can induce kinesin-mediated cell polarization. Proc Natl Acad Sci U S A. 2009;106:2194–9. doi: 10.1073/pnas.0808602106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee JS, Hale CM, Panorchan P, Khatau SB, George JP, Tseng Y, Stewart CL, Hodzic D, Wirtz D. Nuclear lamin A/C deficiency induces defects in cell mechanics, polarization, and migration. Biophys J. 2007;93:2542–52. doi: 10.1529/biophysj.106.102426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lombardi ML, Jaalouk DE, Shanahan CM, Burke B, Roux KJ, Lammerding J. The interaction between nesprins and sun proteins at the nuclear envelope is critical for force transmission between the nucleus and cytoskeleton. J Biol Chem. 2011;286:26743–53. doi: 10.1074/jbc.M111.233700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khatau SB, Bloom RJ, Bajpai S, Razafsky D, Zang S, Giri A, Wu PH, Marchand J, Celedon A, Hale CM, et al. The distinct roles of the nucleus and nucleus-cytoskeleton connections in three-dimensional cell migration. Sci Rep. 2012;2:488. doi: 10.1038/srep00488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Joseph J, Dasso M. The nucleoporin Nup358 associates with and regulates interphase microtubules. FEBS Lett. 2008;582:190–6. doi: 10.1016/j.febslet.2007.11.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salpingidou G, Smertenko A, Hausmanowa-Petrucewicz I, Hussey PJ, Hutchison CJ. A novel role for the nuclear membrane protein emerin in association of the centrosome to the outer nuclear membrane. J Cell Biol. 2007;178:897–904. doi: 10.1083/jcb.200702026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dupin I, Sakamoto Y, Etienne-Manneville S. Cytoplasmic intermediate filaments mediate actin-driven positioning of the nucleus. J Cell Sci. 2011;124:865–72. doi: 10.1242/jcs.076356. [DOI] [PubMed] [Google Scholar]
- 35.Vallee RB, Stehman SA. How dynein helps the cell find its center: a servomechanical model. Trends Cell Biol. 2005;15:288–94. doi: 10.1016/j.tcb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 36.Tsai JW, Bremner KH, Vallee RB. Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat Neurosci. 2007;10:970–9. doi: 10.1038/nn1934. [DOI] [PubMed] [Google Scholar]
- 37.Wu J, Lee KC, Dickinson RB, Lele TP. How dynein and microtubules rotate the nucleus. J Cell Physiol. 2011;226:2666–74. doi: 10.1002/jcp.22616. [DOI] [PubMed] [Google Scholar]
- 38.Khatau SB, Kim DH, Hale CM, Bloom RJ, Wirtz D. The perinuclear actin cap in health and disease. Nucleus. 2010;1:337–42. doi: 10.4161/nucl.1.4.12331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tomar A, Schlaepfer DD. Focal adhesion kinase: switching between GAPs and GEFs in the regulation of cell motility. Curr Opin Cell Biol. 2009;21:676–83. doi: 10.1016/j.ceb.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maniotis AJ, Chen CS, Ingber DE. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc Natl Acad Sci U S A. 1997;94:849–54. doi: 10.1073/pnas.94.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang N, Tytell JD, Ingber DE. Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nat Rev Mol Cell Biol. 2009;10:75–82. doi: 10.1038/nrm2594. [DOI] [PubMed] [Google Scholar]
- 42.Khatau SB, Kusuma S, Hanjaya-Putra D, Mali P, Cheng L, Lee JS, Gerecht S, Wirtz D. The differential formation of the LINC-mediated perinuclear actin cap in pluripotent and somatic cells. PLoS One. 2012;7:e36689. doi: 10.1371/journal.pone.0036689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dujardin DL, Barnhart LE, Stehman SA, Gomes ER, Gundersen GG, Vallee RB. A role for cytoplasmic dynein and LIS1 in directed cell movement. J Cell Biol. 2003;163:1205–11. doi: 10.1083/jcb.200310097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim DH, Khatau SB, Feng Y, Walcott S, Sun SX, Longmore GD, Wirtz D. Actin cap associated focal adhesions and their distinct role in cellular mechanosensing. Sci Rep. 2012;2:555. doi: 10.1038/srep00555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anderson KI, Wang YL, Small JV. Coordination of protrusion and translocation of the keratocyte involves rolling of the cell body. J Cell Biol. 1996;134:1209–18. doi: 10.1083/jcb.134.5.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fridolfsson HN, Starr DA. Kinesin-1 and dynein at the nuclear envelope mediate the bidirectional migrations of nuclei. J Cell Biol. 2010;191:115–28. doi: 10.1083/jcb.201004118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Friedl P, Wolf K, Lammerding J. Nuclear mechanics during cell migration. Curr Opin Cell Biol. 2011;23:55–64. doi: 10.1016/j.ceb.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mili S, Moissoglu K, Macara IG. Genome-wide screen reveals APC-associated RNAs enriched in cell protrusions. Nature. 2008;453:115–9. doi: 10.1038/nature06888. [DOI] [PMC free article] [PubMed] [Google Scholar]