Summary
How organ-specific metastatic traits arise in primary tumors remains unknown. Here we show a role of the breast tumor stroma in selecting cancer cells that are primed for metastasis in bone. Cancer-associated fibroblasts (CAFs) in triple-negative (TN) breast tumors skew heterogeneous cancer cell populations towards a predominance of clones that thrive on the CAF-derived factors CXCL12 and IGF1. Limiting concentrations of these factors select for cancer cells with high Src activity, a known clinical predictor of bone relapse and an enhancer of PI3K-Akt pathway activation by CXCL12 and IGF1. Carcinoma clones selected in this manner are primed for metastasis in the CXCL12-rich microenvironment of the bone marrow. The evidence suggests that stromal signals resembling those of a distant organ select for cancer cells that are primed for metastasis in that organ, thus illuminating the evolution of metastatic traits in a primary tumor and its distant metastases.
INTRODUCTION
A key question in understanding the origin of metastasis is how cancer cells in a primary tumor acquire the ability to colonize a particular distant organ. Primary tumors release large numbers of cancer cells into the circulation, yet only a small proportion of these cells survive the stress of invading distant organs and progress to metastases (Chambers et al., 2002; Fidler, 2003; Gupta and Massagué, 2006). Moreover, different tumor types metastasize with distinct patterns of organ preference. It was postulated that these metastatic traits are gained through random pro-metastatic mutations in primary tumors, and remain rare until selection in secondary organ sites leading to expansion of the mutant cell clones (Fidler, 1973; Nowell, 1976). In this model, the molecular determinants of metastasis would not be overtly manifest in the bulk cell population of a primary tumor. However, no driver mutations specific for metastasis to particular organs have been identified to date. Growing evidence shows, on the contrary, that the likelihood of metastasis in general, and of metastasis to certain organs in particular, can be predicted from gene expression patterns of primary tumors (Chang et al., 2005; Chiang and Massagué, 2008; Minn et al., 2005; van 't Veer et al., 2002; Weigelt et al., 2003). These findings imply that pro-metastatic activities expressed in large segments of the primary tumor cell population increase the probability that the cancer cells will colonize particular organs.
How organ-specific metastatic traits and their associated gene signatures emerge in primary tumors remains an enigma (Valastyan and Weinberg, 2011). A case in point is the specific association of breast cancer bone metastasis with a gene expression signature (Src response signature, SRS) that denotes activation of Src and Src-dependent enhancement of PI3K-Akt signaling in primary tumors (Zhang et al., 2009). This association is particularly striking in the case of tumors that are triple-negative (TN) for ER, progesterone receptor, and ERBB2 amplification. As a group TN tumors have a strong propensity to metastasize in visceral organs, whereas SRS+ TN tumors additionally have a propensity to metastasize in bone. Although Src can regulate many aspects of cell behavior, its effect in models of breast cancer metastasis is to enhance the survival and incipient outgrowth of metastatic cells that enter the bone marrow, without affecting the rate of entry or the eventual engagement of osteoclasts for osteolytic metastasis. Src promotes the survival of breast cancer cells by amplifying the responsiveness of the PI3K-Akt survival pathway to CXCL12 and IGF1 (Zhang et al., 2009). These cytokines are present in the bone marrow stroma (Méndez-Ferrer et al., 2010) and are more highly expressed in the bone metastasis microenvironment than in other metastatic sites (Zhang et al., 2009). Breast cancer cells that lodge in the bone marrow are more likely to survive in this environment if they possess Src-enhanced responsiveness to these stromal cytokines.
Src hyperactivity however does not confer a net growth advantage in mammary tumors (Zhang et al., 2009), nor is Src frequently mutated or genomically amplified in breast cancer (The Cancer Genome Atlas Network, 2012). These observations raise questions about the mechanisms that lead to the accumulation of Src-hyperactive cells in breast tumors. We addressed these questions by considering two alternative hypotheses. Src hyperactivity in SRS+ breast cancer cells could be biochemically linked to the primary oncogenic alterations driving the tumor or, alternatively, it could result from clonal selection of a Src-dependent growth advantage that is not manifest in the primary tumor. Prior findings, which we confirmed, lent support to the first of these hypotheses in estrogen receptor-positive (ER+) breast tumors (Collins and Webb, 1999) and HER2+ breast tumors (tumors driven by the ERBB2 oncogene) (Ishizawar et al., 2007; Zhang et al., 2011). However, our search for answers in the third major class of breast cancer, TN tumors, revealed a process of stroma-driven selection for clones that are primed for bone metastasis. This process is driven by CXCL12 and IFG1 from mesenchymal stromal cells and it selects for Src-hyperactive cancer cell clones that thrive in environments containing these signals. We delineate this process of metastasis seed preselection in experimental models and provide evidence for the existence of its defining traits in human breast tumors.
RESULTS
Src Activation in Different Subtypes of Breast Cancer
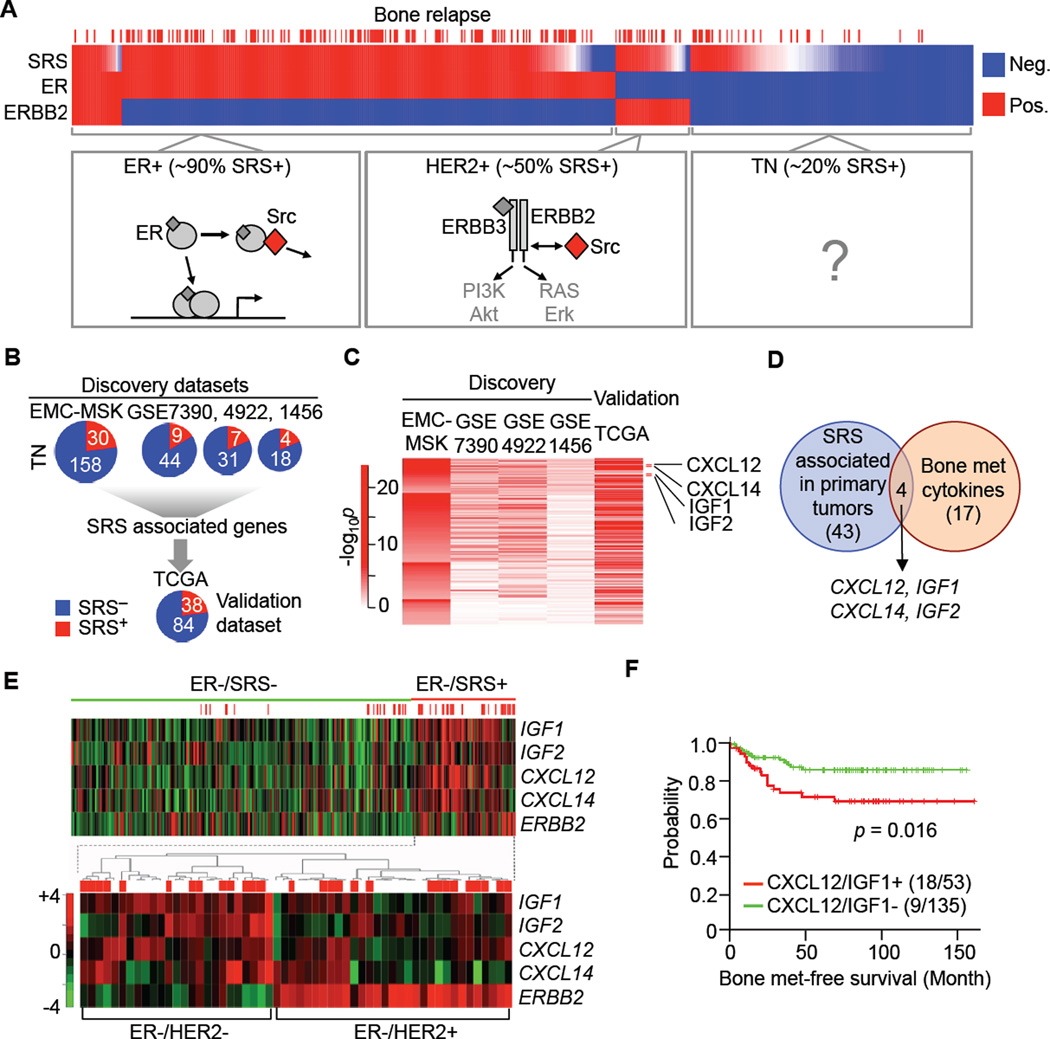
To determine what drives Src activation in primary tumors, we investigated the features associated with SRS+ status in different subtypes of breast cancer. Most ER+ breast tumors and at least half of HER2+ tumors score as SRS+ (Figures 1A, S1A, S1B) (Zhang et al., 2009). Mechanistically this is in line with the ability of Src to interact with ER and activate non-transcriptional tumorigenic signaling (Collins and Webb, 1999), and the capacity of Src and ERBB2/ERBB3 dimers for reciprocal activation (Ishizawar et al., 2007; Zhang et al., 2011). We confirmed these links with biochemical and pharmacologic approaches (Figures S1C-S1H). We concluded that Src activation, SRS+ status, and the propensity to metastasize in bone are intrinsically coupled to the oncogenic drivers in these breast cancer subtypes (Figure 1A, Table S1).
Figure 1. Cytokine Similarities between SRS+ TN Breast Tumors and Bone Metastases.
(A) Src activation in different breast cancer subtypes. Upper panel: hierarchical clustering by SRS, ER and ERBB2 status of 615 primary breast tumors with known bone metastasis outcomes. Red sticks indicate bone metastasis incidence. Lower panels: In ER+ tumors, estrogen receptor (ER) in the cytoplasm interact with Src to trigger cell survival and proliferation (Collins and Webb, 1999). In HER2+ tumors, ERBB2-ERBB3 heterodimers signal cell proliferation and survival by activating the PI3K-Akt pathway. Src potentiates the ERBB2-ERBB3 heterodimers and conversely ERBB2 activates Src (Ishizawar et al., 2007; Zhang et al., 2011). The link to Src in TN tumors is unknown. See also Figure S1.
(B) Comparison of SRS+ and SRS- tumors in the TN subtype to identify genes associated with SRS across independent datasets. Erasmus Medical Center/Memorial Sloan-Kettering (EMC-MSK) (GSE2603, 5327, 2034 and 12276), GSE7390, GSE4922 and GSE1456 datasets were used as discovery datasets and TCGA dataset was used as an independent validation set. The corresponding sample sizes are indicated by pie charts.
(C) Genes that achieve significance (p < 0.001 and fold change > 2) in the EMC-MSK dataset were subjected to further analysis. For the EMC-MSK dataset, p values are log-transformed and represented by the color scale. For other datasets, genes with p < 0.05 are indicated in red. All mapped genes are confirmed in TCGA dataset (p < 0.001 and FDR < 0.05, see Table S2).
(D) Venn diagram showing the overlap between cytokine genes enriched in bone metastases and SRS-associated genes in primary tumors. The number of genes in each category and the four overlapping genes are indicated.
(E) Microarray gene expression heatmap of the four cytokines within all ER- tumors, including HER2+ and TN tumors. The lower heatmap shows unsupervised clustering of ER-/SRS+ tumors using the four cytokines and ERBB2. Red sticks indicate bone metastasis incidence.
(F) Kaplan-Meier plot of the probability of cumulative bone metastasis-free survival in TN patients according to CXCL12/IGF1 expression. Numbers of cases in each category are implicated in brackets. P value was calculated according to log rank test.
Triple-negative (TN) breast tumors have a propensity to metastasize in visceral organs. However, a significant proportion of these patients develop metastasis to bone as the first site (Figure 1A, Table S1) (Dent et al., 2009). SRS+ status in TN tumors is a strong prognostic indicator of bone relapse (Zhang et al., 2009). Unlike the ER+ and HER2+ subtypes, TN tumors are driven by diverse oncogenic mutations. PIK3CA mutations are frequent in breast cancer (The Cancer Genome Atlas Network, 2012). In the TCGA breast cancer cohort, comprising 409 primary tumor samples with matched mutation and expression data, 43% of SRS+ tumors harbor PIK3CA mutations, versus 32% of all tumors (p = 2.7e-5). Mutations in other PI3K pathway components such as PTEN did not correlate with SRS status (p = 0.51). Within TN tumors 16% had PIK3CA mutations, versus 36% in the SRS+ TN subset (p = 0.00027), suggesting a limited contribution of PIK3CA mutations to SRS+ status. Thus, SRS+ status represents an output of diverse upstream mechanisms in TN breast cancer.
CAF Content of TN Tumors is Linked to Bone Metastasis
Seeking additional clues for how Src activity accumulates in SRS+ TN tumors, we compared the gene expression profiles of SRS- and SRS+ TN tumors in independent clinical datasets, and focused on genes that are significantly enriched in SRS+ tumors but are not part of the SRS gene set itself (Figure 1B). Forty-three genes scored as SRS-associated genes, among which 38 were validated in the TCGA dataset (p < 0.001, FDR < 0.05, Figure 1B, Table S2). Notably, these genes included CXCL12, CXCL14, IGF1 and IGF2 (Figure 1C), which are also overexpressed in bone metastasis tissues compared to metastases in lung, liver or brain (Figure 1D) (Zhang et al., 2009). The enriched expression of these four cytokines in ER- tumors was especially pronounced in TN cases compared to HER2+ cases (Figure 1E). A two-gene classifier comprised of CXCL12 and IGF1 alone was also prognostic of bone metastasis (Figure 1F). These observations pointed at an intriguing resemblance in cytokine content between SRS+ TN tumors and the bone marrow where these tumors preferably metastasize.
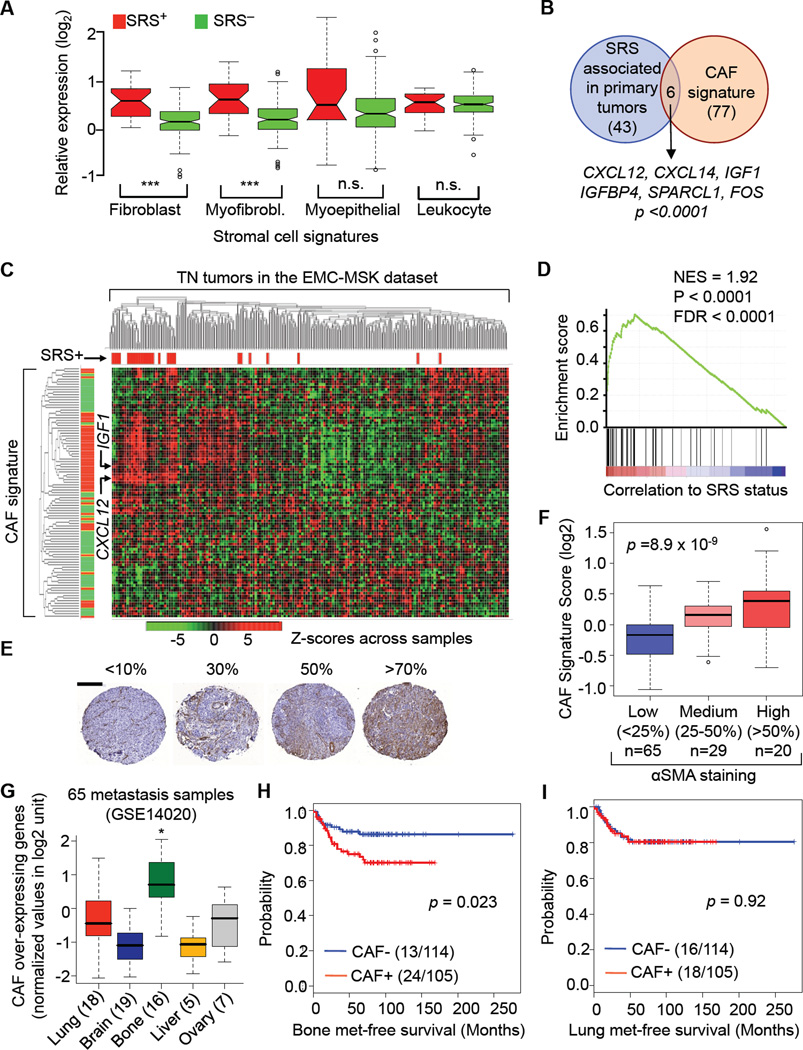
We sought to identify the source of these cytokines in SRS+ TN tumors. None of the TN breast cancer cell lines MDA-MB-231 (MDA231 for short), CN34, BT549, MDA-MB-435 or MDA-MB-436 cells expressed high level of CXCL12, CXCL14, IGF1 or IGF2 (transcriptomic data not shown). To determine whether the association of SRS with CXCL12/14 and IGF1/2 in TN tumors reflects an abundance of a particular cell type, we applied gene-signature classifiers that designate specific breast tumor stromal cell types. These gene signatures were previously derived from purified cell populations from breast tumors of multiple patients (Allinen et al., 2004), and therefore represent consensus profiles of specific stromal types from individually heterogeneous tumors. Notably, SRS+ status in TN tumors was associated with a high score for fibroblast and myofibroblast gene signatures but not leukocyte or myoepithelial gene signatures (Figure 2A). Indeed, tumor-associated mesenchymal cells (fibroblast and myofibroblast), also referred to as cancer associated fibroblasts (CAFs), abundantly express CXCL12 (Orimo et al., 2005). In line with this, previous transcriptional datasets showed no CXCL12 expression and low expression of IGF1 in cancer cells compared to mesenchymal stroma (Allinen et al., 2004).
Figure 2. CAF Content in TN Tumors is Linked to Bone Metastasis.
(A) Box-whisker plots showing the expression of various cancer-associated stromal signatures (Allinen et al., 2004) in TN tumors of the EMC-MSK dataset, according to SRS status. The expression level of each signature in each tumor is represented by the average probe intensity of the genes comprising the signature. Difference between SRS+ and SRS- tumors were tested using Wilcoxon signed rank test. ***: p < 0.0001. “n.s.”: no statistical difference.
(B) Venn diagram showing the overlap between the SRS-associated genes and the CAF signature genes, and identities of the six overlapping genes. p value was calculated by Fisher’s exact test.
(C) Unsupervised hierarchical clustering of TN tumors from the EMC-MSK dataset by CAF signature. SRS+ status is indicated by red sticks above the heatmap. Small arrows point to CXCL12 and IGF1 gene probes. Red and green bars left to the heatmap indicate genes that are over- or under- expressed in CAF signature, respectively.
(D) Gene Set Enrichment Analysis revealed a strong association between SRS status and the enrichment of CAF signature genes.
(E) Representative examples and scores of alpha smooth muscle actin (αSMA) immunohistochemical staining of TN breast tumor samples. Scale bar, 200µm.
(F) Box-whisker plots showing the mean CAF signature score (see Methods) in tumor tissues microarrays with matched microarray data. The definition and sample size of each category are indicated. Statistical significance is assessed using one-way ANOVA.
(G) Box-whisker plots showing the expression of CAF signature in 65 breast cancer metastasis samples from different sites. The number of metastases at each site is indicated in parentheses. The expression in bone metastases was significantly higher (* p < 0.0001, Wilcoxon test) compared to all other sites combined.
(H–I) Kaplan-Meier plots of cumulative bone (H) and lung (I) metastasis-free survival in TN patients according to CAF signature. The median CAF signature score is used as a cutoff to separate CAF+ and CAF- cases. Cases without clinical follow-up information are omitted. The cases included in this analysis are TN samples with matched TMA and microarray data. Numbers of relapse cases over total cases in each category are indicated in brackets. P value was calculated according to log rank test.
See also Figure S2.
The CAF gene set and the SRS-associated gene set included CXCL12, CXCL14 and IGF1 (Figure 2B). Unsupervised hierarchical clustering by the combined mesenchymal stroma gene signature (CAF signature for brevity) identified a set of TN tumors comprising almost exclusively SRS+ cases (Figures 2C, S2A). Gene Set Enrichment Analysis (GSEA) confirmed this association (Figures 2D, S2B). Exclusion of tumors with PIK3CA mutations did not diminish the correlation between CAF+ status and SRS+ status (p=0.0031, versus p=0.004 for all tumors included). Importantly, the CAF signature score of TN breast tumors was tightly correlated with the content of stromal cells staining positive for the CAF marker alpha smooth muscle actin (αSMA) (Figures 2E, 2F). The SRS score was also associated with the abundance of αSMA+ stromal cells (Figure S2C). The CAF signature was overrepresented in bone metastases compared to metastases at other sites (Figure 2G). These results showed a concordance between CAF stromal content, CAF gene signature score, and CAF-derived cytokines in TN SRS+ tumors.
Notably, both the CAF gene signature (Figures 2H, 2I) and CAF cell content in TN breast tumors (Figures S2D, S2E) were associated with bone relapse but not with lung relapse. Thus, by molecular and histological criteria, CAF-rich TN tumors were predisposed to metastasize in bone. CAF signature (p =0.027; hazard ratio 2.36, 95% CI: 1.24–4.47) and SRS (p =0.023; hazard ratio 7.11, 95% CI: 1.87–27.10) as continuous variables also correlated with bone relapse as the end-point. An association of the CAF signature with SRS+ status was also present though less pronounced in ER+ and HER2+ tumors (Figure S2F). Collectively, these results linked the CAF content with the propensity of TN tumors to relapse in bone.
Premises for CXCL12 and IGF1 Driven Selection
The repeated observation of an association between SRS status, CAF gene signature, CXCL12/IGF1 expression, and risk of bone metastasis, led us to consider the possibility of a selection process whereby stroma-derived CXCL12 and IGF1 in breast tumors skew cancer cell populations toward a preponderance of Src-hyperactive clones and thus, toward bone metastasis potential. This “metastasis seed pre-selection” hypothesis required three premises: first, the presence of a selectable trait –Src hyperactivity– in the cancer cells; second, a heterogeneous distribution of this trait in the cancer cell population; and third, the presence of limiting concentrations of CXCL12/IGF1 that would act as a selector of cell fitness to respond to these survival signals.
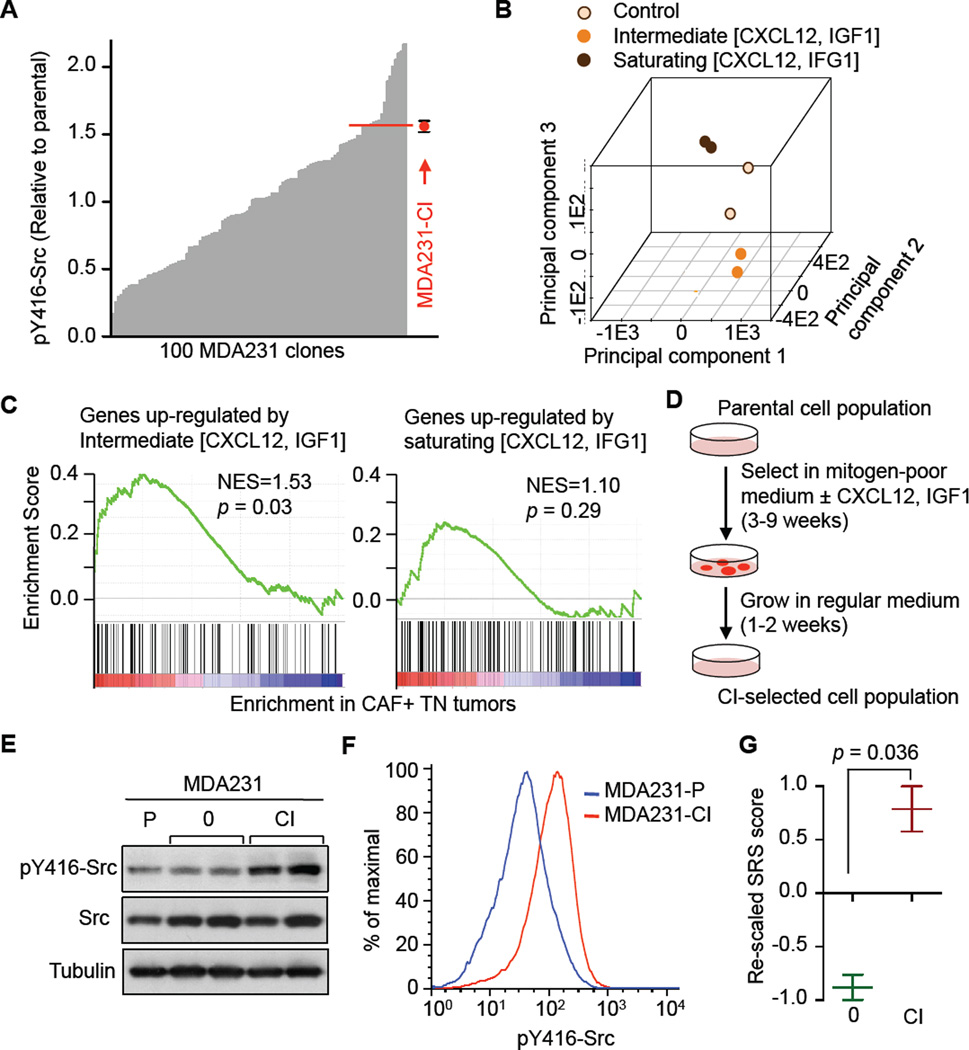
To determine the clonal distribution of Src activity in a heterogeneous breast cancer cell population we analyzed the level of activated Src (Src phosphorylated at Tyr416, pY416-Src) in 100 clonal cell lines isolated from the MDA231 parental population. We quantitated the pY416- Src level in each clone by ELISA. The distribution of values varied over a 12-fold range (Figure 3A). These values remained stable over 25 population doublings in culture (data not shown). Thus, pY416-Src was a heterogeneously distributed, stable trait in the cancer cell population.
Figure 3. CXCL12 and IGF1 Select for Src-hyperactive Cancer Cells.
(A) Clonal heterogeneity of pY416-Src levels in the MDA231 cell population. One hundred random single-cell clones were subjected to pY416-Src ELISA assay. Values were normalized to the value of the parental population and ranked. The pY416-Src level of the in vitro selected subpopulation MDA231-CI is indicated by the red dot.
(B) Principal component analysis of gene expression profiles of MDA231 cells treated with CXCL12 and IGF1 at intermediate (30ng/mL + 10ng/mL) and saturating (300ng/mL + 100 ng/mL) concentrations. The first 3 principal components are shown.
(C) Gene Set Enrichment Analysis of association between gene sets induced by CXCL12/IGF1 at two different concentrations and CAF signature status in TN tumors in EMC-MSK datasets.
(D) Schematic of the in vitro selection procedure. Cell selection in media containing 0.2% serum lasted 3 weeks for MDA231 cells or 9 weeks for CN34 cells. CI, CXCL12/IGF1.
(E) pY416-Src levels of MDA231 subpopulations resulted from in vitro selection as determined by western immunoblotting. MDA231-P, parental cells; MDA231-0, cells selected without CXCL12/IGF1 addition; MDA231-CI, cells selected with 30 ng/ml CXCL12, 10 ng/ml IGF1.
(F) Histograms of pY416-Src distribution at the single cell level as determined by phospho-flow cytometry of the indicated cell lines.
(G) SRS scores derived from gene expression profiles of MDA231-0 and MDA231-CI cells. Scores are −0.76 and −1.0 for two biological replicates of MDA231-0 cells, and 1.0 and 0.58 for MDA231-CI cells.
See also Figure S3.
To define CXCL12 and IGF1 as positive selectors, we determined the Akt response in cancer cells incubated with a range of CXCL12 and IGF1 concentrations (Figure S3A). Based on the sensitivity of Akt activation, we chose intermediate (30 ng/ml CXCL12, 10 ng/ml IGF1) or saturating (300 ng/ml CXCL12, 100 ng/ml IGF1) concentration of these factors and determined their combined effect on MDA231 by RNA-seq. The transcriptomic responses (fold change > 2, p < 0.05) to these two sets of concentrations showed some commonalities but also many differences (Table S3). Each set was qualitatively unique as determined by principal component analysis (Figure 3B). Of the two gene signatures, only that corresponding to the intermediate CXCL12/IGF1 concentration was enriched in CAF+ TN tumors, as determined by GSEA (Figure 3C). These results suggest that endogenous signals in CAF+ tumors elicit a response similar to that of cancer cells stimulated with limiting concentrations of CXCL12 and IGF1 in vitro.
CXCL12 and IGF1 Select for Src-hyperactive Cancer Cells
With these premises established, we modeled CXCL12/IGF1–driven selection by subjecting cancer cells to long-term culture in mitogen-poor media (0.2% fetal bovine serum) with or without supplementation with CXCL12 and IGF1 (Figure 3D). Without supplementation very few colonies developed. These background clones (MDA231-0 cells) were pooled and expanded in regular media. The level of pY416-Src in MDA231-0 cells was the same as in the parental MDA231 cells (Figure 3E). With CXCL12/IGF1 addition, more colonies grew (Figure S3B). After expansion in regular growth media, the resulting cell population (CXCL12/IGF1-selected MDA231 cells, MDA231-CI) showed a gain in pY416-Src as determined by immunoblotting (Figure 3E) and phospho-flow cytometry (Figure 3F). The level of pY416-Src in MDA231-CI cells matched the upper quartile of the parental MDA231 population (Figure 3A), and had a high SRS score (Figure 3G). Both parameters were stable over at least 25 population doublings in regular culture conditions without CXCL12 or IGF1 additions (data not shown).
The gain in pY416-Src was verified by immunoprecipitation with Src-specific antibody followed by anti-pY416-Src immunoblotting (Figure S3C). Src was not activated by CXCL12 and IGF1 but it rather enhanced the activation of the PI3K-Akt pathway by these cytokines, and this enhancement was inhibited by the broad spectrum Src kinase inhibitor dasatinib (Figure S3D) (Zhang et al., 2009). In MDA231-CI cells half-maximal activation of Akt occurred at lower concentrations of CXCL12 and IGF1 (Figure S3E). In sum, under restrictive growth conditions the presence of limiting concentrations of CXCL12 and IGF1 skewed the cancer cell population towards a preponderance of clones with an intrinsically high level of activated Src.
CXCL12 and IGF1 Select for Bone Metastatic Clones
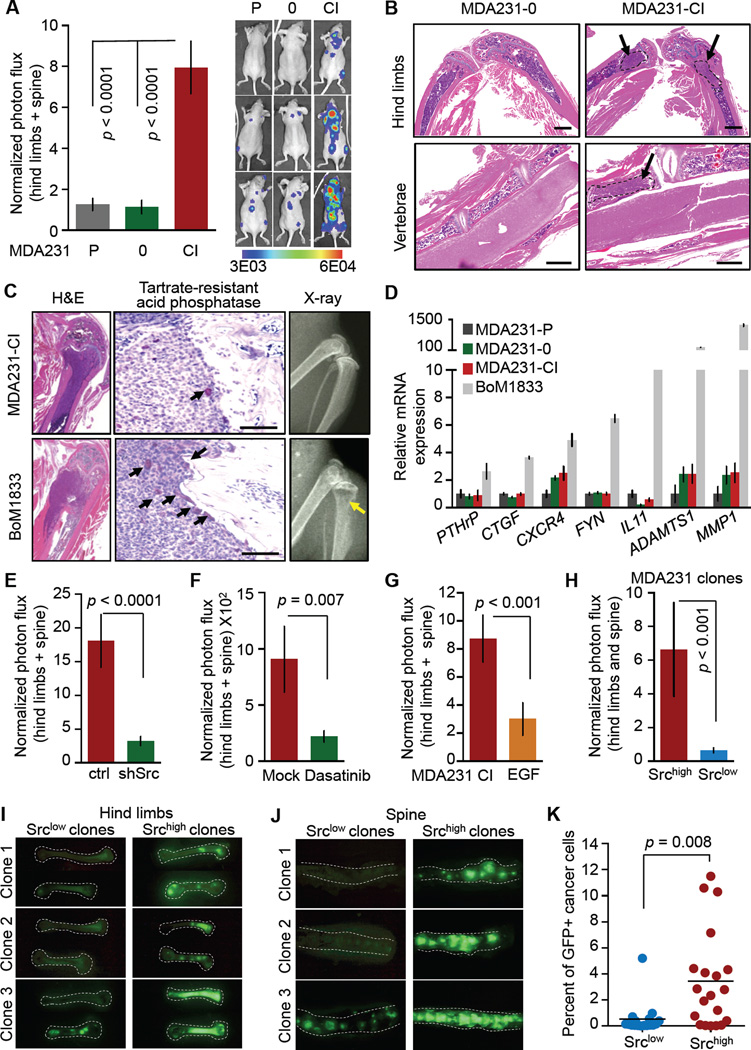
A prediction from the above findings is that breast cancer cells with a high level of pY416-Src have a particular survival advantage in the bone marrow because CXCL12 and IGF1 are relatively more abundant in this tissue than in other organ sites (Zhang et al., 2009). Indeed, when inoculated into the arterial circulation of mice, the MDA231-CI cells were significantly more metastatic to bone compared to parental MDA231 and MDA231-0 cells (Figure 4A). Bone lesions formed by MDA231-CI typically affected femora and vertebrae (Figure 4B). MDA231-CI showed no gain in lung metastatic activity (Figures S4A, S4B). The lesions formed by MDA231-CI cells were mostly restricted to the bone marrow cavity, with little engagement of osteoclasts or bone matrix destruction, as compared to lesions formed by the aggressive bone metastatic derivative MDA231-BoM1833 (Kang et al., 2005; Kang et al., 2003; Lu et al., 2009) (Figure 4C). In line with this result, MDA231-CI showed little or no gain in the expression of typical proosteolytic genes (Figure 4D). Thus, MDA231-CI cells spontaneously presented a survival and outgrowth advantage in the bone marrow prior to the onset of the osteolytic stage.
Figure 4. CXCL12/IGF1-selected Cells are Highly Metastatic to Bone.
(A) Bone metastasis formation after intracardiac inoculation of the indicated MDA231 subpopulations as monitored by quantitative luciferase bioluminescence. Representative images of mice in each group are shown. Error bars = SEM, n = 8–10 in each group.
(B) H&E staining of bone lesions derived from experiments in (A). Lesions from hind limbs and vertebral bones are shown. Scale bars are 1mm.
(C) Bone lesions formed by MDA231-CI cells or the highly osteolytic population MDA231-BoM1833 cells (Kang et al., 2003). Left panels: low magnification H&E staining. Middle panels: high magnification haematoxylin staining showing the interface between metastasis and bone matrix. Small arrows indicate active osteoclasts as revealed by tartrate-resistant phosphatase staining. Scale bars, 100µm. Right panels: Radiographic images of representative bone lesions. Arrow, a large osteolytic lesion.
(D) Expression of bone osteolytic genes (Kang et al., 2003; Lu et al., 2009) in the indicated cell lines. Error bars = SEM.
(E) Effect of Src knockdown on the bone metastasis activity of MDA231-CI cells. Error bars = SEM, n = 8–10 in each group.
(F) Dasatinib treatment inhibits bone metastasis by MDA231-CI cells. Dasatinib (10mg/kg) was administrated daily by oral gavage starting 14 days after injection. Error bars = SEM, n = 8–10 in each group.
(G) Bone metastasis assays of EGF-selected MDA231 subpopulations and MDA231-CI cells. Error bars = SEM, n = 8–10 in each group.
(H) Bone metastasis activity of pooled Srchigh and Srclow clones isolated from MDA231 parental population. Error bars = SEM, n = 8–10 in each group.
(I–J) Spontaneous bone metastases from mammary tumors formed by different Srchigh and Srclow clones. Representative GFP fluorescence images of hind limbs (I) and spine (J) are shown. GFP+ signals indicate metastasis foci.
(K) Quantification of bone metastases from (I) by flow cytometry of bone marrow flush. Percentages of cancer cells out of total bone marrow content are shown.
See also Figure S4.
Several lines of evidence supported the specificity of these effects. Src knockdown in MDA231-CI (Figures 4E, S4C, S4D) or administration of dasatinib two weeks after MDA231-CI inoculation (Figure 4F) inhibited bone metastases. Bone metastasis inhibition by dasatinib in MDA231 cells is specifically mediated by inhibition of Src in the cancer cells (Zhang et al., 2009). No EGF family members were present in the SRS+ associated gene set or in the CAF signature, and MDA231 cell populations selected with EGF in low-serum media showed little gain in pY416-Src content (Figure S4E) or bone metastatic activity (Figures 4G, S4F).
Cancer cell clones with high Src activity (Srchigh, refer to Figure 3A) showed an intrinsically high bone metastatic ability independently of osteolytic gene expression (Figures 4H, S4G, S4H), thus matching the phenotype of the CXCL12/IGF1-selected MDA231-CI population. To test whether Src-hyperactive cells show elevated bone metastatic ability from the orthotopic site, we labeled Srchigh cells with turbo-GFP, which generates a strong fluorescence signal that facilitates visualization of lesions. Nine weeks after mammary tumor implantation, no significant difference was observed in the size of primary tumors formed by Srchigh or Srclow cells (Figure S4I) or in the accumulation of cancer cells in the lungs (Figure S4J). Notably, Srchigh mammary tumors showed a much higher rate of spontaneous bone metastases compared to Srclow tumors (Figures 4I, 4J), and the proportion of GFP+ cancer cells in bone marrow aspirates subjected to flow cytometry (Figure S4K) was much higher in mice bearing Srchigh tumors (Figure 4K).
CXCL12 and IGF1 Select for Clones that Persist in the Bone Marrow
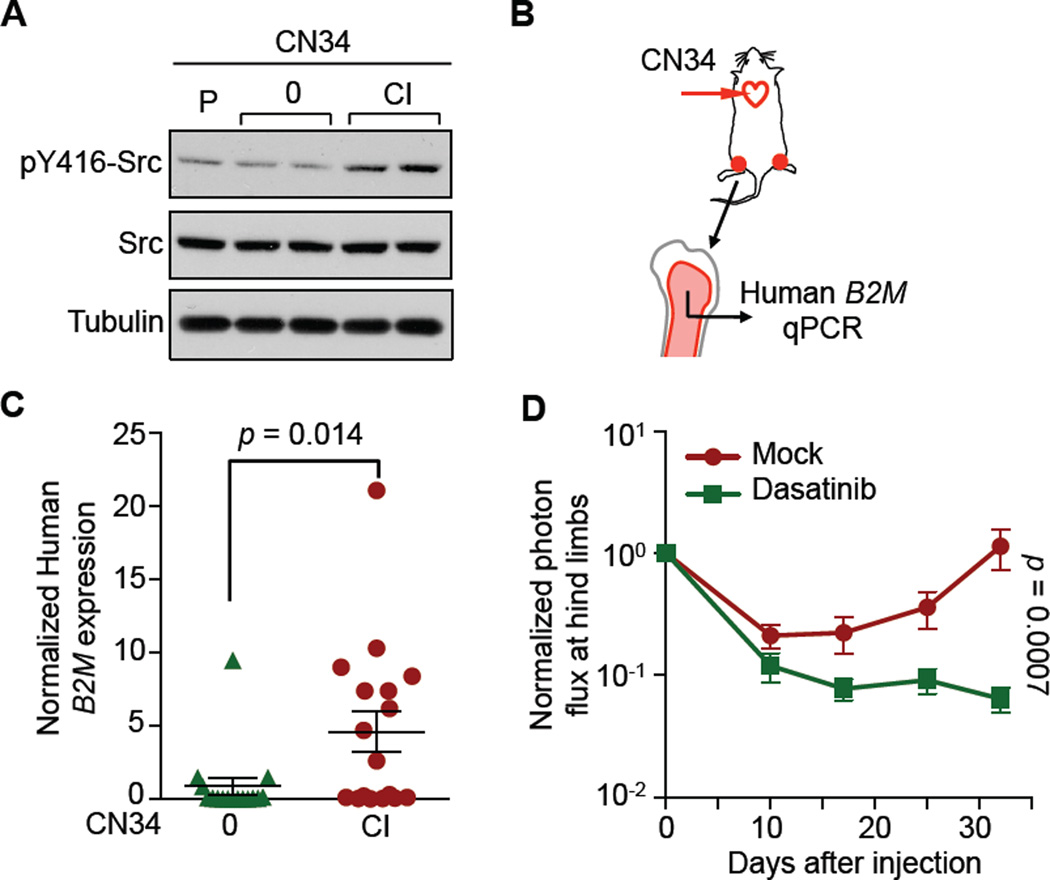
To further characterize the phenotype selected by CXCL12 and IGF1 we applied the in vitro selection protocol to the CN34 model, which is more indolent than MDA231 and allows analysis of the latent metastasis state (Zhang et al., 2009). Similar enrichment of Src-hyperactive clones was seen after long-term culture of CN34 cells in CXCL12/IGF1-supplemented media (Figures 5A, S5A). We determined the load of human cells in the bone marrow by quantitation of human β2-microglobulin (B2M) mRNA levels (Figure 5B), which can detect as few as 10 human cancer cells in 5×106 mouse cells (Figure S5B). Eight weeks after inoculation of the cells into the arterial circulation of mice, the CXCL12/IGF1-selected CN34 cells (CN34-CI) had a superior ability to persist in the bone marrow (Figure 5C). Under experimental conditions that allow the incipient outgrowth of these cells in the marrow, administration of dasatinib suppressed the outgrowth (Figure 5D).
Figure 5. CXCL12 and IGF1 Select for Clones that Persist in the Bone Marrow.
(A) Sub-populations of CN34 cells were selected and examined for Src activity using the same procedures as described in Figure 2D, 2E.
(B–C) CN34-0 and -CI cells were inoculated into the left ventricle of 7-week Bg/nu mice (105 cells per mouse). Eight weeks later, mice were sacrificed and bone marrow samples were prepared. RT-qPCR was performed to quantify human B2M mRNA. Mouse-specific β-actin (Actb) was used for normalization. Each dot in the plot represents one hind limb.
(D) Dasatinib treatment inhibits the outgrowth of CN34-CI bone micrometastases. Cells were injected into 4-week Bg/nu mice (105 cells per mouse). Dasatinib (10mg/kg) was administrated daily by oral gavage, starting 3 days after injection. Error bars = SEM, n = 8–10 in each group.
See also Figure S5.
Transcriptional Shift of Mammary Tumors Under a Mesenchymal Stroma
We investigated whether supplementing experimental mammary tumors with mesenchymal cells would lead to the selection of Src-hyperactive cancer cells and a gain in bone metastatic activity. To this end, we used bone marrow-derived human mesenchymal stem cells (MSCs) as a source of mesenchymal stromal signals. MSCs express CXCL12 (Méndez-Ferrer et al., 2010), undergo CAF-like differentiation in response to cancer cell signals (Mishra et al., 2008), and contribute to the stroma in primary tumors (Karnoub et al., 2007; Quante et al., 2011). In their basal state, MSCs secreted CXCL12 and, when stimulated with cancer cell-conditioned media, MSCs additionally secreted IGF1 (Figures S6A, S6B). In contrast, the breast cancer cells did not secrete CXCL12 or IGF1. Transcript levels for CXCL12 and IGF1 were in line with these results (Figure S6C).
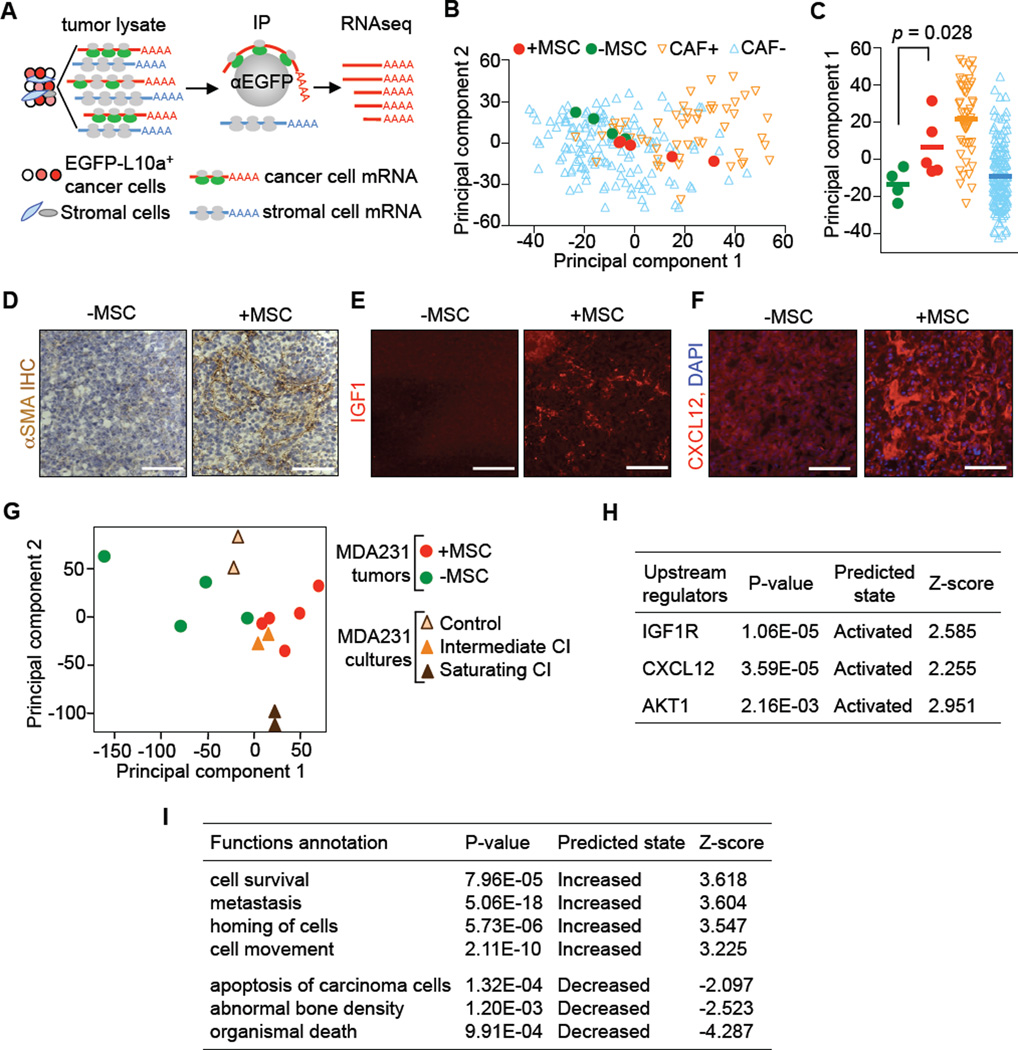
To assess whether MSC-supplementation would recapitulate the key features of human CAF+ breast tumors, we transcriptionally profiled cancer cells in MSC-supplemented experimental tumors in situ. This was achieved by engineering MDA231 cells to express a ribosomal protein L10a fused to enhanced green fluorescent protein (EGFP-L10a). The expressed EGFP-L10a is incorporated into the cancer cell ribosomes, allowing the specific retrieval of cancer cell transcripts from tumor lysates by polysome immunoprecipitation with anti-EGFP antibody (Figure 6A, translating ribosome affinity purification, TRAP). TRAP (Heiman et al., 2008) enables RNA-seq analysis of the cancer cell transcriptome from freshly excised tumors without intervening cell isolation steps that could confound the results.
Figure 6. Transcriptional Shift in Mammary Tumors Under a Mesenchymal Stroma.
(A) Isolation of cancer cell-specific, polysome-associated transcripts by TRAP for RNA-seq analysis.
(B–C) Principal component analyses of polysome-associated transcriptomic data of MDA231 from xenograft tumors and transcriptomic data of TN tumors from a clinical cohort. MDA231 mammary tumors were formed with MSC supplementation (+MSC) or without (-MSC) supplementation; TN tumors scoring as CAF+ or CAF- based on the CAF signature were from the EMC-MSK cohort. Principal component 1 maximally separates CAF+ from CAF- tumors and was used to generate the plot in (C). p value was calculated with t-test.
(D–F) Histological analysis of mammary tumors generated with or without admixed MSCs. (D): Immunohistochemical (IHC) staining of αSMA. (E): Immunofluorescent (IF) staining of IGF1. (F): IF staining of CXCL12, and DAPI nuclear staining. Scale bars, 100 µm.
(G) Principal component analysis using combined data of TRAP RNA-seq of MDA231 in vivo mammary tumors and that of in vitro CXCL12+IGF1 treatment (Figure 3C). The results show that intermediate CI samples fall closest to MSC+ tumors. Saturating CI samples, on the other hand, are dissimilar. Intermediate CI, intermediate [CXCL12, IGF1]; saturating CI, saturating [CXCL12, IGF1], refer to Figure 3C.
(H–I) Ingenuity pathway analysis of the differentially expressed genes from polysome-associated transcriptomic data of MDA231 mammary tumors in (A–C). IGF1, CXCL12 and AKT signaling are predicted to be activated based on upstream regulator analysis (H). Metastasis-related biological processes are enriched (I).
See also Figure S6.
MDA231 cells expressing EGFP-L10a were implanted alone as a control, or MDA231 admixed with MSCs at a 1:1 ratio into the mammary fat pads of mice. RNA-seq analysis of the TRAP samples showed that tumor supplementation with MSCs caused a transcriptomic shift in the cancer cells that resembled the shift associated with CAF+ status in clinical tumor samples (Figures 6B, 6C). Consistently, MSC-supplemented tumors showed a 3- to 10-fold increase in the proportion of cells staining positive for the CAF marker αSMA (Figure 6D), and a higher level of CXCL12 and IGF1 immunostaining (Figures 6E, 6F). The TRAP RNA-seq data revealed no ribosome-associated CXCL12 or IGF1 transcripts in cancer cells (Figure S6D).
The gene expression profile of cancer cells from MSC-supplemented tumors resembled the profile of MDA231 cells treated with limiting CXCL12 and IGF1 concentrations (Figure 6G). This similarity was further validated by Ingenuity pathway analysis of the differentially expressed genes. Upstream regulator analysis revealed CXCL12- and IGF1-induced signaling and their downstream PI3K-Akt signaling in the cancer cells (Figure 6H). These genes are enriched in biological processes associated with metastasis and bone biology (Figure 6I). Thus, the model tumors co-transplanted with MSCs gained a bioactive CXCL12/IGF1 microenvironment and response features that are characteristic of CAF+ TN tumors.
A Mesenchymal Stroma Selects for Bone Metastatic Cells in Mammary Tumors
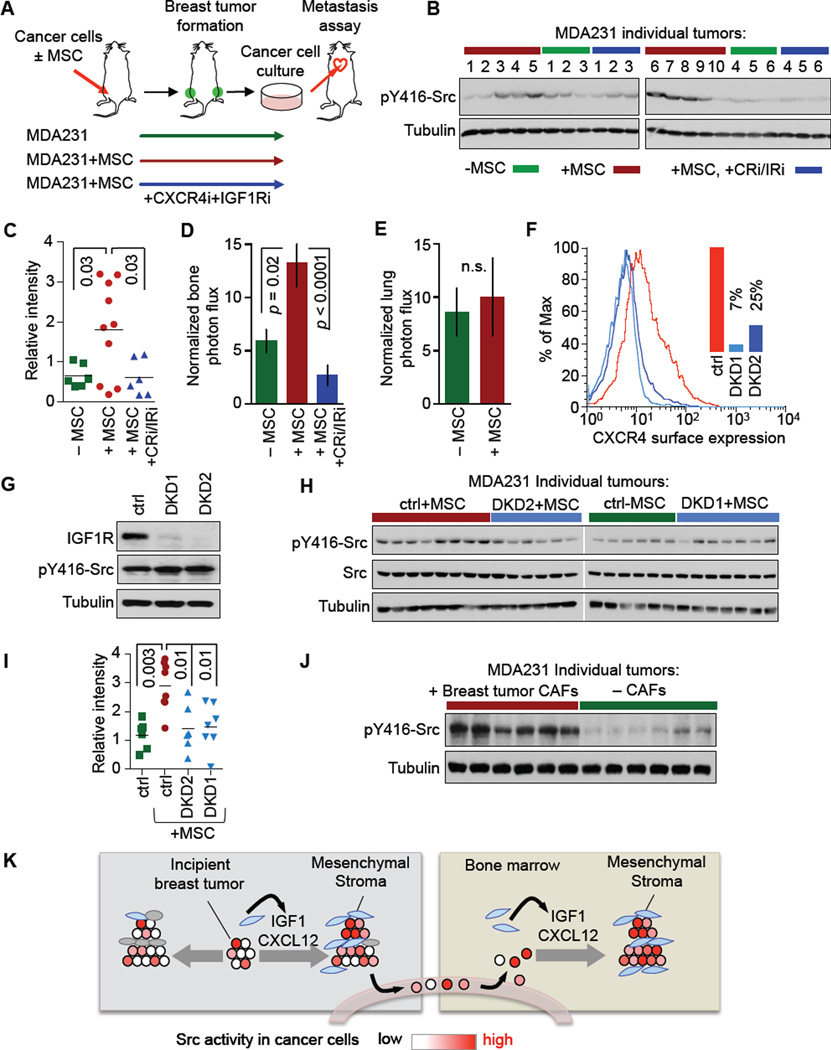
Cancer cells were purified from MDA231 mammary tumors, expanded in culture for several passages, and characterized (Figure 7A). Cancer cells recovered from MSC-supplemented tumors showed a high level of pY416-Src compared to populations derived from control tumors (Figures 7B, 7C). Treatment of the mice with inhibitors of the CXCL12 receptor CXCR4 (AMD3100) (Hatse et al., 2002) and of the IGF1 receptor IGF1R (BMS754807) (Carboni et al., 2009) (Figure S7A) during tumor growth prevented the accumulation of pY416-Src-high cells (Figures 7B, 7C), without significantly affecting overall tumor growth. Importantly, cancer cell populations derived from the MSC-supplemented tumors were significantly more metastatic to bone (Figure 7D), without a gain in lung colonizing ability (Figure 7E).
Figure 7. A Mesenchymal Stroma Selects for Bone Metastatic Cells in Mammary Tumors.
(A) Experimental strategy: MDA231 cells alone or admixed with MSCs were implanted in mammary fat pads. One set of mice was treated with CXCR4 inhibitor (CRi, AMD3100) and IGF1R inhibitor (IRi, BMS754807). After reaching 1cm3, tumors were excised and cancer cells were recovered and expanded in culture for testing.
(B–C) pY416-Src levels in cancer cell lines from individual mammary tumors formed by MDA231 under the indicated conditions; immunoblot (B) and quantification (C).
(D–E) Bone metastasis (D) and lung colonization (E) activity of the pooled cell lines derived in (B). Normalized bioluminescence signals were quantified at 21 days (D) or 30 days (E) after inoculation. Error bars = SEM, n = 8–10 in each group.
(F) Flow cytometry analysis of CXCR4 surface expression in MDA231 with two independent double-knockdown (DKD1, DKD2) of CXCR4 and IGF1R compared to control (ctrl).
(G) Western immunoblot analysis of IGF1R and pY416-Src protein levels in two DKD cell lines.
(H–I) pY416-Src levels in cancer cell lines from mammary tumors formed by control MDA231 (ctrl) or two DKD cell lines with or without MSC supplementation; immunoblot (H) and quantification (I).
(J) pY416-Src levels of cancer cell lines from individual mammary tumors formed by MDA231 cells with or without admixed CAFs from a human breast tumor.
(K) A model of bone metastasis seed pre-selection by a mesenchymal-rich stroma in the mammary tumor. Blue and grey cells represent mesenchymal and non-mesenchymal stromal cells, respectively. As the tumor grows in a mesenchymal-rich microenvironment, the cancer cell population is skewed towards a preponderance of clones that thrive on mesenchymal cytokines such as CXCL12 and IGF1, which selects for a predisposition to metastasize in CXCL12- and IGF1-rich bone marrow.
See also Figure S7.
Cancer cell populations derived from mice treated with IGF1R and CXCR4 inhibitors were poorly metastatic to bone (Figure 7D). To confirm the specificity of these effects, we knocked down CXCR4 and IGF1R simultaneously in MDA231 cells (Figures 7F, 7G, S7B). Knockdown of the two receptors did not affect the level of pY416-Src in MDA231 cells (Figure 7G) but it prevented MSCs from selecting for carcinoma clones with high pY416-Src content (Figures 7H, 7I). The ability of admixed MSCs to select for clones with high pY416-Src and bone metastatic ability was confirmed in the CN37 cell line (Figures S7C, S7D). Furthermore, primary CAFs isolated from a human breast tumor secreted CXCL12 and IGF1 (12.6 ±0.2 ng/ml and 0.44 ±0.02 ng/ml, respectively after 96 h in culture) and selected for a pY416-Src enriched cancer cell population when admixed with MDA231 cells forming mammary tumors in mice (Figure 7J). The evidence therefore supports a model in which CAFs in mammary tumors cause an accumulation of stromal CXCL12 and IGF1, and these factors act directly on cancer cells to drive the selection of Src-hyperactive clones that are automatically primed for adaptation to the bone metastatic microenvironment.
DISCUSSION
Pre-selection of Bone Metastatic Seeds by the Primary Tumor Stroma
Triple negative breast cancer is a major and particularly aggressive subtype of the disease. Unlike hormone receptor-positive breast tumors, which have a marked tendency to metastasize mainly in bone, TN tumors as a whole prominently metastasize in non-skeletal organs including lungs, liver and brain (Dent et al., 2009). We have focused here on a subset of TN tumors that deviates from this trend in that it also metastasizes prominently in bone. Cancer cells from this subset are characterized by a high level of Src activity and a high sensitivity to PI3K-Akt pathway activation by CXCL12 and IGF1 (Zhang et al., 2009). By identifying the primary tumor stroma as a driver of the emergence of this phenotype, the present work illuminates the larger question of how organ-specific metastatic traits may arise in cancer.
Our evidence suggests that when the stroma of a TN breast tumor is rich in mesenchymal cells it selects for cancer clones that are fit to thrive on the CAF-derived cytokines CXCL12 and IGF1 (Figure 7K). This selection process leads to a gain in the predisposition of cancer cells for metastasis to bone, a site that is relatively rich in stromal CXCL12 and IGF1 compared to other metastasis target sites. In other major subtypes of breast cancer Src activation is integral to the oncogenic drivers like ERα (Collins and Webb, 1999) and ERBB2 (Ishizawar et al., 2007; Zhang et al., 2011), and might similarly contribute to bone metastatic tropism of these tumors.
The clinical evidence also links the CAF content with a predisposition for bone metastasis in TN tumors. In multiple human breast cancer cohorts, TN tumors that score high for cancer cell Src signature present a mesenchymal-rich stroma both based on a CAF gene expression signature and on marker αSMA immunostaining. These tumors are also characterized by the expression of CXCL12 and IGF1, factors that are mainly of mesenchymal origin in the tumors. Each of these traits in turn is individually associated with an elevated rate of bone relapse.
Our experimental evidence suggests that CXCL12 and IGF1 can drive TN cancer cell populations towards enrichment for clones that have a constitutively high level of Src activity and bone metastatic ability. Three essential premises for the clonal evolution of a cancer cell population are fulfilled under these conditions. First, Src activity in the cancer cells acts as a selectable trait that confers heightened sensitivity to CXCL12 and IGF1 as activators of PI3K/Akt survival signaling. Second, this trait ranges widely in the clonally heterogeneous cancer cell population that we examined. Third, the positive selectors for this trait, CXCL12 and IGF1, are present at limiting concentrations in the tumor microenvironment, as determined by analysis of breast tumor transcriptomic data using dose-specific CXCL12/IGF1 response classifiers. When modeled in vitro, the end result of this process is the emergence of Src-hyperactive clones with an intrinsically high ability to survive and outgrow in the CXCL12/IGF1- rich environment of the bone marrow.
Further support for this model comes from our experiments with orthotopically implanted mammary tumors in mice supplemented with MSCs or CAFs. We show that mesenchymal supplementation shifts the transcriptome of the cancer cell population in a tumor towards a profile resembling that of CAF+ TN human tumors. Moreover, enriching tumors with MSCs caused CXCL12/IGF1-like stimulation in the cancer cells, and the level of this stimulation at the transcriptome reflected limiting concentrations of the cytokines and was comparable to that observed in CAF+ human tumors. These conditions skew the cancer cell population towards clones with intrinsically high levels of Src activity and a heightened capacity for bone metastasis. The accumulation of these Src hyperactive, bone metastatic cells was dependent on the CXCL12 and IGF1 receptor expression in the cancer cells and was inhibited by administration of pharmacologic inhibitors of these receptors.
The Primary Tumor Stroma as a Determinant of Metastatic Tropism
The evidence supports a model in which a TN tumor stroma rich in mesenchymal cytokines CXCL12 and IGF1 skews the carcinoma population toward a preponderance of clones with a high level of activated Src and a predisposition to grow in the bone marrow, a soil that is rich in CXCL12 and IGF1 compared to lung, brain and other metastasis target tissues. Thus, similarities between the cytokine microenvironment of CAF-rich primary tumors and that of the bone marrow make the pre-selected metastatic seeds compatible with this target tissue (Figure 7K). Stromal cells that produce CXCL12 and IGF1 in a primary tumor are not necessarily superior to other stromal components in supporting primary tumor growth, but are superior in pre-selecting bone metastatic seeds. Our present findings are limited to the case of bone metastasis tropism in TN breast cancer. It will be important to determine whether these phenomena occur also in other bone metastatic tumor types, and whether other stromal components such as leukocytes may similarly select cancer cells with metastatic tropism for other organs.
The ability of the primary tumor stroma to drive the selection of organ-specific metastatic cells is distinct from previously described roles of stromal signals in tumor progression (Chung et al., 2010; Hanahan and Coussens, 2012; Hanahan and Weinberg, 2011; Joyce and Pollard, 2009), including roles in the resistance to stress and chemotherapy (Acharyya et al., 2012; Calón et al., 2012; DeNardo et al., 2011; Gocheva et al., 2010; Qian et al., 2011). These previously described stromal signals do not explain how organ-specific metastatic traits and their associated gene signatures emerge in primary tumors. The formation of pre-metastatic niches by tumor-derived systemic signals that condition distant organs for seeding by disseminated cancer cells also does not entail an acquisition of specific metastatic traits in the primary cancer cell population itself (Kaplan et al., 2005). The mechanism of metastasis seed pre-selection identified here may coexist with other effects of the tumor stroma, but is distinct in that it selects for organ-specific metastatic traits.
The interplay between a primary tumor and a specific metastatic site revealed by the present work expands current views on the dynamic nature of metastasis. Coupled with processes of tumor self-seeding whereby previously disseminated cells may repopulate and expand in their tumor of origin (Kim et al., 2009), seed pre-selection at the primary site may further sway the organotropic metastatic traits of a primary tumor mass with clinically manifest consequences. Our results also imply that breast cancer cases with a fibroblast-rich stroma or with incipient bone metastasis may benefit from therapies that target the PI3K-Akt cell survival pathway or its amplifier Src for the eradication of disseminated disease.
EXPERIMENTAL PROCEDURES
Computational Analyses of Clinical Data Sets
SRS was derived as previously described (Zhang et al., 2009). Stratification of the 615 patients with SRS, ER and ERBB2 in the Erasmus Medical Centre/Memorial Sloan-Kettering Cancer Center (EMC-MSK) cohort (GSE2603, GSE5327, GSE2034 and GSE12276 data sets) was done with unsupervised clustering implemented by the “heatmap.2” command of the “gplots” package of R statistical programming software. ER+, HER2+ (ER-/HER2+), and triple-negative (TN, ER-/HER2-) subtypes were determined either using the clinical annotation associated with the dataset (if applicable) or by the transcript levels of ESR1 and ERBB2, respectively. The continuous scores of SRS were computed with the sum of z-scores of 159 SRS genes and the DART algorithm (Jiao et al., 2011).
To identify genes associated with SRS status, SRS was applied to EMC-MSK, TRANSBIG (GSE7394), UPPSALA (GSE4922) and STOCKHOLM (GSE1456) datasets. SRS+ and SRS-tumors were compared within the TN subset of each dataset. Genes that are expressed at significantly higher level in SRS+ tumors (p < 0.001 and fold change > 2 for EMC-MSK and p < 0.05 for the other three, t tests with Welch correction) were identified. To compute the combined p value for each gene across all four datasets, we employed Fisher’s combination (see SUPPLEMENTAL INFORMATION). To further evaluate the significance of these genes, we utilized the TCGA cohort as a validation dataset (https://tcgadata.nci.nih.gov/tcga/tcgaDownload.jsp). We examined the p values and FDR of the 43 genes by comparing TN/SRS+ tumors with TN/SRS- tumors. Both p values and FDR were computed using “siggene” package in R statistical software based on SAM algorithm (Tusher et al., 2001). All 38 genes that can be mapped to TCGA platform reach significance (Table S2).
Gene signatures associated with CAFs were obtained from a previous study (Allinen et al., 2004). We used unsupervised hierarchical clustering, the mean expression of CAF genes, and Gene Set Enrichment Analyses (GSEA) to determine the CAF status of a tumor. Other stromal cell types were analyzed using the same approaches (see SUPPLEMENTAL INFORMATION for details).
For analyses of correlation between somatic PIK3CA mutation status and SRS, somatic mutation data from the TCGA dataset were matched with mRNA expression data. The overlap between PIK3CA mutation and SRS+ status was assessed with Fisher’s exact test.
To examine the correlation between CAF gene expression signature, SRS, and, αSMA IHC, we divided the IHC staining into three categories depending on the percentage of αSMA+ fibroblastic cells (see Figure 2E). CAF and SRS signatures were then evaluated as continuous scores (described above) in different categories (Figures 2F, S2C). The statistical difference among the three groups was assessed by one-way ANOVA implemented by R.
Cancer Cell-specific Polysome-associated Transcript Profiling
MDA231 cells were engineered to express an EGFP-L10a construct (Heiman et al., 2008). For the analysis of cytokine responses in vitro, cancer cells were treated for 6h with 30 ng/ml CXCL12 and 10 ng/ml IGF1, or with 300 ng/ml CXCL12 and 100 ng/ml IGF1. For the analysis of gene responses to MSCs in vivo, breast cancer cells were injected into the mammary fat pads of mice with or without a 1:1 admixture of MSCs, and tumors were harvested 3 weeks later. Cultured cells or tumors were lysed and immediately subjected to TRAP as previously described (Heiman et al., 2008) with modifications described in Supplementary Information.
For RNA-seq data analysis, the raw sequence data (FASTQ files) were mapped using TopHat (v1.4.0) (Trapnell et al., 2009) to the human genome (hg19 build) available from UCSC. 89% of reads were uniquely mapped on average. Gene level expression counts were determined using HTSeq and normalized with DESeq (Anders and Huber, 2010). The normalized counts were then log-transformed for analysis. Genes with zero counts were still assigned as zeros after log-transformation to avoid exceptions in programming. The approaches of subsequent analyses were described in SUPPLEMENTAL INFORMATION.
Animal Studies
All animal work was done in accordance with a protocol approved by the MSKCC Institutional Animal Care and Use Committee. Intracardiac injections of cancer cells for bone metastasis assays, intravenous injections for lung colonization assays, and orthotopic injections for mammary tumor formation were performed as previously described (Kang et al., 2003; Minn et al., 2005). Animals were imaged weekly by bioluminescence imaging (IVIS Xenogen). For quantification of bioluminescence signal, all signals were normalized to the signals at Day 0 right after injection. For bone metastases, signals at hind limbs and vertebrae were jointly quantified. Osteolytic lesions were visualized by X-ray radiography and were identified on radiographs as demarcated radiolucent lesions in the bone. Cancer cells from mammary tumors were retrieved by enzyme digestion (0.125% collagenase III and 0.1% hyaluronidase in DMEM/F12 medium) and GFP sorted for further experiments or inoculation into mice. Dasatinib (Bristol-Myers Squibb) was administered by daily oral gavage with a dose of 10 mg/kg in PBS. AMD3100 (Tocris, Cat. No: 3299), a CXCR4 inhibitor, was administered by subcutaneous injection twice a day at 2.5 mg/kg in PBS. BMS754807 (ChemieTek, Cat. No: CT-BMS75), an IGF1R inhibitor was administered by daily oral gavage at 6.25 mg/kg in polyethyleneglycol:H2O (4:1).
Supplementary Material
HIGHLIGHTS.
The primary tumor stroma can determine organ-specific metastatic tropism.
Cancer-associated fibroblasts in breast tumors select for bone metastatic cells.
CAF-rich tumors mimic the CXCL12-rich microenvironment of the bone marrow.
CAF-derived CXCL12 and IGF1 select for high Src activity, a bone metastatic trait.
ACKNOWLEDGEMENTS
We would like to thank E. Holland, N.X. Rosen, A. Ventura, S. Vanharanta for helpful input, A. Viale and N. Socci for support with RNA-seq analysis, N. Fan and M. Turkekul for advice on immunohistochemistry, and R. Somwar for reagents. This work was supported by NIH grants CA151293 (X.Z.) and CA94060 (J.M.). X.Z. is a McNair Scholar and the recipient of a grant from Breast Cancer Research Foundation. S.M. is an American Cancer Society postdoctoral fellow. J.M. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Acharyya S, Oskarsson T, Vanharanta S, Malladi S, Kim J, Morris PG, Manova- Todorova K, Leversha M, Hogg N, Seshan VE, et al. A CXCL1 paracrine network links cancer chemoresistance and metastasis. Cell. 2012;150:165–178. doi: 10.1016/j.cell.2012.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allinen M, Beroukhim R, Cai L, Brennan C, Lahti-Domenici J, Huang H, Porter D, Hu M, Chin L, Richardson A, et al. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 2004;6:17–32. doi: 10.1016/j.ccr.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calón A, Espinet E, Palomo-Ponce S, Tauriello DV, Iglesias M, Cespedes MV, Sevillano M, Nadal C, Jung P, Zhang XH, et al. Dependency of colorectal cancer on a TGF-beta-driven program in stromal cells for metastasis initiation. Cancer Cell. 2012;22:571–584. doi: 10.1016/j.ccr.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carboni JM, Wittman M, Yang Z, Lee F, Greer A, Hurlburt W, Hillerman S, Cao C, Cantor GH, Dell-John J, et al. BMS-754807, a small molecule inhibitor of insulin-like growth factor-1R/IR. Mol Cancer Ther. 2009;8:3341–3349. doi: 10.1158/1535-7163.MCT-09-0499. [DOI] [PubMed] [Google Scholar]
- Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2:563–572. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- Chang HY, Nuyten DS, Sneddon JB, Hastie T, Tibshirani R, Sorlie T, Dai H, He YD, van't Veer LJ, Bartelink H, et al. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc Natl Acad Sci U S A. 2005;102:3738–3743. doi: 10.1073/pnas.0409462102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang AC, Massagué J. Molecular basis of metastasis. N Engl J Med. 2008;359:2814–2823. doi: 10.1056/NEJMra0805239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung AS, Lee J, Ferrara N. Targeting the tumour vasculature: insights from physiological angiogenesis. Nat Rev Cancer. 2010;10:505–514. doi: 10.1038/nrc2868. [DOI] [PubMed] [Google Scholar]
- Collins P, Webb C. Estrogen hits the surface. Nat Med. 1999;5:1130–1131. doi: 10.1038/13453. [DOI] [PubMed] [Google Scholar]
- DeNardo DG, Brennan DJ, Rexhepaj E, Ruffell B, Shiao SL, Madden SF, Gallagher WM, Wadhwani N, Keil SD, Junaid SA, et al. Leukocyte complexity predicts breast cancer survival and functionally regulates response to chemotherapy. Cancer Discov. 2011;1:54–67. doi: 10.1158/2159-8274.CD-10-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent R, Hanna WM, Trudeau M, Rawlinson E, Sun P, Narod SA. Pattern of metastatic spread in triple-negative breast cancer. Breast Cancer Res Treat. 2009;115:423–428. doi: 10.1007/s10549-008-0086-2. [DOI] [PubMed] [Google Scholar]
- Fidler IJ. Selection of successive tumour lines for metastasis. Nat New Biol. 1973;242:148–149. doi: 10.1038/newbio242148a0. [DOI] [PubMed] [Google Scholar]
- Fidler IJ. The pathogenesis of cancer metastasis: the 'seed and soil' hypothesis revisited. Nat Rev Cancer. 2003;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- Gocheva V, Wang HW, Gadea BB, Shree T, Hunter KE, Garfall AL, Berman T, Joyce JA. IL-4 induces cathepsin protease activity in tumor-associated macrophages to promote cancer growth and invasion. Genes Dev. 2010;24:241–255. doi: 10.1101/gad.1874010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta GP, Massagué J. Cancer metastasis: building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21:309–322. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hatse S, Princen K, Bridger G, De Clercq E, Schols D. Chemokine receptor inhibition by AMD3100 is strictly confined to CXCR4. FEBS Lett. 2002;527:255–262. doi: 10.1016/s0014-5793(02)03143-5. [DOI] [PubMed] [Google Scholar]
- Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, Suarez-Farinas M, Schwarz C, Stephan DA, Surmeier DJ, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizawar RC, Miyake T, Parsons SJ. c-Src modulates ErbB2 and ErbB3 heterocomplex formation and function. Oncogene. 2007;26:3503–3510. doi: 10.1038/sj.onc.1210138. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Lawler K, Patel GS, Purushotham A, Jones AF, Grigoriadis A, Tutt A, Ng T, Teschendorff AE. DART: Denoising Algorithm based on Relevance network Topology improves molecular pathway activity inference. BMC Bioinformatics. 2011;12:403. doi: 10.1186/1471-2105-12-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009;9:239–252. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, He W, Tulley S, Gupta GP, Serganova I, Chen CR, Manova-Todorova K, Blasberg R, Gerald WL, Massagué J. Breast cancer bone metastasis mediated by the Smad tumor suppressor pathway. Proc Natl Acad Sci U S A. 2005;102:13909–13914. doi: 10.1073/pnas.0506517102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, Guise TA, Massagué J. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- Kaplan RN, Riba RD, Zacharoulis S, Bramley AH, Vincent L, Costa C, MacDonald DD, Jin DK, Shido K, Kerns SA, et al. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature. 2005;438:820–827. doi: 10.1038/nature04186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnoub AE, Dash AB, Vo AP, Sullivan A, Brooks MW, Bell GW, Richardson AL, Polyak K, Tubo R, Weinberg RA. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449:557–563. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- Kim MY, Oskarsson T, Acharyya S, Nguyen DX, Zhang XH, Norton L, Massagué J. Tumor self-seeding by circulating cancer cells. Cell. 2009;139:1315–1326. doi: 10.1016/j.cell.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Wang Q, Hu G, Van Poznak C, Fleisher M, Reiss M, Massagué J, Kang Y. ADAMTS1 and MMP1 proteolytically engage EGF-like ligands in an osteolytic signaling cascade for bone metastasis. Genes Dev. 2009;23:1882–1894. doi: 10.1101/gad.1824809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez-Ferrer S, Michurina TV, Ferraro F, Mazloom AR, Macarthur BD, Lira SA, Scadden DT, Ma'ayan A, Enikolopov GN, Frenette PS. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466:829–834. doi: 10.1038/nature09262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massagué J. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra PJ, Mishra PJ, Humeniuk R, Medina DJ, Alexe G, Mesirov JP, Ganesan S, Glod JW, Banerjee D. Carcinoma-associated fibroblast-like differentiation of human mesenchymal stem cells. Cancer Res. 2008;68:4331–4339. doi: 10.1158/0008-5472.CAN-08-0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- Orimo A, Gupta PB, Sgroi DC, Arenzana-Seisdedos F, Delaunay T, Naeem R, Carey VJ, Richardson AL, Weinberg RA. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121:335–348. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- Qian BZ, Li J, Zhang H, Kitamura T, Zhang J, Campion LR, Kaiser EA, Snyder LA, Pollard JW. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 2011;475:222–225. doi: 10.1038/nature10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quante M, Tu SP, Tomita H, Gonda T, Wang SS, Takashi S, Baik GH, Shibata W, Diprete B, Betz KS, et al. Bone marrow-derived myofibroblasts contribute to the mesenchymal stem cell niche and promote tumor growth. Cancer Cell. 2011;19:257–272. doi: 10.1016/j.ccr.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147:275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- Weigelt B, Glas AM, Wessels LF, Witteveen AT, Peterse JL, van't Veer LJ. Gene expression profiles of primary breast tumors maintained in distant metastases. Proc Natl Acad Sci U S A. 2003;100:15901–15905. doi: 10.1073/pnas.2634067100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Huang WC, Li P, Guo H, Poh SB, Brady SW, Xiong Y, Tseng LM, Li SH, Ding Z, et al. Combating trastuzumab resistance by targeting SRC, a common node downstream of multiple resistance pathways. Nat Med. 2011;17:461–469. doi: 10.1038/nm.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XH, Wang Q, Gerald W, Hudis CA, Norton L, Smid M, Foekens JA, Massagué J. Latent bone metastasis in breast cancer tied to Src-dependent survival signals. Cancer cell. 2009;16:67–78. doi: 10.1016/j.ccr.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.