Background: Non-segmented negative-strand RNA (NNS) virus polymerase initiation and elongation are targets for intervention.
Results: 2′ modifications to the template and NTP inhibit elongation more than initiation by the vesicular stomatitis virus polymerase.
Conclusion: Polymerase is sensitive to template and substrate modifications, and its active site is directly influenced by the nucleoprotein.
Significance: Nucleoprotein influences the substrate utilization of an NNS RNA virus polymerase.
Keywords: Enzyme Inhibitors, Negative-strand RNA Viruses, Nucleoside Nucleotide Analogs, RNA, RNA Polymerase
Abstract
The RNA synthesis machinery of non-segmented negative-sense RNA viruses comprises a ribonucleoprotein complex of the genomic RNA coated by a nucleocapsid protein (N) and associated with polymerase. Work with vesicular stomatitis virus (VSV), a prototype, supports a model of RNA synthesis whereby N is displaced from the template to allow the catalytic subunit of the polymerase, the large protein (L) to gain access to the RNA. Consistent with that model, purified L can copy synthetic RNA that contains requisite promoter sequences. Full processivity of L requires its phosphoprotein cofactor and the template-associated N. Here we demonstrate the importance of the 2′ position of the RNA template and the substrate nucleotide triphosphates during initiation and elongation by L. The VSV polymerase can initiate on both DNA and RNA and can incorporate dNTPs. During elongation, the polymerase is sensitive to 2′ modifications, although dNTPs can be incorporated, and mixed DNA-RNA templates can function. Modifications to the 2′ position of the NTP, including 2′,3′-ddCTP, arabinose-CTP, and 2′-O-methyl-CTP, inhibit polymerase, whereas 2′-amino-CTP is incorporated. The inhibitory effects of the NTPs were more pronounced on authentic N-RNA with the exception of dGTP, which is incorporated. This work underscores the sensitivity of the VSV polymerase to nucleotide modifications during initiation and elongation and highlights the importance of the 2′-hydroxyl of both template and substrate NTP. Moreover, this study demonstrates a critical role of the template-associated N protein in the architecture of the RNA-dependent RNA polymerase domain of L.
Introduction
The RNA synthesis machinery of non-segmented negative-sense (NNS)2 RNA viruses is a ribonucleoprotein complex extensively studied for vesicular stomatitis virus (VSV). The VSV ribonucleoprotein comprises an 11,161-nt RNA completely coated by the viral nucleocapsid protein (N) to form the N-RNA template for the RNA-dependent RNA polymerase (RdRP). The RdRP resides within the 241-kDa large (L) protein that also contains the catalytic activities necessary for cap addition, namely a GDP:polyribonucleotidyltransferase and a dual specificity mRNA cap methyltransferase (1–5). VSV L has the appearance of a core ring-like domain to which the RdRp maps and an appendage of three globules involved in mRNA capping (6). L alone cannot, however, copy the N-RNA template and, instead, requires a 29-kDa phosphoprotein (P) to bridge interactions with the template. In addition to this structural role, P also induces a conformational change in L and renders the RdRP more processive. Full processivity of L also depends on the template-associated N (7, 8).
N-RNA structures have been solved for several NNS RNA viruses, including VSV (9–11). In each case, the RNA is sequestered between N- and C-terminal domains of the protein. Those structural data led to the model that N molecules must transiently dissociate from the RNA to allow the polymerase access to its template. Further support for this model comes from recent work that demonstrates that L alone can copy naked RNA in vitro (7). This latter assay has been replicated to study the biology of other negative-strand RNA viruses, including the significant human pathogens Machupo and respiratory syncytial virus. This is an achievement that holds promise in studies to define the activities of polymerase inhibitors that could serve as important start points for antiviral development (12, 13).
The mechanistic basis of RNA synthesis initiation in NNS RNA viruses has been inferred from atomic structures of other RdRPs as well as in vitro analyses of polymerase activity. Landmark studies with the polymerases of double-strand and positive-strand RNA viruses (14–18) show that the incoming nucleotide contacts the polymerase active site via the triphosphate and ribose moieties, with the base interacting mainly with the primer and template. In the vicinity of a nucleotide-binding pocket, the 2′-hydroxyl group of the ribose of the incoming nucleotide forms critical hydrogen bonds with the side chain of essential amino acids that leads to correct RNA synthesis. In the same manner, the RNA template engages in specific interactions with the active site. Irrespective of whether the RdRP depends on a primer to initiate RNA synthesis or initiates synthesis de novo, interactions between the template and active site and the incoming NTPs are critical (17, 19–21).
Earlier work with VSV demonstrates that L can incorporate some modified nucleotides into mRNA in vitro (22). Among the molecules tested were dGTP, which was efficiently incorporated into mRNA. In this study, we investigate the role of the 2′-hydroxyl of both the incoming nucleotide and the template on RNA synthesis by VSV L. We observe that initiation is most efficient when the first two nucleotides incorporated contain a 2′-hydroxyl group. Similarly, the presence of the 2′-hydroxyl group on nucleotides incorporated at subsequent positions facilitates elongation. VSV L can, however, initiate RNA synthesis on a template devoid of a 2′-hydroxyl group but fails to elongate beyond position 5 on this template. Distinctions between the inhibitory effect of modified nucleotides on N-RNA versus naked RNA templates underscore the role of the template-associated N protein in the organization of the RdRP domain.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Recombinant VSV L and P were expressed and purified as described in Ref. 6. Briefly, 6× His-tagged L was expressed in Spodoptera frugiperda 21 (Sf21) cells and affinity-purified by nickel-nitrilotriacetic acid-agarose (Qiagen), followed by MonoS (GE Healthcare) chromatography. 10× His-tagged P was expressed in Escherichia coli BL21 (DE3) cells and affinity-purified with nickel-nitrilotriacetic acid-agarose (Qiagen), followed by gel filtration (Superdex 200 HR 10/30, GE Healthcare).
In Vitro Polymerase Assay
Naked RNA, DNA, and DNA-RNA templates were chemically synthesized and purified (Integrated DNA Technologies).
Polymerase assays were carried out as described in Ref. 7 using 0.4 μm template (RNA, DNA, or DNA-RNA) with 0.1 μm VSV L and 0.3 μm VSV P in a reaction mixture containing 20 mm Tris base (pH 8), 50 mm NaCl, 6 mm MgCl2, 200 μm UTP or dUTP, 1.5 mm ATP or dATP, 1.5 mm CTP or dCTP, and 165 nm of [α-32P]GTP (3000 Ci/mmol). Assays studying the effects of dGTP incorporation were carried out using 200 μm GTP or dGTP with either [α-32P]GTP (3000 Ci/mmol) or [α-32P]dGTP (3000 Ci/mmol). The standard polymerase assay was supplemented with 250 μm GTP when N-RNA was used as a template. Reactions were incubated at 30 °C for 3 h and stopped by addition of EDTA/formamide. Reactions products were resolved using denaturing polyacrylamide/urea gel electrophoresis in TBE buffer and analyzed by autoradiography. The sizes of the products were determined by comparison to a 19-nt marker RNA labeled by T4 polynucleotide kinase (New England Biolabs) using [γ-32P]ATP (3000 Ci/mmol). To determine the effects of modified CTP nucleotides on RNA synthesis by L, CTP was replaced by either 2′-amino CTP, 2′,3′-ddCTP, arabinose-CTP, and 2′-O-methyl-CTP (TriLink).
RESULTS
Effect of the 2′-Hydroxyl Group of the Initiating Nucleotide on RNA Synthesis
During the early steps of transcription, VSV L initiates RNA synthesis by forming a phosphodiester bond between the first two nucleotides, ATP and CTP, respectively. Those initiating NTPs are required at high concentrations (7). To study the role of the 2′-hydroxyl group of the ribose of the initiating nucleotides on RNA synthesis, we compared initiation in the presence of dATP and dCTP using an in vitro polymerase assay. Briefly, purified recombinant L and a chemically synthesized RNA corresponding to the first 19 nt of the leader (Fig. 1B) are incubated in the presence of NTPs, and the products of RNA synthesis are analyzed by electrophoresis on acrylamide gels (7). Replacement of ATP by dATP results in the synthesis of RNAs ranging up to ∼11 nt (Fig. 1A). This result demonstrates that L can initiate with and incorporate dATP, albeit less efficiently than ATP (Fig. 1, B and C). Incorporation of dATP could also be visualized by the slight difference in the migration position of every product compared with their RNA homologues. Although L is able to initiate RNA synthesis with dATP, almost no products with a size longer than 12 nt were synthesized, suggesting that the 2′-hydroxyl group of the ribose may facilitate elongation. Replacement of CTP with dCTP also results in initiation, although less readily than the substitution of ATP for dATP (Fig. 1, B and C), although the polymerase can elongate further in the presence of dCTP (Fig. 1B). As the concentration of dATP or dCTP increases from 1 to 3 mm, the inhibition becomes more pronounced (data not shown). This result suggests that the 2′-hydroxyl group of the second nucleotide also plays an important role in initiation.
FIGURE 1.
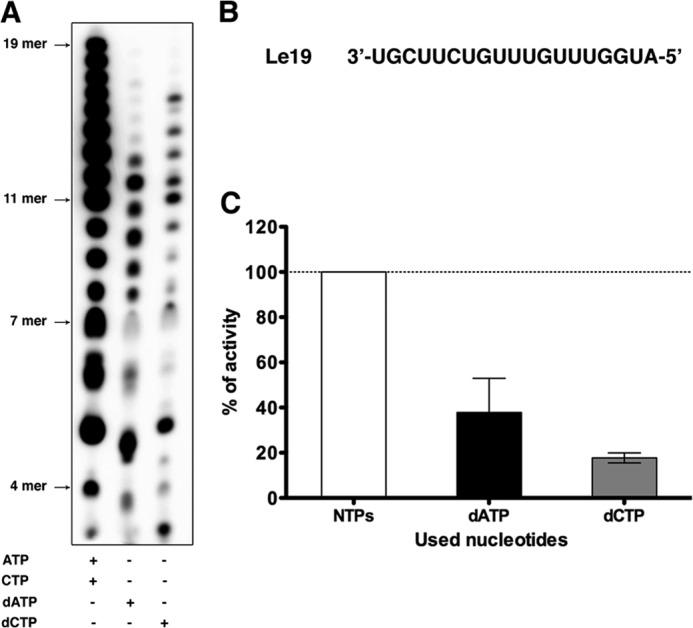
VSV L can initiate with dATP and dCTP. A, de novo RNA synthesis by VSV L-P on Le19 with all the NTPs and either ATP replaced by dATP or CTP replaced by dCTP. Activity assays were set up as described under “Experimental Procedures” using 0.1 μm Le19, 0.4 μm proteins, and [α-32P]GTP. Reactions were quenched by the addition EDTA/formamide and analyzed on a 20% polyacrylamide/7 M urea gel. De novo initiation product sizes are indicated on the left. B, sequence of the Le19 RNA. C, the total amount of product synthesized in A was quantified with a PhosphorImager, normalized to the reaction setup with all the NTPs, and graphed. Error bars represent the mean ± S.D. of independent experiments.
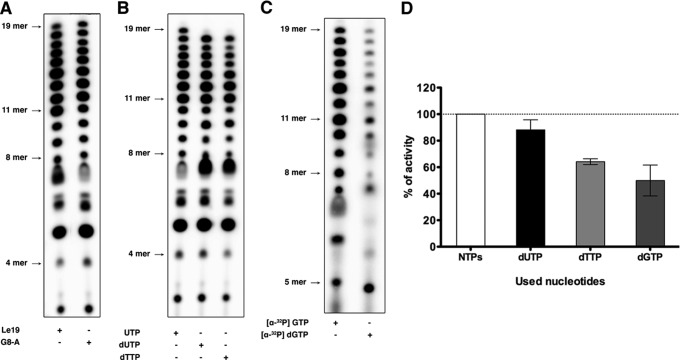
VSV L Incorporates dGTP, dUTP, and dTTP
We demonstrated previously that GTP and UTP are required at a lower concentration by VSV L to initiate RNA synthesis (7). The above experiments indicate that the 2′-hydroxyl of the incorporated nucleotide can influence the processivity of the polymerase. We therefore evaluated the effects of inclusion of dUTP, dTTP, and dGTP on RNA synthesis. Because UTP is normally the 19th nt incorporated, we elected to use an altered RNA template that specifies incorporation of UTP at position 8 (G8A). Copying of this G8A template results in the synthesis of less 7-mer product (Fig. 2A), which may naturally reflect a more efficient incorporation of UTP than CTP at position 8 or an interaction of the base of the template at position 8. Irrespective of the explanation for this small difference, dUTP was efficiently incorporated in place of UTP at position 8, but its incorporation was barely detected at position 19 (Fig. 2, B and D). Moreover, more 7-nt product is present when UTP is replaced by dUTP, reflecting less efficient incorporation of dUTP than UTP in position 8. In DNA, uridine is replaced by a thymidine that differs only by the presence of a methyl group in position 5 of the base in the latter. Thus, we compared the incorporation of dUTP and dTTP by VSV L on the G8A template. We found that L incorporates dTTP less efficiently than dUTP, indicating that the presence of the methyl group of the thymidine base may obstruct the incoming nucleotide tunnel or the extraction of the newly synthesized RNA. Although dUTP is well incorporated in position 8, its incorporation at position 19 is very weak, reflecting a possible role of the 2′-hydroxyl group in RNA synthesis elongation. We then compared the incorporation of UTP and dUTP on a double mutant template where both the G8 and U13 positions were replaced by an A nucleotide (G8A/U13A). Although synthesis on G8A and G8A/U13A with UTP is very similar, synthesis with dUTP induces a major stop at position 12 (Fig. 3). Although incorporation of dUTP at position 8 was not critical for RNA synthesis, incorporation of dUTP in further positions blocks RNA synthesis. This result underscores the importance of the 2′-hydroxyl group of the incorporated nucleotides during elongation. The VSV polymerase was also able to efficiently incorporate dGTP into products (Fig. 2, C and D). Collectively, these data demonstrate that the VSV polymerase can initiate using dNTPs and incorporate them into product. The efficiency of incorporation varied with dNTP so that dCTP < dATP < dGTP < dTTP < dUTP. Because ATP and CTP are critical for initiation, these data imply that the 2′-hydroxyl group on the incoming NTP plays a key role in initiation. The 2′-hydroxyl group also influences elongation, as demonstrated by the blocks observed in the presence of each dNTP. However, the impact of the 2′-hydroxyl on elongation is not fully clear from these studies because their impact was influenced by their number and location and by the specific identity of the nucleotide.
FIGURE 2.
VSV L can use dGTP, dUTP, and dTTP. A, de novo RNA synthesis by VSV L-P on Le19 or the G8A mutant. B, de novo RNA synthesis by VSV L-P on G8A with all NTPs and either UTP replaced by dUTP or dTTP. C, de novo RNA synthesis by VSV L-P on Le19 with all the NTPs using [α-32P]GTP or GTP replaced by dGTP using [α-32P]dGTP. D, the total amounts of product synthesized in B and C were quantified with a PhosphorImager, normalized, and graphed. Error bars represent the mean ± S.D. of independent experiments.
FIGURE 3.
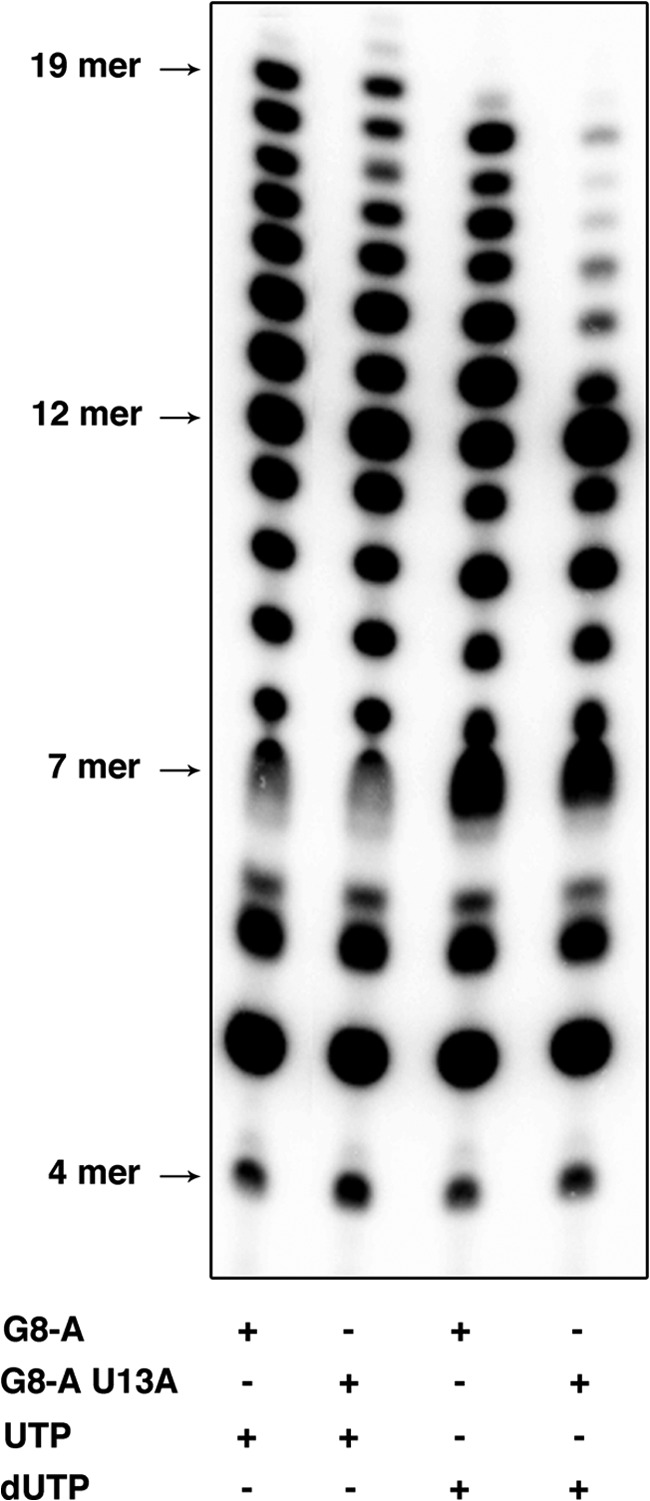
dUTP incorporation by VSV L. De novo RNA synthesis by VSV L-P on G8A and G8A/U13A mutants with either UTP or dUTP.
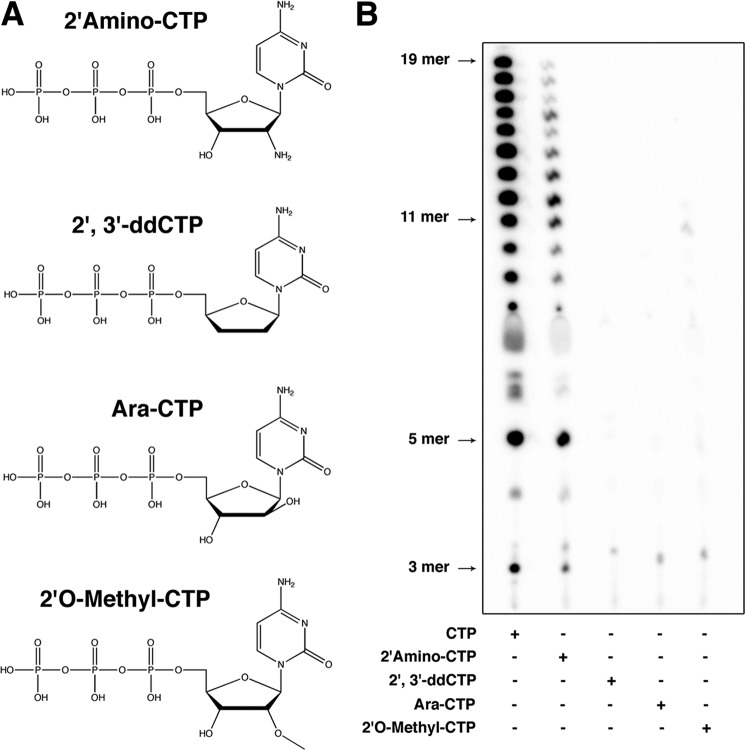
Effects of 2′-modified Nucleotides on RNA Synthesis by L
To investigate the role of 2′ modification on the incoming NTP, we elected to replace CTP with four different 2′-modified analogs: 2′-amino-CTP, 2′,3′-ddCTP, arabinose-CTP, and 2′-O-methyl-CTP (Fig. 4A). During RNA synthesis by L on Le19, CTP is incorporated at the second position. As expected, no RNA synthesis is observed when the CTP is replaced by 2′,3′-ddCTP because of the lack of a hydroxyl group at position 3′ (Fig. 4B). Neither arabinose-CTP nor 2′-O-methyl-CTP were incorporated by the polymerase because we did not observe the expected pppApCpG trinucleotide. By contrast, 2′-amino-CTP was incorporated by L, and the polymerase was able to elongate the RNA chain. This result may reflect the fact that the space use by the amino group located at the 2′ position of the ribose is similar to the space use by the hydroxyl group, with the exception of the presence of a second hydrogen atom. The decrease in RNA synthesis with 2′-amino-CTP could reflect a disruption of this extra hydrogen in the vicinity of the active site. The modification nitrogen/oxygen could also alter interactions with specific amino-acids in L that bind or discriminate the 2′ position of the incoming NTP. The presence of a methyl group bound to the oxygen located at the 2′ position of the ribose most probably generates a steric obstruction that would block RNA synthesis by L. Surprisingly, when the ribose is replaced by arabinose, no RNA synthesis was observed. Because dCTP could be used by VSV L, the fact that no RNA synthesis is observed with the arabinose-CTP indicates that the position of the hydroxyl in a space that was originally devoid of any atoms blocks RNA synthesis by L. These results suggest that the vicinity surrounding the 2′ position of the incoming nucleotide is essential for correct RNA synthesis by VSV L.
FIGURE 4.
Effects of 2′-modified CTP on RNA synthesis by VSV L. A, structure of the 2′-modified nucleotides tested for RNA synthesis by VSV L. B, de novo RNA synthesis by VSV L-P with either CTP, 2′-amino-CTP, 2′,3′-ddCTP, arabinose-CTP, and 2′-O-methyl-CTP.
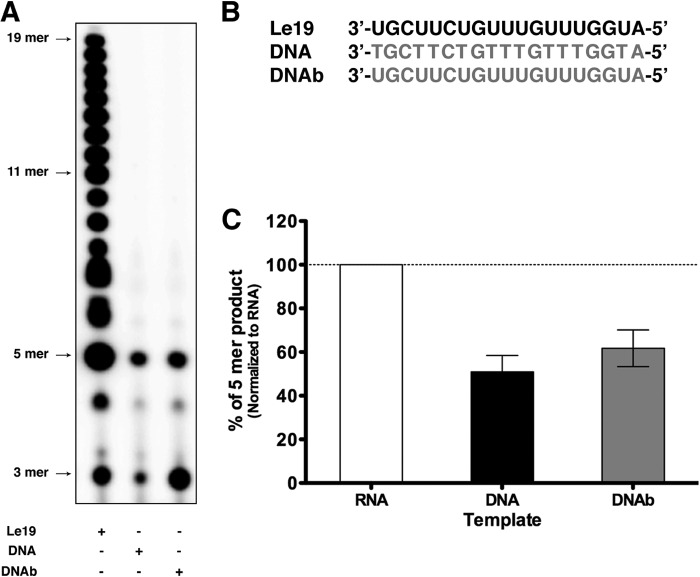
VSV L Initiates RNA Synthesis on a DNA Template
We next investigated the role of the 2′-hydroxyl group of the template during RNA synthesis by VSV L using a DNA corresponding to Le19 sequence (DNAb) and a similar DNA where the U bases are replaced by T (DNA) (Fig. 5B). Both DNAs served as template for synthesis of a 5-nt-long product. Longer products were not detected (Fig. 5A). This result demonstrates that L can initiate RNA synthesis with a template lacking the 2′-hydroxyl group but cannot use these templates for elongation of RNA synthesis.
FIGURE 5.
VSV L initiates RNA synthesis on DNA templates. A, de novo RNA synthesis by VSV L-P with Le19, DNA, and DNAb. B, sequences of Le19, DNA, and DNAb. Gray letters indicate deoxyribonucleotides. C, the total amount of 5-mer product synthesized in A was quantified with a PhosphorImager, normalized, and graphed. Error bars represent the mean ± S.D. of independent experiments.
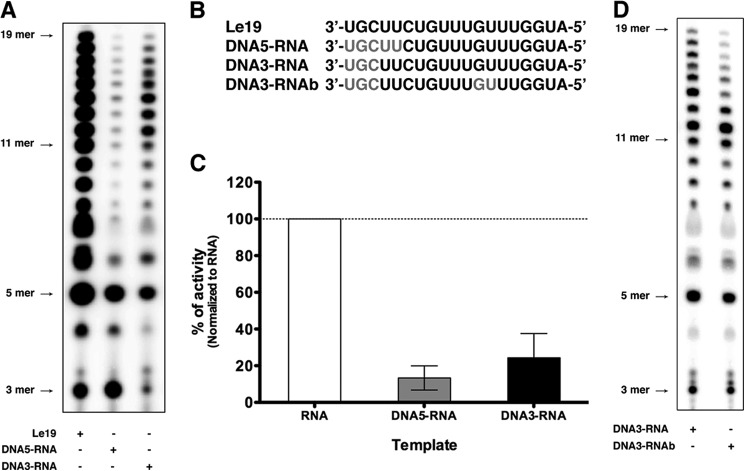
To further study the effect of the 2′-hydroxyl group of the template in RNA synthesis, we employed two templates corresponding to Le19 with either three or five nucleotides (DNA3-RNA or DNA5-RNA, respectively) replaced by deoxynucleotides at their 3′ end (Fig. 6B). Although total RNA synthesis was reduced, products up to 19 nt were detected from both templates (Fig. 6A). More products were observed for the DNA3-RNA template (Fig. 6, A and C), suggesting that the transition from initiation to elongation is more efficient in the presence of the 2′-hydroxyl group. We next examined the impact of introducing two additional deoxynucleotide sites at subsequent positions in the template. Comparing RNA synthesis by L-P on DNA3-RNA and on the same template where two nucleotides were replaced by deoxynucleotides in positions 12 and 13 (DNA3-RNAb), we found an inhibition of products longer than 13 nt with DNA3-RNAb (Fig. 6D). These results show that the active site of L can use a template devoid of the 2′-hydroxyl group to initiate RNA synthesis but dramatically influences the transition from initiation to elongation. The result also confirms that subsequent sites of dNTP incorporation are more efficiently tolerated. This may reflect a critical role of interactions between the 2′-hydroxyl group and a presumed exit tunnel through which the template must transit.
FIGURE 6.
The RNA template is required by VSV L to elongate RNA synthesis. A, de novo RNA synthesis by VSV L-P with Le19, DNA5-RNA, and DNA3-RNA. B, sequences of Le19, DNA5-RNA, DNA3-RNA, and DNA3-RNAb. Gray letters indicate deoxyribonucleotides. C, the total amount of product synthesized in A was quantified with a PhosphorImager, normalized, and graphed. Error bars represent the mean ± S.D. of independent experiments. D, de novo RNA synthesis by VSV L-P with DNA3-RNA and DNA3-RNAb.
The Template-associated N Protein Alters the Sensitivity of the Polymerase to Different Nucleotide Analogs
The template-associated N protein is essential for full processivity of the polymerase complex (7). Although the mechanistic underpinning of this effect is not understood, we hypothesized that the RdRP domain may undergo structural alterations in the presence of N. We therefore investigated whether the template-associated N alters the ability of L to incorporate 2′-modified nucleotides. During transcription of the N-RNA template, a 47-nt leader RNA is synthesized (Fig. 7). In the presence of dATP, no products of RNA synthesis were observed, indicating that L cannot initiate (Fig. 7). In contrast, dCTP, dTTP, and dUTP permit initiation and elongation, and dGTP was well tolerated (Fig. 7). As observed with the naked RNA template, the only 2′-modified CTP incorporated is 2′-amino-CTP (Fig. 7). This result confirms that the space and charge of the amino group can fit in the vicinity of the active site. However, the low ratio of RNA synthesis with this modified CTP suggests that the interaction of the amino group with the active site is less efficient than the interaction of the hydroxyl. As observed previously, none of the other 2′-modified CTPs can be incorporated by L. Where incorporated, dNTPs also result in defects in polymerase processivity, as evident from the premature termination products observed for dUTP, dTTP, dCTP, and even dGTP (Fig. 7, note the increase in the abundance of products around 20 nt). The differences observed in RNA synthesis on the naked and encapsidated template demonstrate that the active site of the RdRP domain of L is ordered by the template-associated N protein.
FIGURE 7.
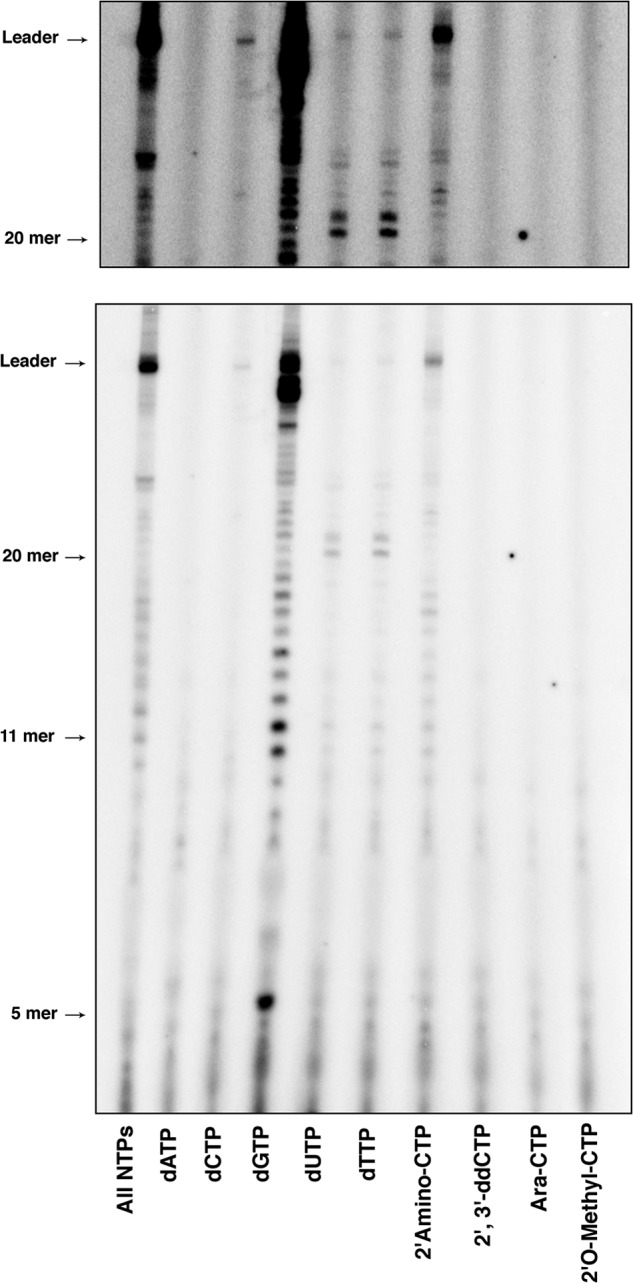
Effects of N on incorporation of 2′-modified NTPs during RNA synthesis by VSV L. Shown is de novo RNA synthesis by VSV L-P on the N-RNA template with either all NTPs; ATP replaced by dATP; GTP replaced by dGTP; UTP replaced by dUTP or dTTP; and CTP replaced by either dCTP, 2′-amino-CTP, 2′,3′-ddCTP; arabinose-CTP, or 2′-O-methyl-CTP. The top panel shows an eight times longer exposure of the products between 20 nucleotides and the leader.
DISCUSSION
Several classes of polymerase inhibitors are attractive candidates for antiviral therapy. They include nucleoside and nucleotide inhibitors that serve as alternate substrates for the polymerase as well as non-nucleoside inhibitors that serve as allosteric inhibitors of polymerase function. Here we evaluated the impact of a set of 2′ substitutions in either the template or substrate nucleotides on the activity of the VSV polymerase in vitro. Several conclusions are apparent from our studies: (i) 2′ modifications can serve to inhibit polymerase activity in vitro; (ii) polymerase alters insensitivity to such inhibition during the initiation versus elongation phases of RNA synthesis, a transition that appears to occur at a nascent chain length of 5-nucleotides; and (iii) the template associated N protein alters the sensitivity of the polymerase to specific nucleotide analogs. This study has implications for our understanding of polymerase function for VSV and perhaps other NNS RNA viruses and is instructive for further consideration of modified nucleotides as potential antivirals.
Although atomic level structural data do not yet exist for the NNS RNA virus polymerases, the conserved properties of all polymerases for which structures are available provide a useful framework for the interpretation of our data. Those studies show that RNA synthesis requires several steps of alignment and realignment between the template substrate and enzyme. The template interacts with the catalytically active site of the enzyme, the initiating nucleotide interacts specifically with both the template and the active site, the polymerase undergoes conformational changes to be in an elongation stage, and the nascent RNA is released from the polymerase. RdRPs, DNA-dependent RNA polymerases, reverse transcriptases, and DNA-dependent RNA polymerases all share a right hand-like shape with fingers, palm, and thumb domains (for a review, see Refs. 20, 23–25). The similarities between the polymerases extend to their mechanism of catalysis, in which they also share a two-metal mechanism. The most conserved domain between polymerases is the palm region that contains the active site. In addition to the common features shared with all polymerases, RdRPs possess specific features, including a closed hand shape compared with the “open” or U conformations for other polymerases. During RNA synthesis, the RNA enters and binds the active site via a template tunnel well defined by the fingertip region. The incoming nucleotides access the active site via a small, positively charged tunnel. The most divergent part of RdRPs is the thumb, which differs in size and shape between the RdRPs from several viral families.
Substitutions at the 2′ position of the ribose can block the activity of several RdRP molecules in vitro, including the NS5B protein of the hepatitis C virus (26). Although we did not screen an exhaustive list of such modified nucleotides, we show that most substitutions at the 2′ position inhibit VSV polymerase activity in vitro. Of the set of modifications that we tested, the best tolerated by the VSV polymerase was alteration of the hydroxyl to a methyl group, as found for dNTPs, or to an amino group. During elongation, the 2′-hydroxyl group of the incoming nucleotide likely undergoes specific interactions in the newly formed nucleotide tunnel that are essential for its correct transportation to the active site. Biochemical and structural data on RNA polymerases support that polymerases transit from an initiation complex to an elongation complex for full processivity (18, 20, 25, 27–29). The poliovirus RdRP also incorporate dNTPs, but at reduced efficiency (15).
We also found that the polymerase could initiate on DNA as a template but that elongation was blocked. This work suggests that 2′-modified nucleotides might be considered as an antiviral approach. There are, however, challenges to the use of such modified nucleotides. Invariably, the nucleoside analog must first be taken up by the cell and converted by the cellular kinases to the triphosphate form. Consequently the modification cannot inhibit those cellular enzymes. Moreover, the resulting NTP must have a high selectivity index for the viral enzyme with little off-target inhibition of other polymerases. We did not examine any of those aspects in this study.
Our finding that the VSV polymerase can use dNTPs is not without precedent (22). A prior study demonstrated that VSV polymerase could incorporate dGTP and, to a lesser extent, dCTP but not dATP, dUTP, or dTTP. In addition, dGTP could serve as a substrate in the mRNA-capping reaction, which is catalyzed by an unconventional GDP::polyribonucleotidyltransferase in L (2, 4). In this study, we found that dATP was also a poor substrate for the polymerase but observed some incorporation of dUTP, dTTP, and dCTP and efficient incorporation of dGTP. There were, however, marked differences in their incorporation during copying of a naked RNA template so that each of the dNTPs could serve as a substrate. This result underscores the fact that the template-associated N protein must play a role in the organization of the architecture of the catalytic domain of the RdRP so that, in the absence of the N protein, the polymerase is generally more tolerant of altered nucleotides. Thus, the presence of N protein on the template facilitates an arrangement of the RdRP domain of L that results in a greater discrimination of the incoming NTPs. Such a discrimination could be manifested by alterations to the nucleotide entrance tunnel, the template tunnels, the catalytic site itself, or a combination of all three. The ability to catalyze RNA synthesis on naked RNA has been established for negative-sense RNA viruses that are pathogenic for humans, including respiratory syncytial virus (13). Such assays represent a significant advance in our ability to interrogate whether nucleotide analogs can block polymerase activity, but it will be important to consider how the template-associated N protein influences such results.
In addition to the use of dNTPs as substrate for the VSV polymerase, we establish that DNA can also serve as template. In copying DNA, however, the polymerase is unable to transcribe a product greater than 5 nt in length. The polymerase that synthesizes a 5-nt RNA product from a DNA template can, however, continue to copy its template provided the remainder is RNA. We interpret that finding as reflecting a critical structural transition in L that occurs shortly following initiation that renders the 2′-hydroxyl of the template critical. By analogy with RdRPs for which atomic structural data exist, we anticipate that the template would interact specifically with the exit tunnel via the 2′-hydroxyl groups. Other RdRPs can also utilize DNA as template. For example, brome mosaic virus, a positive-strand RNA virus, can initiate synthesis of a full-length RNA product using a DNA promoter template with almost the same efficiency as the RNA template (30, 31). The nonstructural protein 5B from bovine viral diarrhea virus (32) can also use DNA. However, bovine viral diarrhea virus RNA synthesis requires a primer to initiate, which indicates that VSV and bovine viral diarrhea virus polymerases have different initiation mechanism, and the transition initiation elongation likely differs between these two polymerases. All template-dependent polynucleotide polymerases likely derive from a common ancestor, which likely accounts for the ability of the various RdRPs to copy DNA templates. The relative differences in the efficiency of this may reflect distinctions in the evolution of positive- and negative-strand RNA virus polymerases with respect to requirements of the 2′-hydroxyl group for template binding and transit through the exit tunnel. The mechanism by which some 2′ substitutions inhibit the polymerase may also reflect the impact of such modifications on the sugar itself. For example, DNA can adopt alternate conformations, reflecting differences in the sugar pucker. We did not examine how dNTP incorporation or the use of DNA templates influence polymerase but note that the effects are more pronounced as the length of a DNA template increases or as the number of incorporated dNTPs in the nascent chain increases. Further studies will be required to discern the mechanism of inhibition.
In summary, this study demonstrates the critical nature of the 2′-hydroxyl position of the NTP and RNA template in the process of RNA synthesis by a prototype NNS RNA virus. This work shows that substituted nucleotides do inhibit polymerase activity and demonstrates that the polymerase is much more tolerant of incorporating modified nucleotides during the process of initiation. Importantly, this work provides evidence for the influence of the template-associated N protein on the RdRP activity of L, a finding that has important considerations in testing antivirals against polymerase and in structural studies of the protein.
Acknowledgments
We thank Robin Ross for assistance with protein purification and Amal Rahmeh for review of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants AI059371 and AI057159.
- NNS
- non-segmented negative-sense
- VSV
- vesicular stomatitis virus
- N
- nucleocapsid protein
- L
- large protein
- P
- phosphoprotein
- RdRP
- RNA-dependent RNA polymerase
- TBE
- Tris borate-EDTA
- nt
- nucleotide(s).
REFERENCES
- 1. Li J., Fontaine-Rodriguez E. C., Whelan S. P. (2005) Amino acid residues within conserved domain VI of the vesicular stomatitis virus large polymerase protein essential for mRNA cap methyltransferase activity. J. Virol. 79, 13373–13384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Li J., Rahmeh A., Morelli M., Whelan S. P. (2008) A conserved motif in region V of the large polymerase proteins of nonsegmented negative-sense RNA viruses that is essential for mRNA capping. J. Virol. 82, 775–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li J., Wang J. T., Whelan S. P. (2006) A unique strategy for mRNA cap methylation used by vesicular stomatitis virus. Proc. Natl. Acad. Sci. U.S.A. 103, 8493–8498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ogino T., Banerjee A. K. (2007) Unconventional mechanism of mRNA capping by the RNA-dependent RNA polymerase of vesicular stomatitis virus. Mol. Cell 25, 85–97 [DOI] [PubMed] [Google Scholar]
- 5. Sleat D. E., Banerjee A. K. (1993) Transcriptional activity and mutational analysis of recombinant vesicular stomatitis virus RNA polymerase. J. Virol. 67, 1334–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rahmeh A. A., Schenk A. D., Danek E. I., Kranzusch P. J., Liang B., Walz T., Whelan S. P. (2010) Molecular architecture of the vesicular stomatitis virus RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 107, 20075–20080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Morin B., Rahmeh A. A., Whelan S. P. (2012) Mechanism of RNA synthesis initiation by the vesicular stomatitis virus polymerase. EMBO J. 31, 1320–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rahmeh A. A., Morin B., Schenk A. D., Liang B., Heinrich B. S., Brusic V., Walz T., Whelan S. P. (2012) Critical phosphoprotein elements that regulate polymerase architecture and function in vesicular stomatitis virus. Proc. Natl. Acad. Sci. U.S.A. 109, 14628–14633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Albertini A. A., Wernimont A. K., Muziol T., Ravelli R. B., Clapier C. R., Schoehn G., Weissenhorn W., Ruigrok R. W. (2006) Crystal structure of the rabies virus nucleoprotein-RNA complex. Science 313, 360–363 [DOI] [PubMed] [Google Scholar]
- 10. Green T. J., Zhang X., Wertz G. W., Luo M. (2006) Structure of the vesicular stomatitis virus nucleoprotein-RNA complex. Science 313, 357–360 [DOI] [PubMed] [Google Scholar]
- 11. Tawar R. G., Duquerroy S., Vonrhein C., Varela P. F., Damier-Piolle L., Castagné N., MacLellan K., Bedouelle H., Bricogne G., Bhella D., Eléouët J. F., Rey F. A. (2009) Crystal structure of a nucleocapsid-like nucleoprotein-RNA complex of respiratory syncytial virus. Science 326, 1279–1283 [DOI] [PubMed] [Google Scholar]
- 12. Kranzusch P. J., Whelan S. P. (2011) Arenavirus Z protein controls viral RNA synthesis by locking a polymerase-promoter complex. Proc. Natl. Acad. Sci. U.S.A. 108, 19743–19748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Noton S. L., Deflubé L. R., Tremaglio C. Z., Fearns R. (2012) The respiratory syncytial virus polymerase has multiple RNA synthesis activities at the promoter. PLoS Pathog. 8, e1002980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Arnold J. J., Gohara D. W., Cameron C. E. (2004) Poliovirus RNA-dependent RNA polymerase (3Dpol). Pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mn2+. Biochemistry 43, 5138–5148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gohara D. W., Arnold J. J., Cameron C. E. (2004) Poliovirus RNA-dependent RNA polymerase (3Dpol). Kinetic, thermodynamic, and structural analysis of ribonucleotide selection. Biochemistry 43, 5149–5158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gohara D. W., Crotty S., Arnold J. J., Yoder J. D., Andino R., Cameron C. E. (2000) Poliovirus RNA-dependent RNA polymerase (3Dpol). Structural, biochemical, and biological analysis of conserved structural motifs A and B. J. Biol. Chem. 275, 25523–25532 [DOI] [PubMed] [Google Scholar]
- 17. Ng K. K., Arnold J. J., Cameron C. E. (2008) Structure-function relationships among RNA-dependent RNA polymerases. Curr. Top. Microbiol. Immunol. 320, 137–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zamyatkin D. F., Parra F., Alonso J. M., Harki D. A., Peterson B. R., Grochulski P., Ng K. K. (2008) Structural insights into mechanisms of catalysis and inhibition in Norwalk virus polymerase. J. Biol. Chem. 283, 7705–7712 [DOI] [PubMed] [Google Scholar]
- 19. Butcher S. J., Grimes J. M., Makeyev E. V., Bamford D. H., Stuart D. I. (2001) A mechanism for initiating RNA-dependent RNA polymerization. Nature 410, 235–240 [DOI] [PubMed] [Google Scholar]
- 20. Ferrer-Orta C., Arias A., Escarmís C., Verdaguer N. (2006) A comparison of viral RNA-dependent RNA polymerases. Curr. Opin. Struct. Biol. 16, 27–34 [DOI] [PubMed] [Google Scholar]
- 21. Tao Y., Farsetta D. L., Nibert M. L., Harrison S. C. (2002) RNA synthesis in a cage. Structural studies of reovirus polymerase λ3. Cell 111, 733–745 [DOI] [PubMed] [Google Scholar]
- 22. Schubert M., Lazzarini R. A. (1982) In vitro transcription of vesicular stomatitis virus. Incorporation of deoxyguanosine and deoxycytidine, and formation of deoxyguanosine caps. J. Biol. Chem. 257, 2968–2973 [PubMed] [Google Scholar]
- 23. Kao C. C., Singh P., Ecker D. J. (2001) De novo initiation of viral RNA-dependent RNA synthesis. Virology 287, 251–260 [DOI] [PubMed] [Google Scholar]
- 24. Steitz T. A. (1998) A mechanism for all polymerases. Nature 391, 231–232 [DOI] [PubMed] [Google Scholar]
- 25. van Dijk A. A., Makeyev E. V., Bamford D. H. (2004) Initiation of viral RNA-dependent RNA polymerization. J. Gen. Virol. 85, 1077–1093 [DOI] [PubMed] [Google Scholar]
- 26. Sofia M. J., Chang W., Furman P. A., Mosley R. T., Ross B. S. (2012) Nucleoside, nucleotide, and non-nucleoside inhibitors of hepatitis C virus NS5B RNA-dependent RNA-polymerase. J. Med. Chem. 55, 2481–2531 [DOI] [PubMed] [Google Scholar]
- 27. Ferrer-Orta C., Arias A., Perez-Luque R., Escarmís C., Domingo E., Verdaguer N. (2004) Structure of foot-and-mouth disease virus RNA-dependent RNA polymerase and its complex with a template-primer RNA. J. Biol. Chem. 279, 47212–47221 [DOI] [PubMed] [Google Scholar]
- 28. Gong P., Peersen O. B. (2010) Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 107, 22505–22510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yin Y. W., Steitz T. A. (2002) Structural basis for the transition from initiation to elongation transcription in T7 RNA polymerase. Science 298, 1387–1395 [DOI] [PubMed] [Google Scholar]
- 30. Siegel R. W., Bellon L., Beigelman L., Kao C. C. (1999) Use of DNA, RNA, and chimeric templates by a viral RNA-dependent RNA polymerase. Evolutionary implications for the transition from the RNA to the DNA world. J. Virol. 73, 6424–6429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tayon R., Jr., Kim M. J., Kao C. C. (2001) Completion of RNA synthesis by viral RNA replicases. Nucleic Acids Res. 29, 3576–3582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhong W., Gutshall L. L., Del Vecchio A. M. (1998) Identification and characterization of an RNA-dependent RNA polymerase activity within the nonstructural protein 5B region of bovine viral diarrhea virus. J. Virol. 72, 9365–9369 [DOI] [PMC free article] [PubMed] [Google Scholar]