Abstract
The Runx family genes encode transcription factors that play key roles in hematopoiesis, skeletogenesis and neurogenesis and are often implicated in diseases. We describe here the cloning and characterization of Runx1, Runx2, Runx3 and Runxb genes in the elephant shark (Callorhinchus milii), a member of Chondrichthyes, the oldest living group of jawed vertebrates. Through the use of alternative promoters and/or alternative splicing, each of the elephant shark Runx genes expresses multiple isoforms similar to their orthologs in human and other bony vertebrates. The expression profiles of elephant shark Runx genes are similar to those of mammalian Runx genes. The syntenic blocks of genes at the elephant shark Runx gene loci are highly conserved in human, but represented by shorter conserved blocks in zebrafish indicating a higher degree of rearrangements in this teleost fish. Analysis of promoter regions revealed conservation of binding sites for transcription factors, including two tandem binding sites for Runx that are totally conserved in the distal promoter regions of elephant shark Runx1-3. Several conserved noncoding elements (CNEs), which are putative cis-regulatory elements, and miRNA binding sites were identified in the elephant shark and human Runx gene loci. Some of these CNEs and miRNA binding sites are absent in teleost fishes such as zebrafish and fugu. In summary, our analysis reveals that the genomic organization and expression profiles of Runx genes were already complex in the common ancestor of jawed vertebrates.
Introduction
The Runt domain transcription factor, known as the polyomavirus enhancer-binding protein 2 (PEBP2) or core-binding factor (CBF) is a heterodimer of α and β subunits. In humans, the α-subunit comprises three proteins, RUNX1, RUNX2 and RUNX3 that contain an evolutionarily conserved 128 amino acid Runt domain responsible for DNA binding and heterodimerization with the β-subunit. The β-subunit includes a single protein, RUNX β (also known as PEBP2β or CBFβ) that does not contain a DNA-binding domain but allosterically enhances the DNA-binding activity of the α-subunit and regulates its turnover by protecting it from ubiquitin-proteasome-mediated degradation [1], [2].
RUNX1-3 are key transcriptional regulators involved in several major developmental pathways including hematopoiesis, neurogenesis and skeletogenesis. RUNX1 is among the most frequently mutated genes in human leukemias [3], [4], [5]. In mice, Runx1 is critical for the generation and maintenance of hematopoietic stem cells (HSC) [6], [7]. In addition, Runx1 is involved in the development of skeletal muscle [8], neurons [9] and hair follicles [10]. On the other hand, Runx2 is indispensable for bone development, as evidenced by Runx2-/- mice which lack ossified skeleton and therefore die from respiratory failure shortly after birth [11]. In humans, haploinsufficiency of RUNX2 is associated with cleidocranial dysplasia (CCD), an autosomal dominant skeletal disorder [12]. Runx3 is expressed in a wide range of tissues and has diverse biological functions. It plays roles in the regulation of epithelial homeostasis in the gastrointestinal tract [13], [14], T-cell development during thymopoiesis [15] and differentiation of immune cells including natural killer cells [16], dendritic cells [17] and B cells [18]. Runx3 is also crucial for the differentiation of proprioceptive dorsal root ganglion (DRG) neurons [19] and chondrocyte maturation during skeletogenesis [20]. In human, RUNX3 has been implicated in a multitude of cancers where it can function as a tumor suppressor or oncogene [21], [22].
Given the importance of Runx genes in major developmental pathways and human diseases, orthologs of Runx have been characterized in phylogenetically diverse organisms to facilitate better understanding of their origin and functions. Runx genes have undergone duplications independently in some invertebrate lineages (e.g., fruit fly, mosquito) and in the stem vertebrate lineage followed by acquisition of specialized functions such as hematopoiesis and eye development in Drosophila and bone development in vertebrates [23], [24]. Tetrapods contain three Runx genes that are orthologs of human RUNX1-3. Among teleost fishes, pufferfishes (fugu and Tetraodon) contain four Runx genes [25], [26], of which three are orthologs of mammalian Runx1-3. The fourth gene, called frRunt, is hypothesized to represent an ancestral vertebrate Runx gene that was subsequently lost in tetrapods [25]. In zebrafish, besides Runx1 and Runx3, there are two copies of Runx2 (Runx2a and Runx2b) which likely arose from the whole-genome duplication event in the teleost fish ancestor [27]. In contrast to multiple copies of α-subunit encoding Runx genes, Runxβ is present as a single copy in all metazoans analysed [24]. Studies into the co-evolution of Runxα and Runxβ subunits have reported similar evolutionary rates of these interacting proteins and evolutionary conservation of the structure of the Runxα-Runxβ-DNA complex, suggesting that these proteins have co-evolved to maintain their ability to interact and to coregulate the transcription of target genes [24].
Cartilaginous fishes (Chondrichthyes) are phylogenetically the oldest living group of jawed vertebrates (Gnathostomes) that diverged from bony vertebrates (Osteichthyes) approximately 450 million years (My) ago [28]. By virtue of their phylogenetic position, cartilaginous fishes are a useful reference for the study of the origin and evolution of jawed vertebrate genes and their regulation. Cartilaginous fishes are split into two groups: elasmobranchs comprising sharks, rays and skates; and holocephalans represented by chimaeras such as elephant shark (Callorhinchus milii). Coding sequences of Runx genes were previously cloned in an elasmobranch, the dogfish (Scyliorhinus canicula) [29]. Three Runx genes, orthologous to mammalian Runx1-3 were found to be expressed in the developing cartilage, teeth and placoid scales suggesting that they may be involved in the ancient processes of vertebrate skeletogenesis in this cartilaginous fish [29]. To improve our understanding of the evolution, function and regulation of Runx genes, we have now cloned Runx genes from the elephant shark and analysed their genomic organization. The elephant shark has the smallest genome among cartilaginous fishes and has been proposed as a model cartilaginous fish genome [30], [31]. Recently the whole genome sequence of the elephant shark was completed [32]. Its comparison with human and other vertebrate genomes indicated that elephant shark is the slowest evolving known vertebrate genome [32]. Furthermore, human and elephant shark were found to share twice as many conserved noncoding elements (CNE), which are putative cis-regulatory elements, as human and teleosts fishes [33]. Thus, elephant shark is a valuable reference genome for understanding the evolution of gene families and cis-regulatory networks in jawed vertebrates.
In this study, we have characterized three members of Runx family genes in the elephant shark, as well as the gene encoding the β-subunit, CmRunxb, by cloning of cDNA and mining the elephant shark genome database. We have determined the tissue-specific mRNA expression of CmRunx and showed that these patterns reflect those in mammalian tissues, which may point to evolutionarily conserved gene functions and developmental pathways. Comparisons of noncoding sequences in the elephant shark and other jawed-vertebrate Runx loci were able to identify CNEs that are conserved over 450 million years of vertebrate evolution and are likely to be cis-regulatory elements.
Results and Discussion
Cloning and characterization of elephant shark α-subunit Runx family members
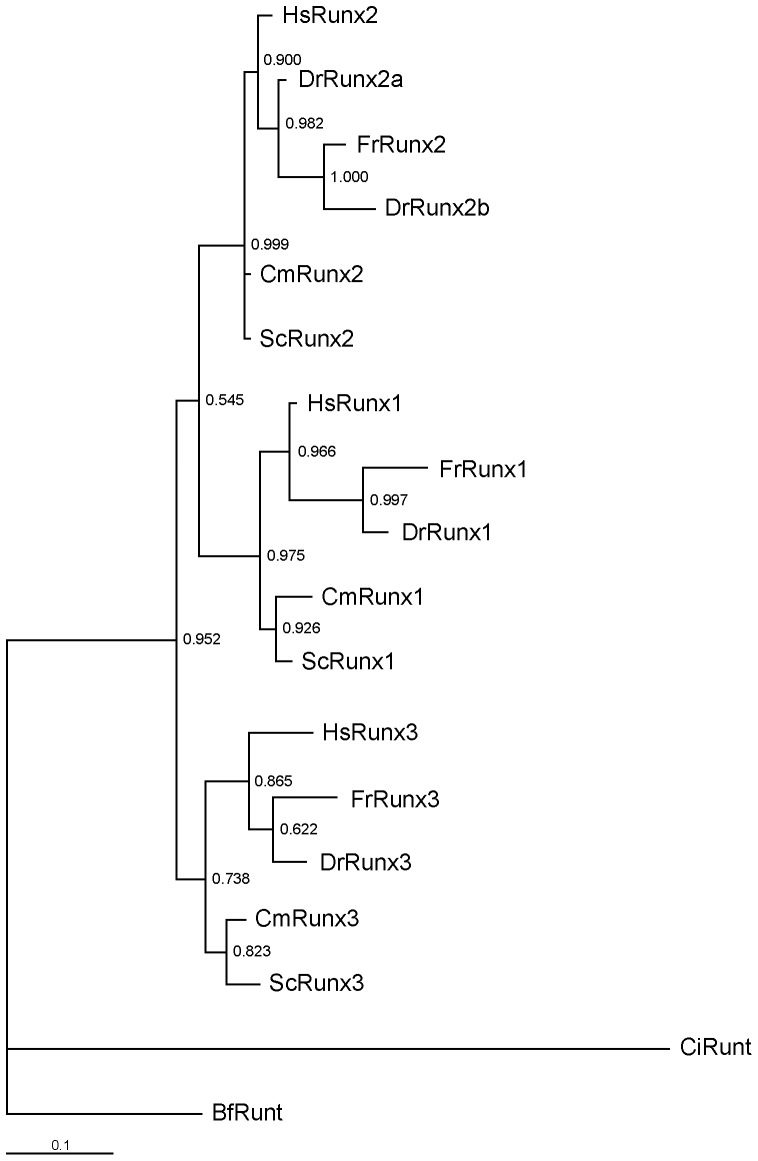
To identify Runx genes in the elephant shark, we Blast-searched the elephant shark 1.4× coverage sequence assembly [31] using human RUNX protein sequences and identified scaffolds containing fragments of Runx genes. By designing primers complementary to selected exons of Runx and carrying out RT-PCR and RACE using gill and kidney cDNA as templates, we were able to obtain full-length coding sequences for three elephant shark Runx α-subunit encoding genes. These sequences were then mapped to the whole-genome assembly of the elephant shark [32], and their precise exon-intron boundaries, transcription start sites (TSS) and UTRs were determined (Fig. 1). To confirm the orthology of the three elephant shark genes, we carried out phylogenetic analysis together with Runx sequences from representative chordates. The phylogenetic analysis identified the three genes as Runx1, Runx2 and Runx3 (Fig. 2) that are located on scaffolds #152, #106 and #121, respectively.
Figure 1. Exon-intron organization of elephant shark (Cm) Runx genes.
Schematic representation of the gene structures and transcript isoforms of (A) CmRunx1, (B) CmRunx2 and (C) CmRunx3. Exons are indicated by boxes. The vertical dashed lines indicate internal splice sites located within the coding exon. Exons constituting the Runt domain are indicated in grey. The two alternative promoters are denoted as P1 and P2. Crosshatched boxes indicate 5′- and 3′ UTRs. The asterisk (*) indicates an exon in CmRunx1 that is absent in mammalian Runx1. Not drawn to scale.
Figure 2. Phylogenetic tree (Bayesian inference) of chordate Runx sequences.
Values adjacent to the nodes represent branch support (Bayesian posterior probability). Lancelet (Branchiostoma floridae) Runt (BfRunt) was used as the outgroup. Hs, human; Dr, Danio rerio; Fr, Fugu (Takifugu) rubripes; Cm, Callorhinchus milii; Sc, Scyliorhinus canicula; Ci, Ciona intestinalis.
All three elephant shark Runx genes encode a highly conserved Runt domain (Fig. 1). Additionally, like all mammalian Runx genes characterized to date, each of the elephant shark Runx genes contain two alternative promoters, P1 (distal) and P2 (proximal), that are separated by a characteristically large intron. The exon-intron boundaries of the three elephant shark Runx genes are largely conserved compared to their orthologs in human and other jawed vertebrates. However, CmRunx1 was found to contain an extra exon (exon 4.1) (Fig. 1A) which has not been characterized in any of the known Runx1 genes, except that of the dogfish, S. canicula, suggesting that this exon might be a chondrichthyes-specific feature. In addition, CmRunx2 lacks the equivalents of exons 5.2 and 6.1 found in the mammalian Runx2 gene [34] (Fig. 1B). These exons are also absent in chicken, frog and teleost fishes [26], [35], indicating that they were recruited in a mammalian ancestor and likely to perform functions that are specific to mammals. Among the three elephant shark Runx genes, Runx3 is the shortest (CmRunx1, 114 kb; CmRunx2, 139 kb; CmRunx3, 74 kb) similar to its orthologs in mammals (HsRunx1, 261 kb; HsRunx2, 223 kb; HsRunx3, 65 kb; MmRunx1, 225 kb; MmRunx2, 211 kb; MmRunx3, 57 kb). The short sequence of the Runx3 genes is likely due to the loss of an exon equivalent to exon 5.1 in Runx1 and Runx2 genes (Fig. 1).
Mammals express a variety of isoforms for each of the Runx genes arising from transcription initiation at two alternative promoters (P1 and P2) and alternative splicing of exons. Isoforms generated from the P1 promoter possess unique N-terminal sequences arising from transcription initiation in exon 1 and splicing that bypasses the P2 initiator ATG on exon 2. The use of alternative promoters contributes to the transcriptional and functional complexity of the Runx genes, as evidenced by expression studies and analyses of P1 and P2 promoter knockout mice wherein these transcripts display distinct patterns of expression and exert divergent biological functions during development [36], [37]. We could identify 2 to 4 isoforms for each of the elephant shark Runx genes that are homologous to the isoforms in mammals, indicating that the genomic organization and transcriptional profile of Runx genes were already complex in the common ancestor of jawed vertebrates.
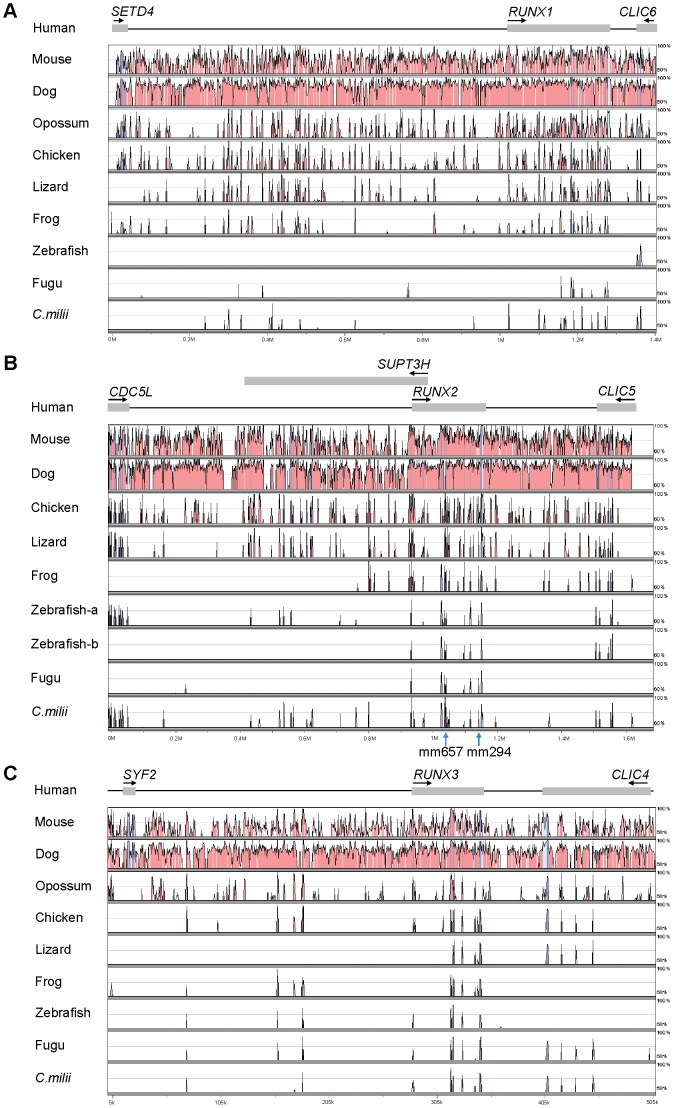
The availability of the whole-genome sequence of elephant shark enabled us to compare the synteny of genes at Runx loci in elephant shark and other sequenced vertebrate genomes. The synteny of genes at the elephant shark Runx1, Runx2 and Runx3 loci is highly conserved in tetrapods (Fig. 3). A comparison of syntenic genes across the three Runx gene loci indicates that paralogs of Clic and Rcan genes are present in all three Runx gene loci. This indicates that three Runx gene loci in the jawed vertebrates are the result of duplication of an ancestral locus that comprised Runx, Clic and Rcan genes in that order. Of note are the characteristic “interlocked” gene structures of Runx2 and Supt3h in all the jawed vertebrates, except the duplicated locus in zebrafish (Runx2b locus) in which Supt3h has been lost. Indeed, a Supt3h gene resides next to Runt gene in the genome of the most basally branching chordate, the amphioxus (chrUn: 270,089,714–270,297,951; Mar 2006-JGI1.0/braFlo1) indicating that their linkage is an ancient feature. It can therefore be inferred that linkage of Supt3h to Runt gene was retained in the jawed vertebrate Runx2 locus whereas the paralogs of Supt3h in Runx1 and Runx3 loci were lost secondarily.
Figure 3. Synteny of genes in the Runx loci of elephant shark and selected bony vertebrates.
(A) Runx1 locus, (B) Runx2 locus and (C) Runx3 locus. Genes are represented by arrows. Genes with conserved synteny are coloured. Clusters of some non-syntenic genes are represented as white boxes and labelled in brackets. The gene order is from Ensembl (www.ensembl.org) and the elephant shark genome assembly (http://esharkgenome.imcb.a-star.edu.sg/).
In contrast to the relatively larger blocks of conserved syntenic genes in elephant shark and tetrapod Runx loci, the conserved syntenic blocks in zebrafish Runx loci are shorter (Fig. 3). This indicates that Runx loci in zebrafish have experienced a higher degree of rearrangements, and that the arrangement of genes in human Runx loci is more similar to that of elephant shark than zebrafish, underscoring the importance of elephant shark as a useful model reference genome for studying the origin and evolution of human gene loci.
Comparison of elephant shark Runx α-subunit protein sequences
Alignment of elephant shark Runx1, 2 and 3 protein sequences with human RUNX1, 2 and 3 sequences revealed several highly conserved protein domains. Among these is the 128 amino acid Runt domain which is almost totally conserved across elephant shark and human Runx proteins (Fig. 4). Within this domain, amino acid residues for DNA-binding as well as sequence motifs that interact with the β-subunit [38], [39] are well conserved. In addition, the nuclear localization signal (NLS), PY and VWRPY motifs, and sites of phosphorylation by Erk2/cdc2 (Fig. 4B) are also highly conserved. The PY and VWRPY motifs mediate transcriptional activity of Runx proteins by recruiting different interaction proteins. The PY motif in the transactivation domain mediates the binding of Runx proteins to WW domain-containing proteins, such as YAP and TAZ [40], [41]. Found invariably in all known vertebrate Runx proteins, the C-terminal pentapeptide VWRPY, is responsible for the repressive function of Runx proteins, through the recruitment of transcriptional co-repressors TLE/Groucho [42]. In human RUNX1, the serine residues S249 and S273 are each followed by a proline residue and acts as phosphorylation sites for ERK [43], [44]. These phosphorylation sites are also conserved in human RUNX2 (S280 and T305) and elephant shark Runx1 and Runx2. Additionally, the consensus phosphorylation site for CDC2, (S/T)PX(R/K), at which serine residue S451 of human RUNX2 was reported to be phosphorylated [45], is remarkably conserved in all human and elephant shark Runx proteins (Fig. 4B).
Figure 4. Runx α-subunit proteins Runx1, Runx2 and Runx3 in elephant shark and human.
(A) Schematic representation of Runx α-subunit proteins. The characteristic domains found in all Runx proteins are indicated to show their relative positions along the protein. (B) Alignment of elephant shark and human Runx α-subunit proteins. The first block shows the amino-terminal part of the protein derived from the P1 promoter that differs from that derived from the P2 promoter. The highly conserved Runt domain is boxed with a red line. Within the Runt domain, surfaces involved in DNA contact and interaction with the β-subunit are denoted by black and blue lines, respectively. Cysteine residues involved in the redox regulation of DNA-binding activity are indicated with asterisks. Nuclear localization signal (NLS) is demarcated by a green dashed box. The PY and VWRPY motifs are indicated by orange and blue boxes, respectively. The transactivation domain (TAD) is highlighted in blue and the inhibitory domain (ID) is boxed by purple dotted lines. The nuclear matrix translocalization signal (NMTS) is boxed with a brown line. Minimal consensus sequences for phosphorylation by Erk or cdc2 are boxed by dashed and solid black lines, respectively. The residue targeted for phosophorylation is indicated by ℗. Cm, Callorhinchus milii; Hs, Homo sapiens.
The N-termini of the P1 and P2 isoforms beginning with MAS(N/D)S and MR(I/V)PV, respectively, are highly conserved across all three Runx proteins between elephant shark and their mammalian orthologs. A distinctive feature present only in CmRunx1 (and its dogfish ortholog) is a short stretch of 18 amino acids downstream of the Runt domain (Fig. 4). This unique sequence is encoded by an extra exon 4.1 that is alternatively spliced in one of the isoforms of CmRunx1 (Fig. 1). Whether these additional amino acids alter the structure and function of the CmRunx1 isoform is unknown, although one might speculate that the presence of this stretch within the nuclear localization signal (Fig. 4B, boxed in green dotted lines) might affect the isoform's nuclear translocation.
Of the three mammalian Runx genes, unique to Runx2 is the Q/A stretch, an activation domain located N-terminal to the Runt domain that is composed of successive polyglutamine and polyalanine amino acids. Whilst human RUNX2 contains a stretch of 23 glutamine residues followed by 17 alanine residues, no glutamine repeats but a stretch of 10 alanine residues are found in elephant shark Runx2 (Fig. 4). The QA domain has been proposed to contribute to the osteoblast-specific function of Runx2. This domain controls transactivation function of the Runx2 and also inhibits the heterodimerization of Runx2 with the β-subunit [46]. In carnivoran mammals, the ratio of the glutamines to alanines (QA ratio) in Runx2 is strongly correlated with facial length [47]. In human, insertion in the alanine tract (23Q/27A) was observed in a CCD patient, suggesting that QA variations of RUNX2 may influence skeletal phenotype [48]. Indeed, glutamine repeat variants in RUNX2 have been recently found to be associated with a lower bone mineral density in the general human population [49]. Therefore, given the significance of the QA domain for skeletal functions of Runx2, it would be of interest to study the physiological relevance of this domain of Runx2 in cartilaginous fishes like the elephant shark which lack ossified endoskeleton.
Altogether, our comparisons indicate that protein domains of elephant shark Runx proteins are well conserved with those of human and demonstrate the highly conserved nature of Runx proteins in all jawed vertebrates.
Expression profile of elephant shark α-subunit Runx genes
We investigated the expression patterns of Runx genes in various tissues of adult elephant shark by quantitative RT-PCR. CmRunx1 is highly expressed in the gills, muscle, testis, skin and spleen (Fig. 5A). High CmRunx1 expression in the gill, a tissue enriched in blood cells, as well as the spleen, a lymphomyeloid tissue of the shark, is concordant with the integral function of Runx1 in hematopoiesis. In addition, high expression of CmRunx1 in the muscle reflects its known role in mammalian skeletal muscle development. Although Runx1 is not known to be expressed at high levels in testis of mammals, high expression of Runx1 has been reported in another cartilaginous fish, the dogfish. Thus, Runx1 might have a function in testis that is specific to cartilaginous fishes. However, this hypothesis remains to be verified. Furthermore, a significant level of Runx1 expression in the skin of the elephant shark is consistent with previous reports of Runx expression in placoid scales which are small conical structures in the skin of cartilaginous fishes [29].
Figure 5. Expression patterns of elephant shark Runx genes.
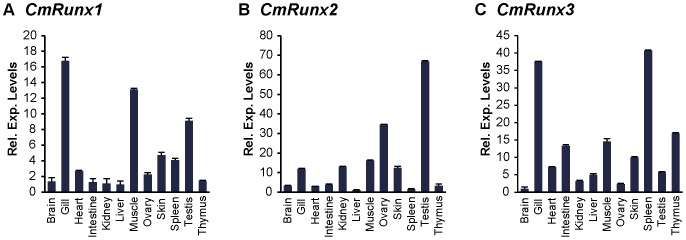
Relative expression levels of (A) CmRunx1, (B) CmRunx2 and (C) CmRunx3 in various tissues of the elephant shark determined by qRT-PCR.
Particularly striking is the significant expression of Runx2 in the gonads (ovary and testis) of the elephant shark (Fig. 5B). This expression pattern is reminiscent of that in mice, in which ovary and testis were reported to be the predominant sites of non-skeletal tissue Runx2 expression [50]. In human, recent investigations into the role of RUNX2 in the ovary have highlighted its importance in ovulation and luteinisation [51], [52]. These congruent patterns of expression may suggest a role for Runx2 in similar reproductive processes in the elephant shark. Given the indispensable role of Runx2 in osteogenesis, analysis of Runx2 expression in the cartilaginous/skeletal elements of the elephant shark would have been informative, but due to the unavailability of cartilage tissue from elephant shark, we could not verify expression of Runx genes in the cartilage. However, in the dogfish, all three Runx genes were found highly expressed in the visceral cartilage [29], indicating the possible prominent role of Runx genes in the unossified skeleton of cartilaginous fishes.
The immune system of cartilaginous fishes share several features common to other jawed vertebrates. These include tissue sites for immune cell production (thymus and spleen), specialized cell types for innate and adaptive immunity and genes encoding proteins for immune function, such as various immunoglobulin subtypes, TCR, MHC and cytokine-like molecules [53], [54]. The elephant shark Runx3 is highly expressed in the gill, spleen and thymus (Fig. 5C), which are all considered to be lymphomyeloid tissues in cartilaginous fishes. Based on the well-established roles of Runx3 in the development and function of diverse immune related cell types in mammals, it appears that Runx3 may have already established roles in the development of the cellular immune system in the common ancestor of jawed vertebrates.
Analysis of elephant shark α-subunit Runx promoter regions
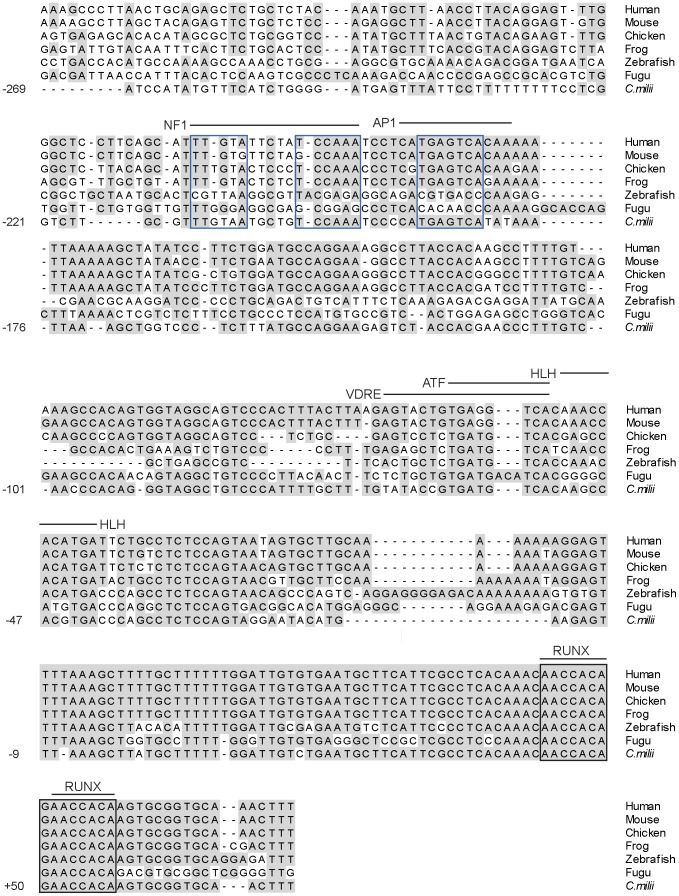
The P1 promoter regions of mammalian Runx1, 2 and 3 genes harbour binding sites for transcription factors that are critical for their transcriptional regulation. In particular, P1 promoters of mammalian Runx1, 2 and 3 contain two tandem binding sites for Runx. These sites have been implicated in auto and cross regulation of the three Runx genes [55], [56]. To verify if these binding sites are conserved in elephant shark, we compared the P1 promoter regions of Runx1-3 in elephant shark, fugu/zebrafish and tetrapods. Remarkably, the tandem pair of Runx binding sites is totally conserved in P1 promoter regions of elephant shark Runx1 (Fig. S1A), Runx2 (Fig. 6) and Runx3 (Fig. S1B) loci. The conservation of these binding sites in elephant shark and tetrapods indicate that the auto/cross regulation of the three Runx genes is an ancient feature of Runx genes and was present in the single ancestral Runx locus that gave rise to the three Runx loci in jawed vertebrates. Interestingly, in contrast to total conservation of these binding sites in elephant shark, they are less conserved in Runx1 and 3 loci of fugu and zebrafish (Fig. S1 and 6). It remains to be seen if the divergent binding sites in teleosts are still functional and whether Runx genes in teleosts are subject to auto/cross regulation similar to that of elephant shark and tetrapods.
Figure 6. P1 promoter region of Runx2 gene from elephant shark and selected bony vertebrates.
Alignment of the P1 promoter regions is shown. The numbers to the left of the alignment indicate positions relative to the TSS (+1) of elephant shark. Sequences between –124 and -102 that contain insertions/deletions in some species are not shown. The conserved tandem Runx binding sites are boxed by black lines. Putative transcription factor binding sites are indicated by black solid lines and labelled. Core sites for NF1 and AP1 binding are boxed by blue lines.
In addition to the two Runx binding sites, the P1 promoter of the mammalian Runx2 locus contains well-characterized binding sites for other transcription factors that regulate the expression of Runx2. These include (i) a vitamin D response element (VDRE) with an overlapping ATF motif, that binds the VDR/RXR heterodimer and mediates the suppression of RUNX2 transcription by the steroid hormone 1,25-(OH)2-vitamin D3 [57]; (ii) a Helix-loop-Helix consensus motif that is critical for the basal transcription of Runx2; and (iii) NF1 and AP1 motifs required for osteoblast-specific activity of the Runx2 P1 promoter [58] (Fig. 6). All these binding sites, except the VDRE site, are conserved highly in the elephant shark Runx2 gene, but to a lesser extent in the zebrafish and fugu Runx2 genes (Fig. 6).
Conserved noncoding elements in human and elephant shark α-subunit Runx loci
Runx genes exhibit highly specific expression in several tissues and cell lineages that is critical for their diverse roles in development and disease. However, the transcriptional regulation of Runx genes has not been well characterized. Comparative genomics is a powerful strategy for identifying evolutionarily conserved cis-regulatory elements. Since functional elements evolve slowly compared to their flanking sequences, cis-regulatory elements can be identified as conserved noncoding elements (CNEs) in the genomes of related species. Functional assay of CNEs in transgenic systems has indicated that many of them drive reporter gene expression in specific tissues [59]. Indeed, one such CNE predicted in the first intron of Runx1 and conserved in human, mouse, rat, dog, horse and opossum, was shown to drive highly-specific expression to hematopoietic stem cells (HSCs) of mouse [60], indicating that it is an HSC enhancer.
The common ancestor of human and elephant shark diverged about 450 million years ago. Any CNEs conserved in these vertebrates over 900 million years of divergent evolution are likely to be functional elements and must be playing a fundamental role in the regulation of their associated genes. In order to identify such evolutionarily conserved cis-regulatory elements, we aligned the three human Runx loci with their corresponding orthologs from elephant shark and other jawed vertebrates and predicted CNEs that are >65% identical across 50 bp or more. Several CNEs that met these criteria were identified in the three Runx loci of human and elephant shark (Fig. 7) (Table S2). Notably, two of the CNEs located in the intronic regions of human RUNX2 (Runx2_CNE7 and Runx2_CNE8) overlap two functionally characterized enhancers, denoted as mm657 and mm924 (Fig. 7B), that drive reporter gene expression in the branchial arches and facial mesenchyme respectively of transgenic mouse embryos [61]. The overlap of functional enhancers with the predicted human-elephant shark CNEs provides further support to the notion that CNEs conserved in human and elephant shark are likely to be functional cis-regulatory elements. Thus, they are strong candidates for functional assays that can help to identify enhancers of human RUNX genes.
Figure 7. Conserved noncoding elements (CNEs) in Runx loci.
VISTA plots obtained from the global alignment between the human, mouse, dog, opossum, chicken, lizard, frog, zebrafish, fugu and elephant shark (A) Runx1, (B) Runx2 and (C) Runx3 loci are shown. Sequence for opossum Runx2 locus is not available. Human sequence is used as the reference sequence. Conserved sequences were predicted at a cut-off of ≥65% identity across >50 bp windows and are represented by peaks. Blue peaks denote conserved coding exons, pink peaks conserved noncoding regions (CNRs) and cyan peaks untranslated regions (UTRs). Experimentally verified CNEs (mm657 and mm924) in the Runx2 locus are indicated with blue arrows below the x-axis and labelled.
Interestingly some of the CNEs conserved in the Runx1-3 loci of human and elephant shark are missing in zebrafish and fugu (Fig. 7A and B), which is consistent with previous observations that CNEs in teleosts have been evolving faster than in elephant shark and other vertebrates [62]. This further emphasizes the importance of elephant shark as a model genome of choice for identifying evolutionarily conserved cis-regulatory elements in the human genome.
miRNA binding sites in elephant shark α-subunit Runx genes
Apart from transcriptional control by alternate promoters and cis-regulatory elements, expression of Runx1-3 is also subjected to post-transcriptional regulation by miRNAs [63], [64], [65]. Several miRNAs have been shown to regulate Runx1 expression levels and consequently its function in hematopoietic differentiation [64]. Among the highly conserved miRNA binding sites in mammalian Runx1 3′UTR, two clustered sites for miR-27a are remarkably well conserved in the 3′UTR of elephant shark Runx1 but not in the teleost fishes, zebrafish and fugu (Fig. 8A). In mammals, miR-27 plays regulatory roles during megakaryocytic and granulocytic differentiation by attenuating Runx1 expression and engaging in feedback loops with Runx1 [64]. Conservation of miR-27 binding sites, coupled with the presence of miR-27 in the elephant shark (GenBank Accession number JX994340) suggests that CmRunx1 may similarly be post-transcriptionally controlled by miR-27 in hematopoietic lineages of the elephant shark.
Figure 8. miRNA binding sites in the 3′UTR of Runx1 and Runx2 genes in elephant shark and selected gnathostomes.
Schematic diagram of (A) Runx1 and (B) Runx2 3′UTRs and miRNA binding sites. The last coding exon is represented by a rectangle and 3′UTR by a grey line. Positions of miRNA binding sites are indicated by vertical lines. Binding sites conserved in human and elephant shark are shown in red. Zebrafish and fugu Runx1 loci are not shown as they do not contain any conserved miRNA binding sites.
Eleven Runx2-targeting miRNAs have been reported to control the physiological levels of Runx2 protein during osteogenesis and chondrocyte maturation [65]. Of these, binding sites for miR-23, miR-218 and miR-338 were found conserved in the 3′UTR of elephant shark Runx2, while only that for miR-28 is present in zebrafish and fugu Runx2 (Fig. 8B). Since miR-23 (JX994633), miR-218 (JX994383) and miR-338 (JX994406) are present in the elephant shark, these conserved miRNAs are likely to be involved in the regulation of Runx2 expression in elephant shark, possibly during chondrogenic differentiation through mechanisms similar to that in other jawed vertebrates.
In contrast to Runx1 and Runx2, no miRNA binding sites were found to be conserved in the rather short 3′UTR of elephant shark Runx3 (data not shown).
Cloning and characterization of elephant shark β-subunit Runx gene
We cloned the full-length coding sequence of the elephant shark gene encoding the β-subunit of the CBF heterodimeric complex, Runxb. The exon-intron organization of elephant shark Runxb (CmRunxb) gene is identical to that of its human ortholog (Fig. 9A) [66]. Furthermore, CmRunxb is transcribed into at least three isoforms that are homologous to the human RUNXB type 1, 2 and 3 isoforms. The CmRunxb type 1, 2 and 3 isoforms encode proteins of 188, 185 and 156 amino acids, respectively (Fig. 9B). CmRunxb Type 1 and Type 3 isoforms are largely similar, except for the absence of exon 5 in the Type 3, as a result of exon skipping. CmRunxb Type 1 and Type 2 isoforms differ in their C-terminal ends, resulting from the use of alternative termination codon and splice donor/acceptor sites in exons 5 and 6 (Fig. 9A). At the protein level, CmRunxβ and human RUNXβ show high conservation of amino acid residues 1-165 (Fig. 9B), of which the N-terminal 135 amino acids required for its heterodimerization with the α-subunit and DNA binding [67] are almost perfectly conserved. We investigated the expression patterns of Runxb isoforms in various tissues of adult elephant shark by quantitative RT-PCR. All isoforms display a ubiquitous pattern of expression, with high levels of expression in the gill, heart, ovary and testis of the elephant shark (Fig. 9C). The functional significance of these isoforms that are conserved in human, elephant shark and other jawed vertebrates remains to be investigated. In an attempt to identify conserved cis-regulatory elements for Runxb locus, we searched for CNEs in the human and elephant shark Runxb loci but found none (data not shown).
Figure 9. Exon-intron organization and protein sequence encoded by the elephant shark Runxβ.
(A) Schematic representation of the genomic structure and the three transcripts cloned (CmRunxb types 1,2 and 3). Exons are indicated by boxes. The vertical dashed lines indicate internal splice sites located within a coding exon. The 5′ and 3′UTRs are represented as crosshatched boxes. (B) Alignment of elephant shark and human RUNXβ amino acid sequences using ClustalW. Conserved residues are shaded grey. Cm, Callorhinchus milii; Hs, Homo sapiens. (C) Expression patterns of elephant shark Runxb transcripts. Relative expression levels of Runxb Types 1+2 and Type 3 in various tissues of the elephant shark determined by qRT-PCR.
Materials and Methods
Ethics statement
Elephant sharks were caught off the coast of Western Port Bay, Victoria (Australia) by licensed commercial fishermen. The samples used in this study were taken from animals that were already dead when the fishermen returned to the fishing jetty. Pieces of tissue were taken from the dead fish, frozen and transported to the laboratory for extraction of DNA and RNA [68], [69]. The extraction of DNA and RNA from frozen elephant shark samples was approved by the Institutional Animal Care and Use Committee (IACUC) of the Institute of Molecular and Cell Biology.
Identification of Runx gene fragments in the elephant shark genome database
The elephant shark 1.4× coverage sequence assembly (http://esharkgenome.imcb.a-star.edu.sg/) was searched with human RUNX1, RUNX2, RUNX3 and RUNXβ protein sequences using ‘TBLASTN’ algorithm. The following contigs that contained fragments of various Runx genes were identified in the assembly: Runx1- AAVX01052077, AAVX01261782, AVX01381831 and AAVX01569464; Runx2 - AAVX01175228, AAVX01545426, AAVX01631487, AAVX01048938, AAVX01307838 and AAVX01048937; Runx3 - AAVX01039927, AAVX01288244, AAVX01157201 and AAVX01598430 and Runxb- AAVX01192354, AAVX01224391 and AAVX01471917.
RT-PCR and 5′ and 3′ RACE
Primers were designed for representative exons of Runx genes identified in the elephant shark scaffolds and the full-length coding sequences of the genes were obtained by RT-PCR, 5′ RACE and/or 3′ RACE (primer sequences available upon request). 5′ and 3′ RACE were performed on single-stranded cDNA prepared from total RNA using the SMART RACE cDNA Amplification kit (Clontech, Palo Alto, CA) in a nested PCR. All RT-PCR and RACE products were cloned into the pGEM-T Easy Vector (Promega), and sequenced completely using the BigDye Terminator Cycle Sequencing Kit (Applied Biosystems, USA) on an ABI 3730xl capillary sequencer (Applied Biosystems, USA). During the course of this study, the whole-genome sequence of elephant shark became available [32]. The full-length cDNA sequences were mapped to the whole-genome assembly and the sequences of scaffolds that contained the genes (scaffolds #152, #106, #121 and #127) were extracted for further annotation and synteny analysis. Sequences for various isoforms of elephant shark Runx genes have been deposited in GenBank (Accession numbers KJ150227-KJ150240).
Amino acid alignment and phylogenetic analysis
The protein sequences for human and other chordate Runx genes were retrieved from the National Center for Biotechnology Information (NCBI) database. Multiple sequence alignments were performed by using ClustalW. For phylogenetic analysis, gaps in the alignments were removed using Gblocks Server (ver. 0.91b) with default parameters [70]. The phylogenetic analysis was performed using MrBayes 3.2, employing Dayhoff+G as a substitution model and running 4 chains for 1,000,000 generations. Trees were sampled every 100 generations and according to a saturation curve of likelihood values, the first 2500 trees were discarded as burn-in. The Runt gene from Branchiostoma floridae was used as the outgroup.
Expression profiling of elephant shark Runx genes by qRT-PCR
Total RNA was extracted from various tissues of adult elephant shark using Trizol reagent (Gibco BRL, Grand Island, NY) according to manufacturer's protocol. Purified total RNA was reverse-transcribed into cDNA with Superscript II (Invitrogen, Carlsbad, CA). The single strand cDNA was used as a template in qRT-PCR reactions with KAPA SYBR FAST qPCR Kit reagents (KAPA Biosystems, Boston, MA). Sequences of primers used in qRT-PCR are given in Table S1. All primer pairs span at least one intron which helps to distinguish cDNA from genomic DNA products. Expression levels of Runx genes were normalized using β-actin gene as the reference.
Prediction of transcription factor binding sites (TFBS)
Analysis of TFBSs was carried out using JASPAR (http://jaspar.binf.ku.dk/cgi-bin/jaspar_db.pl). Only TFBS with prediction scores ≥ 9 were retained as putative TFBSs.
Prediction of conserved noncoding elements (CNEs)
Genomic sequences of the Runx gene loci for the following species were extracted from Ensembl release 73 [71]: human (GRCh37 assembly, February 2009), mouse (GRCm38, December 2011), dog (CanFam3.1, September 2011), opossum (MonDom5, October 2006), chicken (GalGal4, November 2011), lizard (AnoCar2.0, May 2010), frog (JGI 4.2, November 2009), zebrafish (Zv9, July 2010) and fugu (FUGU5, October 2011). Repetitive sequences were masked using CENSOR at default settings [72]. Multiple alignments of Runx gene loci sequences were generated using the global alignment program MLAGAN (http://genome.lbl.gov/vista/index.shtml) with human as the reference sequence. CNEs were predicted using a cutoff of ≥65% identity across 50-bp windows and visualized using VISTA (http://genome.lbl.gov/vista/index.shtml).
Supporting Information
P1 promoter region of Runx1 and Runx3 of elephant shark and selected bony vertebrates. Multiple sequence alignments of the P1 5′UTR regions of (a) Runx1 and (b) Runx3 are shown. The numbers to the left of the alignment indicate positions relative to the TSS (+1) of elephant shark (C. milii) genes. Corresponding regions from other bony vertebrates were aligned using ClustalW. The tandem Runx binding sites are boxed.
(PDF)
Primers used for qRT-PCR.
(PDF)
CNEs in the Runx loci of human and elephant shark.
(PDF)
Acknowledgments
We thank Alison P. Lee and Vydianathan Ravi for critical reading of the manuscript.
Funding Statement
Funding provided by Biomedical Research Council of A*STAR, Singapore. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Huang G, Shigesada K, Ito K, Wee HJ, Yokomizo T, et al. (2001) Dimerization with PEBP2beta protects RUNX1/AML1 from ubiquitin-proteasome-mediated degradation. The EMBO journal 20: 723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Adya N, Castilla LH, Liu PP (2000) Function of CBFbeta/Bro proteins. Semin Cell Dev Biol 11: 361–368. [DOI] [PubMed] [Google Scholar]
- 3. Osato M (2004) Point mutations in the RUNX1/AML1 gene: another actor in RUNX leukemia. Oncogene 23: 4284–4296. [DOI] [PubMed] [Google Scholar]
- 4. Osato M, Ito Y (2005) Increased dosage of the RUNX1/AML1 gene: a third mode of RUNX leukemia? Critical reviews in eukaryotic gene expression 15: 217–228. [DOI] [PubMed] [Google Scholar]
- 5. Speck NA, Gilliland DG (2002) Core-binding factors in haematopoiesis and leukaemia. Nature reviews Cancer 2: 502–513. [DOI] [PubMed] [Google Scholar]
- 6. Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR (1996) AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 84: 321–330. [DOI] [PubMed] [Google Scholar]
- 7. Jacob B, Osato M, Yamashita N, Wang CQ, Taniuchi I, et al. (2010) Stem cell exhaustion due to Runx1 deficiency is prevented by Evi5 activation in leukemogenesis. Blood 115: 1610–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang X, Blagden C, Fan J, Nowak SJ, Taniuchi I, et al. (2005) Runx1 prevents wasting, myofibrillar disorganization, and autophagy of skeletal muscle. Genes Dev 19: 1715–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Inoue K, Shiga T, Ito Y (2008) Runx transcription factors in neuronal development. Neural Dev 3: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Osorio KM, Lee SE, McDermitt DJ, Waghmare SK, Zhang YV, et al. (2008) Runx1 modulates developmental, but not injury-driven, hair follicle stem cell activation. Development 135: 1059–1068. [DOI] [PubMed] [Google Scholar]
- 11. Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, et al. (1997) Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89: 755–764. [DOI] [PubMed] [Google Scholar]
- 12. Otto F, Kanegane H, Mundlos S (2002) Mutations in the RUNX2 gene in patients with cleidocranial dysplasia. Human mutation 19: 209–216. [DOI] [PubMed] [Google Scholar]
- 13. Li QL, Ito K, Sakakura C, Fukamachi H, Inoue K, et al. (2002) Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell 109: 113–124. [DOI] [PubMed] [Google Scholar]
- 14. Brenner O, Levanon D, Negreanu V, Golubkov O, Fainaru O, et al. (2004) Loss of Runx3 function in leukocytes is associated with spontaneously developed colitis and gastric mucosal hyperplasia. Proc Natl Acad Sci U S A 101: 16016–16021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Taniuchi I, Osato M, Egawa T, Sunshine MJ, Bae SC, et al. (2002) Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell 111: 621–633. [DOI] [PubMed] [Google Scholar]
- 16. Ohno S, Sato T, Kohu K, Takeda K, Okumura K, et al. (2008) Runx proteins are involved in regulation of CD122, Ly49 family and IFN-gamma expression during NK cell differentiation. International immunology 20: 71–79. [DOI] [PubMed] [Google Scholar]
- 17. Fainaru O, Woolf E, Lotem J, Yarmus M, Brenner O, et al. (2004) Runx3 regulates mouse TGF-beta-mediated dendritic cell function and its absence results in airway inflammation. The EMBO journal 23: 969–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Watanabe K, Sugai M, Nambu Y, Osato M, Hayashi T, et al. (2010) Requirement for Runx proteins in IgA class switching acting downstream of TGF-beta 1 and retinoic acid signaling. Journal of immunology 184: 2785–2792. [DOI] [PubMed] [Google Scholar]
- 19. Inoue K, Ozaki S, Shiga T, Ito K, Masuda T, et al. (2002) Runx3 controls the axonal projection of proprioceptive dorsal root ganglion neurons. Nature neuroscience 5: 946–954. [DOI] [PubMed] [Google Scholar]
- 20. Soung do Y, Dong Y, Wang Y, Zuscik MJ, Schwarz EM, et al. (2007) Runx3/AML2/Cbfa3 regulates early and late chondrocyte differentiation. Journal of bone and mineral research: the official journal of the American Society for Bone and Mineral Research 22: 1260–1270. [DOI] [PubMed] [Google Scholar]
- 21. Lee CW, Chuang LS, Kimura S, Lai SK, Ong CW, et al. (2011) RUNX3 functions as an oncogene in ovarian cancer. Gynecologic oncology 122: 410–417. [DOI] [PubMed] [Google Scholar]
- 22. Kudo Y, Tsunematsu T, Takata T (2011) Oncogenic role of RUNX3 in head and neck cancer. Journal of cellular biochemistry 112: 387–393. [DOI] [PubMed] [Google Scholar]
- 23. Braun T, Woollard A (2009) RUNX factors in development: lessons from invertebrate model systems. Blood cells, molecules & diseases 43: 43–48. [DOI] [PubMed] [Google Scholar]
- 24. Sullivan JC, Sher D, Eisenstein M, Shigesada K, Reitzel AM, et al. (2008) The evolutionary origin of the Runx/CBFbeta transcription factors—studies of the most basal metazoans. BMC evolutionary biology 8: 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Glusman G, Kaur A, Hood L, Rowen L (2004) An enigmatic fourth runt domain gene in the fugu genome: ancestral gene loss versus accelerated evolution. BMC evolutionary biology 4: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ng CE, Osato M, Tay BH, Venkatesh B, Ito Y (2007) cDNA cloning of Runx family genes from the pufferfish (Fugu rubripes). Gene 399: 162–173. [DOI] [PubMed] [Google Scholar]
- 27. Christoffels A, Koh EG, Chia JM, Brenner S, Aparicio S, et al. (2004) Fugu genome analysis provides evidence for a whole-genome duplication early during the evolution of ray-finned fishes. Molecular Biology and Evolution 21: 1146–1151. [DOI] [PubMed] [Google Scholar]
- 28. Benton MJ, Donoghue PC (2007) Paleontological evidence to date the tree of life. Molecular biology and evolution 24: 26–53. [DOI] [PubMed] [Google Scholar]
- 29. Hecht J, Stricker S, Wiecha U, Stiege A, Panopoulou G, et al. (2008) Evolution of a core gene network for skeletogenesis in chordates. PLoS genetics 4: e1000025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Venkatesh B, Tay A, Dandona N, Patil JG, Brenner S (2005) A compact cartilaginous fish model genome. Current biology: CB 15: R82–83. [DOI] [PubMed] [Google Scholar]
- 31. Venkatesh B, Kirkness EF, Loh YH, Halpern AL, Lee AP, et al. (2007) Survey sequencing and comparative analysis of the elephant shark (Callorhinchus milii) genome. PLoS Biology 5: e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Venkatesh B, Lee AP, Ravi V, Maurya AK, Lian MM, et al. (2014) Elephant shark genome provides unique insights into gnathostome evolution. Nature 505: 174–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Venkatesh B, Kirkness EF, Loh YH, Halpern AL, Lee AP, et al. (2006) Ancient noncoding elements conserved in the human genome. Science 314: 1892. [DOI] [PubMed] [Google Scholar]
- 34. Terry A, Kilbey A, Vaillant F, Stewart M, Jenkins A, et al. (2004) Conservation and expression of an alternative 3′ exon of Runx2 encoding a novel proline-rich C-terminal domain. Gene 336: 115–125. [DOI] [PubMed] [Google Scholar]
- 35. van der Meulen T, Kranenbarg S, Schipper H, Samallo J, van Leeuwen JL, et al. (2005) Identification and characterisation of two runx2 homologues in zebrafish with different expression patterns. Biochimica et biophysica acta 1729: 105–117. [DOI] [PubMed] [Google Scholar]
- 36. Bee T, Swiers G, Muroi S, Pozner A, Nottingham W, et al. (2010) Nonredundant roles for Runx1 alternative promoters reflect their activity at discrete stages of developmental hematopoiesis. Blood 115: 3042–3050. [DOI] [PubMed] [Google Scholar]
- 37. Liu JC, Lengner CJ, Gaur T, Lou Y, Hussain S, et al. (2011) Runx2 protein expression utilizes the Runx2 P1 promoter to establish osteoprogenitor cell number for normal bone formation. The Journal of biological chemistry 286: 30057–30070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Akamatsu Y, Ohno T, Hirota K, Kagoshima H, Yodoi J, et al. (1997) Redox regulation of the DNA binding activity in transcription factor PEBP2. The roles of two conserved cysteine residues. The Journal of biological chemistry 272: 14497–14500. [DOI] [PubMed] [Google Scholar]
- 39. Tahirov TH, Inoue-Bungo T, Morii H, Fujikawa A, Sasaki M, et al. (2001) Structural analyses of DNA recognition by the AML1/Runx-1 Runt domain and its allosteric control by CBFbeta. Cell 104: 755–767. [DOI] [PubMed] [Google Scholar]
- 40. Kanai F, Marignani PA, Sarbassova D, Yagi R, Hall RA, et al. (2000) TAZ: a novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. EMBO J 19: 6778–6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yagi R, Chen LF, Shigesada K, Murakami Y, Ito Y (1999) A WW domain-containing yes-associated protein (YAP) is a novel transcriptional co-activator. EMBO J 18: 2551–2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Javed A, Guo B, Hiebert S, Choi JY, Green J, et al. (2000) Groucho/TLE/R-esp proteins associate with the nuclear matrix and repress RUNX (CBF(alpha)/AML/PEBP2(alpha)) dependent activation of tissue-specific gene transcription. Journal of cell science 113 (Pt 12): 2221–2231. [DOI] [PubMed] [Google Scholar]
- 43. Tanaka T, Kurokawa M, Ueki K, Tanaka K, Imai Y, et al. (1996) The extracellular signal-regulated kinase pathway phosphorylates AML1, an acute myeloid leukemia gene product, and potentially regulates its transactivation ability. Molecular and cellular biology 16: 3967–3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang Y, Biggs JR, Kraft AS (2004) Phorbol ester treatment of K562 cells regulates the transcriptional activity of AML1c through phosphorylation. The Journal of biological chemistry 279: 53116–53125. [DOI] [PubMed] [Google Scholar]
- 45. Qiao M, Shapiro P, Fosbrink M, Rus H, Kumar R, et al. (2006) Cell cycle-dependent phosphorylation of the RUNX2 transcription factor by cdc2 regulates endothelial cell proliferation. The Journal of biological chemistry 281: 7118–7128. [DOI] [PubMed] [Google Scholar]
- 47. Sears KE, Goswami A, Flynn JJ, Niswander LA (2007) The correlated evolution of Runx2 tandem repeats, transcriptional activity, and facial length in carnivora. Evolution & development 9: 555–565. [DOI] [PubMed] [Google Scholar]
- 48. Goodman FR, Mundlos S, Muragaki Y, Donnai D, Giovannucci-Uzielli ML, et al. (1997) Synpolydactyly phenotypes correlate with size of expansions in HOXD13 polyalanine tract. Proceedings of the National Academy of Sciences of the United States of America 94: 7458–7463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Morrison NA, Stephens AA, Osato M, Polly P, Tan TC, et al. (2012) Glutamine repeat variants in human RUNX2 associated with decreased femoral neck BMD, broadband ultrasound attenuation and target gene transactivation. PloS one 7: e42617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jeong JH, Jin JS, Kim HN, Kang SM, Liu JC, et al. (2008) Expression of Runx2 transcription factor in non-skeletal tissues, sperm and brain. Journal of cellular physiology 217: 511–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Park ES, Park J, Franceschi RT, Jo M (2012) The role for runt related transcription factor 2 (RUNX2) as a transcriptional repressor in luteinizing granulosa cells. Molecular and cellular endocrinology 362: 165–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fan HY, Liu Z, Johnson PF, Richards JS (2011) CCAAT/enhancer-binding proteins (C/EBP)-alpha and -beta are essential for ovulation, luteinization, and the expression of key target genes. Molecular endocrinology 25: 253–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dooley H, Flajnik MF (2006) Antibody repertoire development in cartilaginous fish. Dev Comp Immunol 30: 43–56. [DOI] [PubMed] [Google Scholar]
- 54. Boehm T, Iwanami N, Hess I (2012) Evolution of the immune system in the lower vertebrates. Annu Rev Genomics Hum Genet 13: 127–149. [DOI] [PubMed] [Google Scholar]
- 55. Drissi H, Luc Q, Shakoori R, Chuva De Sousa Lopes S, Choi JY, et al. (2000) Transcriptional autoregulation of the bone related CBFA1/RUNX2 gene. Journal of cellular physiology 184: 341–350. [DOI] [PubMed] [Google Scholar]
- 56. Spender LC, Whiteman HJ, Karstegl CE, Farrell PJ (2005) Transcriptional cross-regulation of RUNX1 by RUNX3 in human B cells. Oncogene 24: 1873–1881. [DOI] [PubMed] [Google Scholar]
- 57. Drissi H, Pouliot A, Koolloos C, Stein JL, Lian JB, et al. (2002) 1,25-(OH)2-vitamin D3 suppresses the bone-related Runx2/Cbfa1 gene promoter. Experimental cell research 274: 323–333. [DOI] [PubMed] [Google Scholar]
- 58. Hovhannisyan H, Zhang Y, Hassan MQ, Wu H, Glackin C, et al. (2013) Genomic occupancy of HLH, AP1 and Runx2 motifs within a nuclease sensitive site of the Runx2 gene. Journal of cellular physiology 228: 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, et al. (2006) In vivo enhancer analysis of human conserved non-coding sequences. Nature 444: 499–502. [DOI] [PubMed] [Google Scholar]
- 60. Ng CE, Yokomizo T, Yamashita N, Cirovic B, Jin H, et al. (2010) A Runx1 intronic enhancer marks hemogenic endothelial cells and hematopoietic stem cells. Stem cells 28: 1869–1881. [DOI] [PubMed] [Google Scholar]
- 61. Attanasio C, Nord AS, Zhu Y, Blow MJ, Li Z, et al. (2013) Fine tuning of craniofacial morphology by distant-acting enhancers. Science 342: 1241006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lee AP, Kerk SY, Tan YY, Brenner S, Venkatesh B (2011) Ancient vertebrate conserved noncoding elements have been evolving rapidly in teleost fishes. Molecular biology and evolution 28: 1205–1215. [DOI] [PubMed] [Google Scholar]
- 63. Xu Y, Wang K, Gao W, Zhang C, Huang F, et al. (2013) MicroRNA-106b regulates the tumor suppressor RUNX3 in laryngeal carcinoma cells. FEBS Lett 587: 3166–3174. [DOI] [PubMed] [Google Scholar]
- 64. Rossetti S, Sacchi N (2013) RUNX1: A MicroRNA Hub in Normal and Malignant Hematopoiesis. Int J Mol Sci 14: 1566–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lian JB, Stein GS, van Wijnen AJ, Stein JL, Hassan MQ, et al. (2012) MicroRNA control of bone formation and homeostasis. Nat Rev Endocrinol 8: 212–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ogawa E, Inuzuka M, Maruyama M, Satake M, Naito-Fujimoto M, et al. (1993) Molecular cloning and characterization of PEBP2 beta, the heterodimeric partner of a novel Drosophila runt-related DNA binding protein PEBP2 alpha. Virology 194: 314–331. [DOI] [PubMed] [Google Scholar]
- 67. Kagoshima H, Akamatsu Y, Ito Y, Shigesada K (1996) Functional dissection of the alpha and beta subunits of transcription factor PEBP2 and the redox susceptibility of its DNA binding activity. The Journal of biological chemistry 271: 33074–33082. [DOI] [PubMed] [Google Scholar]
- 68. Davies WI, Tay BH, Zheng L, Danks JA, Brenner S, et al. (2012) Evolution and functional characterisation of melanopsins in a deep-sea chimaera (elephant shark, Callorhinchus milii). PloS one 7: e51276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tan YY, Kodzius R, Tay BH, Tay A, Brenner S, et al. (2012) Sequencing and analysis of full-length cDNAs, 5′-ESTs and 3′-ESTs from a cartilaginous fish, the elephant shark (Callorhinchus milii). PloS one 7: e47174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution 17: 540–552. [DOI] [PubMed] [Google Scholar]
- 71. Flicek P, Ahmed I, Amode MR, Barrell D, Beal K, et al. (2013) Ensembl 2013. Nucleic acids research 41: D48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Jurka J, Klonowski P, Dagman V, Pelton P (1996) CENSOR—a program for identification and elimination of repetitive elements from DNA sequences. Computers & chemistry 20: 119–121. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
P1 promoter region of Runx1 and Runx3 of elephant shark and selected bony vertebrates. Multiple sequence alignments of the P1 5′UTR regions of (a) Runx1 and (b) Runx3 are shown. The numbers to the left of the alignment indicate positions relative to the TSS (+1) of elephant shark (C. milii) genes. Corresponding regions from other bony vertebrates were aligned using ClustalW. The tandem Runx binding sites are boxed.
(PDF)
Primers used for qRT-PCR.
(PDF)
CNEs in the Runx loci of human and elephant shark.
(PDF)