Abstract
Environmental sex determination (ESD) occurs in divergent, phylogenetically unrelated taxa, and in some species, co-occurs with genetic sex determination (GSD) mechanisms. Although epigenetic regulation in response to environmental effects has long been proposed to be associated with ESD, a systemic analysis on epigenetic regulation of ESD is still lacking. Using half-smooth tongue sole (Cynoglossus semilaevis) as a model—a marine fish that has both ZW chromosomal GSD and temperature-dependent ESD—we investigated the role of DNA methylation in transition from GSD to ESD. Comparative analysis of the gonadal DNA methylomes of pseudomale, female, and normal male fish revealed that genes in the sex determination pathways are the major targets of substantial methylation modification during sexual reversal. Methylation modification in pseudomales is globally inherited in their ZW offspring, which can naturally develop into pseudomales without temperature incubation. Transcriptome analysis revealed that dosage compensation occurs in a restricted, methylated cytosine enriched Z chromosomal region in pseudomale testes, achieving equal expression level in normal male testes. In contrast, female-specific W chromosomal genes are suppressed in pseudomales by methylation regulation. We conclude that epigenetic regulation plays multiple crucial roles in sexual reversal of tongue sole fish. We also offer the first clues on the mechanisms behind gene dosage balancing in an organism that undergoes sexual reversal. Finally, we suggest a causal link between the bias sex chromosome assortment in the offspring of a pseudomale family and the transgenerational epigenetic inheritance of sexual reversal in tongue sole fish.
Mechanisms of sex determination can be broadly divided into two major categories: genetic sex determination (GSD) and environmental sex determination (ESD). In organisms with GSD (often based on sex chromosomes), the primary sex of an individual is determined at the moment of fertilization by heritable genetic elements that differ between the sexes, whereas organisms with ESD do not possess a primary sex at fertilization and have their sex induced during ontogeny (Bull 1983; Stelkens and Wedekind 2010; Matsumoto and Crews 2012). Generally, the sex of GSD species will be fixed for life, but in some GSD species, the primary sex can be altered during development, such that individuals can develop into the opposite sex without changing their genotype. This phenomenon is known as environmental sex reversal (ESR) (Stelkens and Wedekind 2010) and is relatively common in insects (Vance 1996; Narita et al. 2007), fishes (Devlin and Nagahama 2002), amphibians (Wallace et al. 1999), and reptiles (Quinn et al. 2007). Diverse environmental triggers for ESR have been documented, including abiotic (e.g., temperature, pH, hormones) and biotic (e.g., crowding, pathogens) factors, of which temperature is the most broadly studied (Kato et al. 2011). Currently, artificial induction of sex reversal is widely used in aquaculture by exposure of fish to exogenous steroids or different rearing temperatures in order to produce the preferred sex (Stelkens and Wedekind 2010). ESD lacks the flexibility of ESR, but may be adaptive because it can preferentially produce the sex that benefits most from the best possible hatching environment in terms of future reproductive success (Janzen 1995; Warner and Shine 2005, 2008). However, a common concern is that ESD systems may become a burden under rapid environmental change, such as global warming and ocean acidification, because it might result in nonadaptive sex ratios that are skewed toward predominantly male or female offspring (Ospina-Alvarez and Piferrer 2008; Mitchell and Janzen 2010; Stelkens and Wedekind 2010). Understanding the mechanisms by which genotype and environment interact to control early sex determination is therefore both of fundamental interest and of relevance for domesticated livestock and natural population conservation.
Half-smooth tongue sole, Cynoglossus semilaevis, is a commercially valuable flatfish that is widely distributed in Chinese coastal waters. In contrast to many other teleost species, classical karyotype analysis and artificial gynogenesis tests have revealed that this species employs a female heterogametic sex determination system (ZW♀/ZZ♂) and has clear sexual dimorphism, with females growing much faster and reaching final body sizes that are 2–4 times those of males (Zhuang et al. 2006; Chen et al. 2009). However, despite the primary determination of sex by chromosome inheritance, ∼14% of ZW genetic females are known to be sex-reversed to phenotypic males when reared under normal conditions (22°C) (Chen et al. 2014). Furthermore, we have recently demonstrated that exposure to relatively high temperatures (28°C) during a sensitive developmental period early in life can increase the sex reversal rate of ZW genetic females to ∼73% (Chen et al. 2014), suggesting that the sexual fate of tongue sole can be overridden by environmental factors. Interestingly, these so-called sex reversed “pseudomales” are fertile and can mate with normal females to produce viable offspring. Furthermore, the F1 generation continues to exhibit an extremely high sex reversal rate (∼94%), even when reared in normal conditions (22°C) (Chen et al. 2014). This implies that the ability of sex reversal can be inherited. Therefore, with its complex sex determination system governed by the interaction between genetic determination and environmental factors, the tongue sole is an excellent model with which to understand the molecular mechanism of sex determination and ESR in fishes. We recently sequenced the genome of this fish and generated a 477-Mb high-quality assembly with up to 93.3% (445 Mb) sequences anchored to 22 chromosomes and 21,516 annotated protein-coding genes (Chen et al. 2014). Comparative genomic analysis revealed that the sex chromosome pair of the tongue sole was derived from the same ancestral vertebrate proto-chromosome that gave rise to the W and Z chromosomes of birds. We also proposed that the sex of tongue sole is determined through a Z-encoded mechanism, with male development being driven by the Z-encoded gene dmrt1 (doublesex and mab-3 related transcription factor 1), which is also the male determining gene in birds (Chen et al. 2014). Thus the coexistence of GSD and ESD (in fact juvenile ESR) within this species, coupled with the availability of its full genome sequence, provides an opportunity to comprehensively investigate the gene expression and gene regulation mechanisms mediating sex determination and ESR at the whole genome level.
It has been proposed that the epigenetic regulation of gene expression during gonad differentiation could provide a basis for adaptive sex reversal in some GSD organisms (Manolakou et al. 2006; Matsumoto and Crews 2012), something that has been demonstrated to date in a few fish examples. For example, experiments in sea bass, Dicentrarchus labrax, suggested that the high temperature incubation during the early development stage increased the DNA methylation level of the promoter region of cyp19a1a (cytochrome P450, family 19, subfamily A, polypeptide 1a), leading to suppressed cyp19a1a expression and male development (Navarro-Martín et al. 2011). However, previous studies have been limited to individual genes, and a comprehensive assessment of epigenetic regulation of sex reversal is still lacking. Here, we performed thorough analyses of whole genome DNA methylome and transcriptome data on half-smooth tongue sole, including gonadal samples collected from different sexes (normal female, normal male, pseudomale) of parental fish and F1 generations of pseudomale, and we offer several novel insights on the roles of epigenetic regulation in sexual reversal of tongue sole fish.
Results
Gonadal DNA methylomes of half-smooth tongue sole
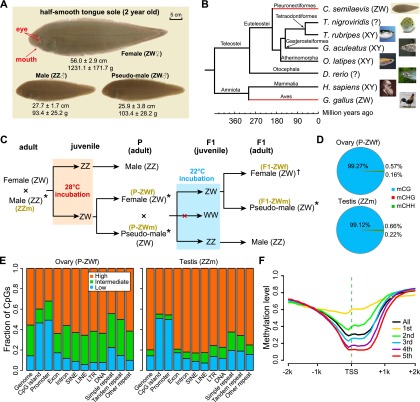
To assess the gonadal DNA methylome patterns across different sexual types of tongue sole, we carried out BS-seq (Xiang et al. 2010) on bisulfite-converted DNA extracted from adult gonads of parental females (P-ZWf), parental pseudomales (P-ZWm), F1 pseudomales (F1-ZWm), and females (F1-ZWf) from a cross between a parental pseudomale and a normal female. We also sampled “normal” male individuals (ZZm) as a control for the ZZm DNA methylation pattern. In addition, we obtained RNA-seq data (Chen et al. 2014) for corresponding samples to quantify gene expression (Fig. 1C; Supplemental Table S1).
Figure 1.
Morphology, phylogeny, and DNA methylation of half-smooth tongue sole. (A) Photos of a female, a normal male, and a pseudomale at 2 yr of age. (B) Genome-based phylogenetic positions and sex chromosome systems of half-smooth tongue sole and other vertebrates. Red lines show that the sex chromosome pairs of tongue sole and chicken were derived from the same ancestral vertebrate proto-chromosome pairs in spite of their distant evolutionary relationship (Chen et al. 2014). (?) No sex chromosomes have been identified so far. (C) Experimental design: The offspring from a normal male (ZZ) and a female (ZW) were exposed to 28°C during the sensitive developmental period, which induced the development of genetic females (ZW) into pseudomales. One of these pseudomales was subsequently crossed with one normal female to produce F1 pseudomales and females. F1 offspring carrying WW sex chromosomes do not exist, as sperms with W instead of Z are not viable. (*) Samples used for both BS-seq and RNA-seq; (†) samples only used for BS-seq. Brown letters in parentheses indicate the symbols for corresponding samples used throughout this paper. (D) Percentage of mCs in the CG, CHG, and CHH contexts. (E) Fraction of CpG in low (methylation level less than 0.25), intermediate (between 0.25 and 0.75), and high (greater than 0.75) methylation levels in different genomic elements. (F) Methylation profile along transcript start sites (TSS) of genes in different expression quintiles. The first quintile is the lowest and the fifth is the highest. Dashed green line indicates the location of TSS. B and C are modified from Figures S16 and 3a in Chen et al. (2014), respectively.
In total, ∼171 Gb methylome data were produced, which yielded an average depth of 22× per strand for each sample (Supplemental Table S2) with ∼90% of genomic cytosines (Cs) being covered by at least two unique reads (Supplemental Table S3). We identified an average of 14.5 million methylated cytosines (mCs) in each sample (see Methods), accounting for 9% of Cs and 86% of CpGs in the reference genome (Supplemental Table S4). More than 99% of mCs were in the CpG context (Fig. 1D; Supplemental Fig. S3A); therefore we decided to focus solely on CpG sites for subsequent analyses. Analysis of the methylation status of CpGs in various genomic elements showed that the majority of CpGs in exons, introns, and repeats were hypermethylated (methylation level >0.75), whereas hypomethylated CpGs (methylated level <0.25) were relatively enriched in gene promoters and CpG islands (CGIs) (Fig. 1E; Supplemental Fig. S3B). Furthermore, ovary harbored more partially methylated CpGs (methylation level between 0.25 and 0.75) compared with testis (Fig. 1E; Supplemental Fig. S3B). As expected, gene promoter methylation was negatively correlated with gene expression (Fig. 1F). In general, the characteristics of DNA methylation as well as the genomic features related to DNA methylation (e.g., CpG o/e, CGIs, and dnmt genes) in tongue sole are similar to those observed in zebrafish (Supplemental Figs. S1, S2, S3; Jiang et al. 2013; Potok et al. 2013).
Genomic methylation patterns are consistent with gonadal differentiation
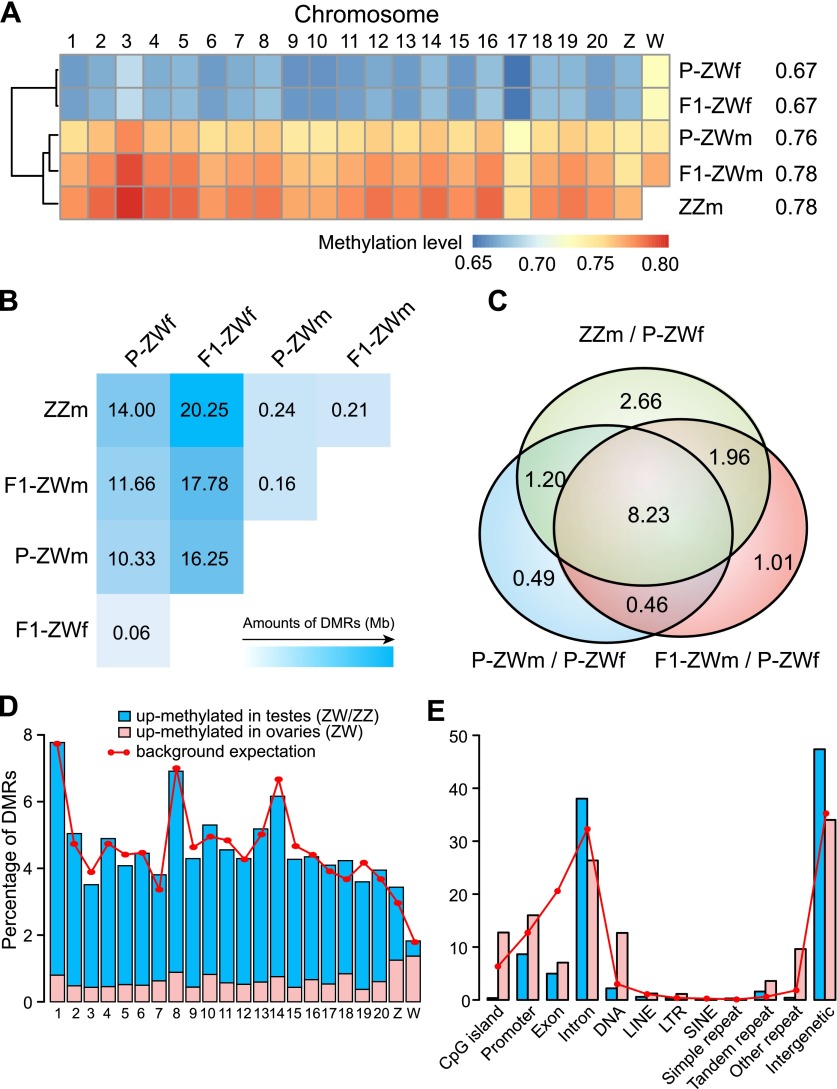
To obtain an overview of DNA methylation patterns for the five types of gonads, we first examined chromosome-wide methylation levels. We found that the methylation patterns of pseudomale testes were similar to those of normal male testes, and all three testis samples were clearly distinguished from the ovary samples by hierarchical clustering analysis (Fig. 2A). Moreover, the overall methylation levels were consistently enhanced by ∼10% in testes compared to ovaries, except for the W chromosome (Fig. 2A; Supplemental Table S5). The relatively high methylation levels of the W chromosome in both ovaries and testes were probably associated with its high repeat content (Supplemental Fig. S4).
Figure 2.
Genome-wide methylation level comparisons. (A) Methylation levels of different chromosomes. Numbers after the sample names represent methylation levels of the whole genome. Only CpGs with ≥10× coverage were used for analysis. (B) Total length (Mb) of DMRs identified in each pairwise comparison. (C) Venn diagrams for DMRs of P-ZWm/P-ZWf, F1-ZWm/P-ZWf, and ZZm/P-ZWf. Numbers represent the total length (Mb) of shared DMRs. (D) Percentage of testis/ovary DMRs on different chromosomes. Background expectation for each chromosome was calculated as the covered length (≥6× in all five samples) of each chromosome divided by the total covered length of all chromosomes. (E) Percentage of testis/ovary DMRs on different genomic elements. Background expectation for each element was calculated as the covered length of each element divided by the total covered length of the genome.
We subsequently investigated the regions that displayed significant methylation changes among samples by performing pairwise comparisons to identify differentially methylated regions (DMRs) (see Methods). In total, only 60 kb of DMRs (0.015% of the genome) were identified between P-ZWf and F1-ZWf, indicating their high concordance in DNA methylation patterns. Similarly, only 160 kb of DMRs (0.040% of the genome) were identified between P-ZWm and F1-ZWm, a figure close to that observed between ZWm and ZZm (∼225 kb; 0.056% of the genome), suggesting that methylation patterns of the three testis samples were also highly similar. In contrast, we identified an average of ∼15 Mb of DMRs (4% of the genome) between testes and ovaries, orders of magnitude more than that between two ovary samples or among three testis samples (Fig. 2B). Taken together, our data suggest that the genome-wide methylation patterns of genetic females have been accurately shaped to the patterns of normal males after phenotypic sex reversal.
Interestingly, up to 86% of the DMRs between P-ZWm and ovaries were maintained in F1-ZWm (F1 pseudomale of P-ZWm) (Fig. 2C; Supplemental Fig. S5), indicating that the overall change in methylation status of the genome after sex reversal had been inherited or re-established by the next generation. Furthermore, ∼95% of these maintained DMRs were also observed between ZZm and ovaries (Fig. 2C; Supplemental Fig. S5), implying that the DNA methylation changes after sex reversal were associated with gonad differentiation in tongue sole.
Gonadal DMGs are associated with development, morphogenesis, and reproduction
Because the overall methylation patterns of pseudomale testes (P-ZWm and F1-ZWm) were very similar to those of normal male testes, we next focused on DMRs where methylation levels were concordant within all three testis samples but different from the two ovary samples (see Methods). In total, we obtained ∼18 Mb DMRs between testes and ovaries. Overall, these DMRs were widely distributed along the genome (Fig. 2D,E). However, in contrast to the overall up-methylated (i.e., the methylation level of a region in one sample is higher than that in the compared sample) bias in testis samples, both sex chromosomes had relatively higher amounts of DMRs up-methylated in the ovary samples, especially for the W chromosome (Fig. 2D). This pattern appears to be related to transposable element (TE) activity, as W and Z chromosomes were shown to have high and moderate percentages of TE components, respectively (Supplemental Fig. S4). Additionally, ovary-up DMRs were relatively enriched in CGIs, promoters, and repeated elements compared with testis-up DMRs, whereas testis-up DMRs tended to target introns and intergenic regions (Fig. 2E) (separate views for Z and W chromosomes are presented in Supplemental Fig. S6A,B).
We identified 2986 genes that harbored testis/ovary DMRs on their promoter regions, which we henceforth referred to as differentially methylated genes (DMGs). Gene ontology (GO) enrichment analysis (see Supplemental Methods) showed that DMGs that are up-methylated in ovaries were overrepresented in biological processes of development and morphogenesis, including reproductive structure development, female gonad development, and oogenesis and spermatogenesis (Supplemental Table S6). In contrast, DMGs that are up-methylated in testes were enriched not only in developmental processes but also in the biological processes of responding to stimulus (e.g., cellular response to steroid hormone stimulus), signal transduction (e.g., steroid hormone mediated signaling pathway), and biological regulations (Supplemental Table S7). Taken together, these results imply that DNA methylation is involved in gonadal differentiation through regulating a series of biological processes.
We then identified DMRs where methylation levels were concordant within P-ZWm and F1-ZWm but different from ZZm, and obtained ∼155 kb DMRs and 71 DMGs, which were relatively enriched in chromosome 9 and Z (Supplemental Fig. S6C,D), nevertheless without significant functional enrichment. We also compared the two pseudomale testis samples (P-ZWm + F1-ZWm) with three other samples (ZZm + P-ZWf + F1-ZWf) to identify regions with pseudomale specific methylation or demethylation after sex reversal; however, no solid DMRs were identified.
The sex determination pathway is the target of DNA methylation regulation
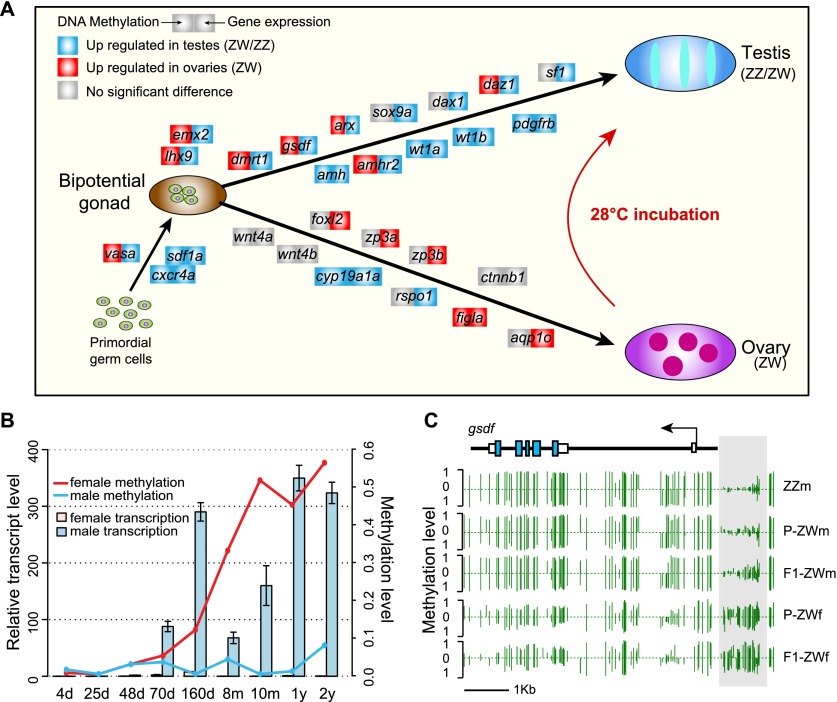
To gain insight into the role of epigenetic regulation in the sex determination pathway, we investigated the methylation status of 58 genomic loci documented to be associated with sex determination in other vertebrates. We observed a remarkable methylation contrast on the sex determination pathway between ZW female versus pseudomale and normal male gonads. In total, 16 of 58 (28%) sex-determination-related genes in tongue sole displayed significantly differential methylation patterns between testes and ovaries (Fig. 3A; Supplemental Table S8), which contrasts to ∼14% (2986/21,516) over the whole genome (Fisher’s exact test: P = 0.0063).
Figure 3.
Differential methylation and sex determination. (A) Differentially methylated and differentially expressed genes in the putative sex determination pathway of tongue sole. For each gene presented in the pathway, the methylation (left square) or expression (right square) changes when comparing testes with ovaries are shown by different colors. (B) DNA methylation and transcription of dmrt1 in different developmental stages after hatching. The methylation levels of different stages were estimated using bisulfite-PCR followed by TA-cloning with a pair of primers targeting the first exon, always using at least 10 randomly selected clones for sequencing for each stage. (C) DNA methylation profiles of gsdf in the five gonadal samples. Green vertical lines indicate the methylation level of cytosines. The light gray box indicates the DMR upstream of gsdf. Profiles of other DMGs in the pathway are presented in Supplemental Figure S8.
One of the DMGs, dmrt1, is a well-described and important gene required for male sexual development in a wide range of invertebrate (e.g., flies and worms) and vertebrate (e.g., birds and human) species (Raymond et al. 2000; Matson and Zarkower 2012). We recently reported that the tongue sole dmrt1 was Z-linked, displayed testis-specific high expression during the critical stage of gonadal differentiation, and harbored a DMR in its promoter region which was up-methylated in ovaries compared with ZW/ZZ testes (Chen et al. 2014). To further examine when the differential methylation patterns emerge during gonad development, we quantified both the DNA methylation levels and the expression levels of this gene in gonad samples ranging from 4 d to 2 yr of age (see Methods). We found that in male gonads, this gene maintained low methylation levels throughout life, whereas in female gonads it underwent increasingly high levels of methylation at the start of the critical sex determination stage (Fig. 3B). Furthermore, its expression was repressed once the methylation had increased during this period. These results raise the possibility that dmrt1 is the critical gene that responds to environmental change and triggers the sex reversal cascade in tongue sole.
In addition, we observed a number of other DMGs that are reported to interact with each other in the sex-determining cascade or display conserved methylation patterns among vertebrates. gsdf (gonadal somatic cell derived factor) was recently reported to be the downstream gene of dmY/dmrt1Y (the Y copy that arose from duplication of the autosomal dmrt1) in the sex-determining cascade of medaka fish (Shibata et al. 2010; Myosho et al. 2012). RT-PCR analysis revealed that the tongue sole gsdf showed a highly similar expression pattern with dmrt1 during sex differentiation (Supplemental Fig. S7; Supplemental Methods), and its promoter region was up-methylated in ovaries compared with ZW/ZZ testes (Fig. 3C), which might correspond to its approximately 23-fold up-regulated expression in testes compared with ovaries. amh (anti-Mullerian hormone) is another well-documented gene that mediates male sexual differentiation, whose expression is consistently higher in males than in females during sex differentiation in many vertebrates (Josso et al. 2005; Wu et al. 2010). The tongue sole amh showed a similar expression pattern (Supplemental Table S8) and harbored a DMR on the promoter region, which was up-methylated in ZW/ZZ testes in comparison with ovaries (Supplemental Fig. S8). Interestingly, we observed that amhr2, the receptor of amh (Wu et al. 2010), and wt1a and wt1b, potential activators of amhr2 (Klattig et al. 2007), also showed differential methylation between ZW/ZZ testes and ovaries (Supplemental Fig. S8). Notably, consistent with the observation in sea bass (Navarro-Martín et al. 2011), the promoter of cyp19a1a was up-methylated in ZW/ZZ testes compared with ovaries (Supplemental Fig. S8), indicating the conserved regulatory role of DNA methylation on this gene.
Local dosage compensation of the Z chromosome in pseudomale testes
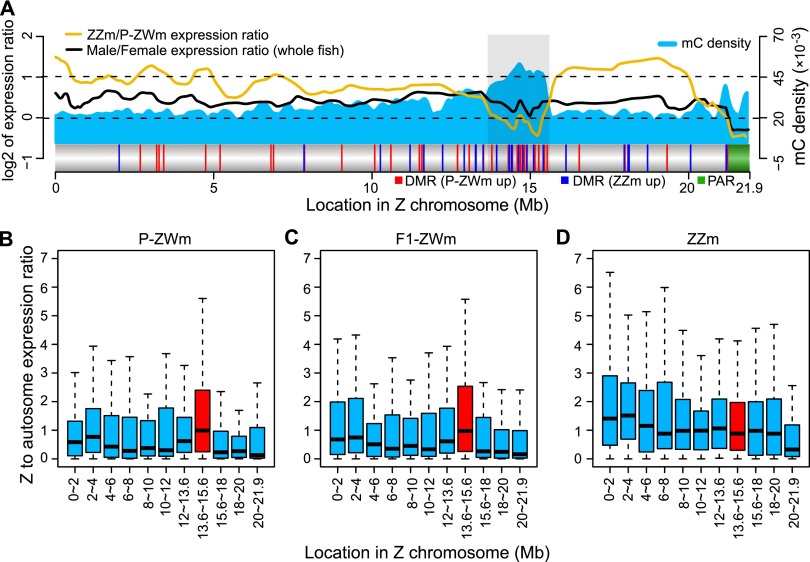
A critical question for animals with both GSD and ESD is how expression of genes in sex chromosomes is balanced after phenotypic sex reversal. We therefore investigated whether there is a specific dosage compensation mechanism on the Z chromosome to balance gene expression in pseudomales by calculating the ZZ testis to ZW testis ratio of gene expression (see Methods). The expected ratio for the complete absence of dosage compensation is two and for the presence of full compensation is one. We found that the expression levels of the Z chromosomal genes (Z-genes) in ZZ testes were on average 1.76 times higher than those in ZW testes (on average 1.73 for P-ZWm and 1.79 for F1-ZWm), a significantly higher ratio than the average male to female expression ratio of around 1.32 (Student’s t-test: P < 10−12 for P-ZW and P < 10−9 for F1-ZW testis) estimated from the normal male and female whole fish samples (excluding gonads) (Fig. 4A; Supplemental Fig. S9). We then mapped the expression ratios along the Z chromosome and observed that a large proportion of the Z chromosome, from 0–12 Mb and 16–20 Mb (covering ∼73% of the entire Z chromosome), pervasively displayed expression ratios of around two in ZZ testes versus ZW testes (Fig. 4A; Supplemental Fig. S9). This is different from the uniform distribution of dosage-compensated genes across the Z chromosome in female versus male whole fish, indicating a lack of dosage compensation for most of the Z chromosome in pseudomale testes.
Figure 4.
Dosage compensation of the Z chromosome in pseudomale testes. (A) Methylated cytosine (mC) density (5-kb window), log2-transformed expression ratios (running averages of 20 genes), and DMR profiles of the Z chromosome. The light gray box indicates the outstanding dosage-compensated region where DMRs were concentrated ([red vertical lines] DMRs that were up-methylated in P-ZWm, [blue vertical lines] DMRs that were up-methylated in ZZm), and the green block indicates the pseudoautosomal region (PAR) where Z and W chromosomes still pair in meiosis. Only 22 genes were annotated in PAR. Z-chromosomal to autosomal gene expression ratios (Z:A) in P-ZWm (B), F1-ZWm (C), and ZZm (D). The dosage compensation region (light gray box in A) is plotted in red. For each Z interval, the expression level of each Z-gene was first divided by the median expression level of all autosomal genes, then the Z:A ratios in each interval were plotted.
Nevertheless, 162 genes, consisting of 17% (162/926) of the Z genes, showed different degrees of dosage compensation in ZW testes (Supplemental Table S9). Of note, there is one region (from 13.6 to 15.6 Mb) that is specifically enriched with dosage-compensated genes in pseudomale testes (Fig. 4A). To answer whether the dosage compensation in this region was the result of down-regulating ZZ testis genes or up-regulating ZW testis genes, we calculated the expression ratio of each Z-gene relative to the median of all autosome genes (Z:A) in ZW testes and ZZ testes separately (see Methods). In ZW testes, the Z:A ratios in this region were significantly higher than those in any other region of the Z chromosome (Mann-Whitney U-test: P < 10−7 for P-ZWm and P < 10−6 for F1-ZWm), displaying a median value close to one and close to most Z regions in ZZ testes (Fig. 4B–D). This indicated that dosage compensation of this region in ZW testes is due to general up-regulation of gene expression. Interestingly, we also found that this dosage-compensated region exhibited a very high density of methylated cytosines (Fig. 4A; Supplemental Fig. S9) and was enriched with Z-chromosomal DMRs between pseudomale and normal male (harbored ∼38% of the DMRs on the entire Z chromosome). These results suggest that DNA methylation might play an important role in regulating dosage compensation at the 13.6- to 15.6-Mb region of the Z chromosome.
The unique dosage compensation region found in pseudomale testes implies the potentially critical role of genes in this region with regard to male development, particularly testis development. As expected, we found that some genes from this region were involved in spermatogenesis. For example, piwil2 (piwi-like 2), a member of the piwi family of genes that exhibit conserved functions relating to transposon silencing during spermatogenesis (Reuter et al. 2009; Wang et al. 2009; De Fazio et al. 2011), displayed a moderate degree of dosage compensation in both ZW testes with ZZ to ZW testis expression ratios of 1.31 and 1.70 in P-ZWm and F1-ZWm, respectively, but no compensation was observed in ovaries (expression ratio of ZZ testis/ZW ovary is 3.69) (Supplemental Table S9). Another example is pik3r1 (phosphoinositide-3-kinase, regulatory subunit 1 [p85 alpha]), a required component in androgen-stimulated PI3K/Akt pathway activation (Castoria et al. 2003; Sun et al. 2003), which plays a central role in the self-renewal division of spermatogonial stem cells (Lee et al. 2007). This gene displayed a high degree of dosage compensation, with ZZ to ZW testis expression ratios of 0.61 and 0.79 in P-ZWm and F1-ZWm, respectively (Supplemental Table S9), whereas highly suppressed expression of this gene was observed in ovaries (expression ratio of ZZ testis/ZW ovary is 8.61). More examples are listed in Supplemental Table S9.
Suppression of female-specific gene expression in pseudomales
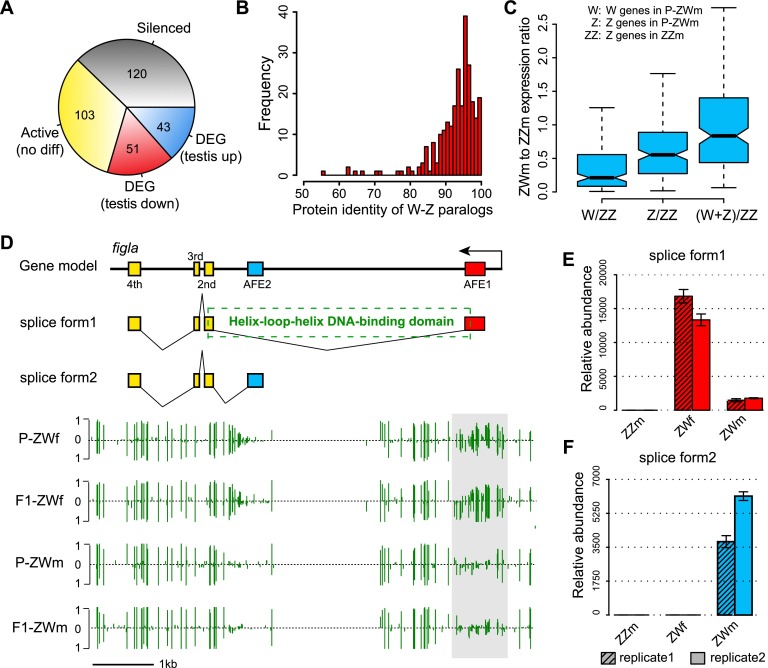
We next investigated the expression of W-chromosomal genes (W-genes) in ovaries and ZW testes and found that among the 317 annotated protein-coding genes on the W chromosome, up to 120 genes (38%) were silenced or had very low expression level (RPKM <1) in both ovaries and ZW testes. Surprisingly, however, up to 94% (185/197) of the 197 remaining genes were still actively transcribed (RPKM ≥1) in either P-ZWm or F1-ZWm. Furthermore, among the 94 genes that were differentially expressed (twofold RPKM change) between ovaries and ZW testes, 46% (43/94) were up-regulated in both ZW testes relative to ovaries (Fig. 5A). Because a large proportion of W-genes are still active in pseudomales, many of the W-genes thus appear to be harmless for male phenotypic development. This may be explained by the young age of the ZW chromosome system in tongue sole, which evolved only around 30 Mya (Chen et al. 2014). We found that for all 317 annotated protein-coding genes on the W chromosome, the paralogs of 272 genes (86%) can be detected on the Z chromosome with high identity (94% in median) (Fig. 5B). This implies that most of the W genes are still too young to develop neofunction in comparison to their counterparts on the Z chromosome; thus, they are generally harmless for male development. Furthermore, their expression may even compensate the gene dosage of their Z counterparts. This is supported by the observation that the expression sum of Z-W paralogous genes in pseudomale testes was close to the dosage of Z-genes in normal males (Fig. 5C; Supplemental Fig. S10).
Figure 5.
W-genes expression pattern in pseudomale testes. (A) Categories of W-genes based on gene expression levels in ovaries and pseudomale testes. (B) Frequency distribution of protein identities for the 272 W-Z paralogous pairs. (C) Pseudomale to normal-male expression ratios calculated from the 272 W-genes and their Z-counterparts. From left to right: The first box represents expression ratios of P-ZWm W-genes to ZZm Z-genes; the second box represents expression ratios of P-ZWm Z-genes to ZZm Z-genes; and the third box represents expression ratios of the sum of P-ZWm W- and Z-genes to ZZm Z-genes, indicating that the expression sum of W-Z paralogous genes in pseudomale testes was close to the dosage of Z-genes in normal males. (D) Alternative splicing and methylation profile of figla. The upper section indicates the gene model and the two splice forms of figla, with the dashed box indicating the position of the basic helix–loop–helix domain, which is coded by first and second exons of splice form1. The lower section shows the DNA methylation profiles in ovaries and pseudomale testes, with the light gray box indicating the position of DMR and green vertical lines indicating methylation levels of cytosines. (E,F) Expression of the two splice forms of figla in testes of normal and pseudomales and female ovaries as determined by RT-PCR. (AFE) Alternative first exon.
Despite the relatively young age of the ZW sex determination system in tongue sole, there are three W-specific genes that do not share any functional homologs on the Z chromosome or autosomes (Chen et al. 2014). Although two of these genes were silenced both in ovaries and ZW testes, the third gene, figla (factor in the germline alpha), which is a germ cell-specific basic helix-loop-helix (bHLH) factor required for ovarian follicle formation (Liang et al. 1997; Soyal et al. 2000; Bayne et al. 2004; Kanamori et al. 2008), displayed interesting expression patterns. RNA-seq data suggested that it could produce two different transcripts by targeting mutually exclusive first exons (Fig. 5D). We found that the transcript with the alternative first exon AFE1 was specifically transcribed in ovaries (Fig. 5E) and was the canonical splicing form that is expressed and is functional in other vertebrate species (Liang et al. 1997; Kanamori et al. 2008). However, the pseudomale testes lack this splicing form, and only express the other transcript with AFE2 (Fig. 5F; see Supplemental Methods). Domain annotation suggested that only the ovary-specific splicing form contains the helix-loop-helix DNA binding domain that is critical to the binding with other proteins (Fig. 5C), so that ovarian follicles can be formed (Liang et al. 1997; Chaudhary and Skinner 1999). It is interesting that the use of different splicing forms by ovaries and pseudomale testes is strongly associated with DNA methylation. There is a DMR between ovaries and pseudomale testes around the AFE1 whose methylation pattern is consistent with the operation of alternative splicing, as this region has a high level of methylation in ovaries (recruiting this exon) but hardly any methylation in pseudomale testes (recruiting another exon) (Fig. 5D). It thus appears that this female-bias gene has been suppressed by alternative splicing via DNA methylation regulation, leading to function loss in pseudomales.
Discussion
Roles of epigenetic regulation in sex determination and sex reversal
In this study, we used half-smooth tongue sole as a model to characterize and compare the epigenetic and transcription pattern of sex-reversed individuals and normal individuals, and to provide insight into the molecular relationship between GSD and ESD. To our knowledge, our study provides the first comprehensive investigation of gene expression and regulation at the whole genome level in a GSD species that exhibits the ESD phenomenon.
Studies on model organisms have revealed that sex development is regulated by a network of genes that have simultaneous input on downstream cascades (Angelopoulou et al. 2012; Munger and Capel 2012). These key master genes in the network are evolutionarily highly conserved and expressed plastically across species (Angelopoulou et al. 2012; Munger and Capel 2012). It has been hypothesized that the functional conserved genetic components involved in gonadal differentiation in GSD animals can also be instrumental in the expression of ESD (Crews and Bull 2009). However, due to the limited genetic resources available for ESD animals, previous analysis has been limited to a few genes. Our results show that a majority of the genes that are involved in sex determination in other vertebrates are highly conserved in the tongue sole genome. Furthermore, their orthologs display substantial differences in gonadal expression or methylation regulation in tongue sole, suggesting a conserved evolutionary core of the sex determination toolkit. We also show that the differences in gonadal expression and methylation pattern of these key genes are consistent between females and the pseudomales that develop from sex-reversed females as a result of environmental influence. This result pinpoints how the master regulatory genes in the sex determination network used in GSD are also utilized during the ESD process to trigger the cascade of alternate gonad development.
It has long been proposed that epigenetic regulation may play an important role in ESD species in response to environmental influences, leading to the gonad developmental change (Manolakou et al. 2006). The cyp19a1a is one of the first cases to show an epigenetic mechanism that mediates temperature effects in sexual reversal (Navarro-Martín et al. 2011). We demonstrate here that regulation by DNA methylation in sexual reversal of tongue sole is not limited to this gene but is applied across the entire sex determination network, which shows an enrichment of DMGs (Fig. 3A). Due to limits in the tissue size required for large-scale sequencing, as well as the challenge of distinguishing the phenotypic sex status of the premature gonad during the sensitive development stage, it is impossible to compare the gene expression and methylation between pre- and post-sexual reversal; thus it is difficult to conclude whether the methylation difference is the cause or consequence of sex reversal. To do so would require more detailed functional experiments. However, our current results provide some insight. For instance, our expression profiling reveals that the first detectable high expression of the dmrt1 gene starts during the period of gonadal sex determination in tongue sole at ∼70 d after hatching and persists at high levels in testes once sex has become differentiated (Fig. 3B). Interestingly, the male specific expression pattern seems to be implemented by the hypermethylation in ovaries on its promoter region. Moreover, this hypermethylation pattern can be eliminated in genotypic females by high temperature incubation and prevent transcript silencing during the critical developmental stage, leading the development of undifferentiated gonad into testes. Of note, medaka also exhibit reduced expression of dmY/dmrt1Y when XY males change into females (Matsuda et al. 2002). It thus appears that ESD can override the standard GSD expression pathways through epigenetic regulation of just a few core genes in the sex determination network, with these affected genes inducing and cascading downstream effects that mediate the expression of sex-specific morphological and physiological traits. As shown in Figure 3, the transcription of several key genes in ovary development is suppressed, whereas the activity of testis development related genes is reactivated after the sexual reversal from genotypic female to phenotypic males by the regulation of DNA methylation.
The role of DNA methylation in mediating gene expression is well documented. It has been proposed recently that the regulation of gene activity by DNA methylation can also be achieved by control of transcript splicing (Shukla et al. 2011). Here, we provide an interesting example, showing that the transcript splicing of a female specific gene figla is associated with DNA methylation. The specific role of methylation in alternative splicing will have to be determined by further experiments, but the case presented here and its important function may shed light on the understanding of this regulatory mechanism.
Resolution of sex-linked gene dosage inequality during sex reversal
Dosage compensation is a process that normalizes the expression level of genes from unequal copy numbers of sex chromosomes in GSD species (Mank 2009, 2013). By comparison of gene expression in females versus males, our previous study suggests that the tongue sole has incomplete dosage compensation in its Z chromosome (Chen et al. 2014). The phenomenon of sexual reversal in tongue sole raises the similar scenario of Z chromosome dosage during the sex reversal from genotypic females to phenotypic males since they have only one copy of Z relative to the normal males. Our result suggests that low dosage compensation on the Z chromosome in the pseudomale is sufficient to achieve normal testis function after sex reversal. Alternatively, the insufficient dosage of Z genes in pseudomales may have been compensated by their undifferentiated paralogs on the W chromosome. Therefore, the dosage compensation is not necessarily fully developed in this differentiating young ZW system, especially when the two sex chromosomes have not been accumulating too many sex-linked mutations and sexually antagonistic genes.
There is one region standing out from the Z chromosome and clustering with a significantly high number of genes depending on dosage compensation in testes (Fig. 4). Consistent with the above hypothesis about dosage compensation, this region is extremely diverged from the W chromosome, containing a significantly low number of paralogous genes in W (19.7%, whole Z chromosome is 31.7%; P = 0.01). This region also bears a high density of methylated cytosine and is significantly hypermethylated (Fig. 4A). It is worth noting that a similar dosage-compensated region has been found on the chicken Z chromosome, where a majority of compensated genes is also located within the so-called male hypermethylated region (MHM) (Teranishi et al. 2001; Melamed and Arnold 2007). It has been suspected that dosage compensation in chicken could be mediated by noncoding RNA in MHM, which recruits a histone acetyltransferase to stimulate up-regulation of genes in that region (Teranishi et al. 2001; Melamed and Arnold 2007). A similar scenario may also apply in pseudomales of tongue sole but through a different trajectory of DNA methylation. Further experiments are needed to elucidate the exact mechanisms. In spite of the similar mechanisms of dosage compensation in chicken and tongue sole, we found no significantly homologous relationships between these two regions, suggesting that they evolved independently.
Another potential dosage dilemma in the combined GSD and ESD system of tongue sole fish concerns the W chromosome of pseudomales, as pseudomales would need to develop a strategy to inhibit the female-beneficial/male-detrimental genes, such as the W-linked figla, during the development of a functional testis instead of an ovary. The tongue sole figla provides an excellent example as to how sex reversed individuals overcome this contradiction. Instead of being completely silenced, this gene is suppressed by an alternative splicing mechanism and associated with DNA methylation regulation. Meanwhile, we also note that only a fraction of the W genes have been repressed in pseudomales, due to the ongoing diverging status of this recently evolved neo-ZW system. We therefore propose that this incompletely differentiated ZW chromosome system, in which neither of the sex chromosomes accumulates sex-specific antagonistic genes, together with epigenetic regulatory mechanisms, may be important in creating the plasticity to develop ESD within a GSD system.
Transgenerational epigenetic inheritance of sex reversal
Transgenerational epigenetic inheritance is the epigenetic effect on the phenotype that can be passed down to subsequent generations and cannot be explained by the changes in the primary DNA sequence and Mendelian genetics (Youngson and Whitelaw 2008; Daxinger and Whitelaw 2010, 2012). It has only rarely been reported in mammals due to the epigenetic reprogramming between generations in mammals that erases and resets most of the epigenetic marks (Daxinger and Whitelaw 2010, 2012; Feng et al. 2010). However, the erasing of epigenetic marks is not a universal rule in animals. For example, DNA methylation is not reset in early development in zebrafish (Macleod et al. 1999), pinpointing the possibility of transgenerational epigenetic inheritance in fishes. This has been confirmed by a recent study in zebrafish, which revealed the heredity of parental DNA methylatome in offspring embryo (Jiang et al. 2013; Potok et al. 2013). A unique advantage of half-smooth tongue sole is that the offspring of pseudomales can spontaneously develop into functional pseudomales without environmental stimuli. An explanation for this is that novel DNA methylation patterns are established in bipotential germ cells when the juvenile fish incubate at high temperature and alter the developmental fate of the germ cells to the opposite sex. These new methylation patterns are then imprinted in the genome and not erased and reset during the development of new generations, so they appear in the offspring. This is supported by our observation that the methylation differences between parental pseudomales versus females are highly consistent with the differences between offspring pseudomales versus females (Fig. 2C; Supplemental Fig. S5).
However, since it is only the methylation patterns of paternal alleles in offspring originating from their pseudomale fathers that are rewritten, while their maternal alleles, which have not been subjected to high temperature exposure, do not contain the novel methylation marks, the following question arises: Why (or how) does the monoallelic methylation change in offspring promote sexual reversal, leading to most or all of the genotypic females’ offspring developing into males? Our previous population survey for a chromosomal inheritance pattern revealed that most or all ZW offspring of the pseudomale inherit their Z chromosomes exclusively from their fathers, and no maternal Z chromosome has been observed in ZW offspring (Chen et al. 2014). This can be explained by the fact that during the spermatogenesis process, no germ cell with only the W chromosome produced by pseudomales can develop to sperm and survive (Chen et al. 2014). Therefore, the ZW offspring of pseudomales only inherit the paternal Z allele, of which the methylation patterns have been altered by environmental stimulation and switch off the ovary developmental pathways. Once the novel methylation marks are inherited, the offspring, which do not have epigenetic reprogramming mechanisms in early developmental stages to reset the epigenetic marks, maintain their paternal methylation pattern through their lifetime, thus directly develop into functional males. Finally, the stable inheritance pattern of the Z chromosome in the pseudomale family offers the possibility of consistent trans-generational epigenetic inheritance of sexual reversal phenomenon in tongue sole fish (Supplemental Fig. S11).
Methods
Sample collection
Normal males (ZZm), parental females (P-ZWf), and induced pseudomales (P-ZWm) were collected from Laizhou Mingbo, Co., from 2008 to 2010 (Fig. 1C). One of the pseudomales (P-ZWm) was crossed with one of the females (P-ZWf) to produce the next generation of females (F1-ZWf) and pseudomales (F1-ZWm), which were collected as F1 samples when they were mature (Fig. 1C). For each of the five samples, two biological replicates were utilized, with each replicate being pooled by five fish. The phenotype and genotype of each selected fish were identified by the histological analysis and PCR validation using the W chromosome specific marker (Chen et al. 2012).
DNA isolation, BS-seq library construction, and sequencing
DNA were isolated from five pooled gonads of the same replicate, then 5 μg DNA was used to perform the bisulfite conversion and BS-seq. The bisulfite conversion of sample DNA was carried out using a modified NH4HSO3-based protocol (Hayatsu et al. 2006). The paired-end library construction and sequencing were carried out using Illumina HiSeq 2000, according to the manufacturer’s instructions (Illumina). We also mixed 25 ng cl857 Sam7 Lambda DNA in each sample to use as a conversion quality control for each library.
BS-seq analysis
The Lambda genome was merged with the reference genome of Cynoglossus semilaevis so that reads originating from the unmethylated control DNA could be aligned. BS-seq reads were mapped to the reference genome using SOAP2 (Li et al. 2009) as described in Bonasio et al. (2012), allowing up to six mismatches for 90-bp paired-end reads. Multiple reads mapping to the same position were regarded as PCR duplicates, and only one of them was kept. Bases with a quality score <20 were not considered for subsequent analysis.
The error rate of each library (sum of the nonconversion rate and T/C sequencing errors) was calculated as the total number of sequenced Cs divided by the total sequencing depth for sites corresponding to Cs in the Lambda genome. The error rate for each library was ∼0.5% (Supplemental Table S2). To distinguish true mCs from false positives, we used a model based on the binomial distribution B(n,p) following Bonasio et al. (2012), and only the mCs with FDR (Benjamini et al. 2001) adjusted P-values < 0.01 were considered true positives.
Methylation level calculation
The methylation level of an individual CpG was determined by the number of reads containing a C at the site of interest divided by the total number of reads covering the site. The methylation level of a specific region was determined by the sum of methylation levels of individual CpGs in the region divided by the total number of covered CpGs in this region.
Differential methylation analysis
Two-way ANOVA was used to identify DMRs between two groups of samples using a 500-bp sliding window with the step length of 250 bp. To ensure the power of statistical tests, only windows with at least six (three per strand) informative CpGs (≥3× coverage) in all replicates of the two compared samples were considered. The two independent variables for ANOVA were sample and cytosine position. For each window, we first calculated the variance between samples and the variance between two replicates of each sample (variance caused by random error), then used F-test to calculate the P-value of each window by comparing the sample variance and random error variance. P-values were then adjusted by FDR (Benjamini et al. 2001). Only windows with adjusted P-value < 0.05 and greater than twofold methylation level change were considered as DMRs. Differentially methylated genes (DMGs) were defined as genes containing DMRs in their putative promoter regions (TSS −2 kb to +500 bp).
Fisher’s exact test and χ2 test (Beißbarth and Speed 2004) were employed to estimate whether the DMGs were enriched in specific GO categories when compared with background genes. P-values were adjusted by FDR (Benjamini et al. 2001), and the adjusted P-value < 0.05 was chosen as the significant threshold (see Supplemental Methods for details).
To track the DNA methylation status of dmrt1 during gonadal development, we performed bisulfite-PCR for genomic DNA extracted from genetic female and male gonads at different developmental stages using a pair of primers targeting the first exon of dmrt1. PCR products were subjected to TA-cloning, and at least 10 clones were randomly selected for Sanger sequencing for each stage (see Supplemental Methods for details).
Gene expression analysis
Gonadal transcriptome data of P-ZWf, P-ZWm, F1-ZWm, and ZZm, as well as transcriptome data of whole fish (excluding gonads) of female and normal male, were from the tongue sole genome project (Chen et al. 2014), and these transcriptome data have been deposited in the NCBI Sequence Read Archive (ZZm: SRX106096; P-ZWf: SRX106097; P-ZWm: SRX106098; F1-ZWm: SRX106099; female whole fish: SRX106100; male whole fish: SRX106103). TopHat v2.0.4 package (Trapnell et al. 2009) was used to map transcriptome reads to the tongue sole genome, with parameters of -a/–min-anchor 8, -m/–splice-mismatches 0, -i/–min-intron-length 50, -I/–max-intron-length 500000,–segment-mismatches 3 and–segment-length 25 for 75-bp reads and 30 for 90-bp reads. Gene expression levels were measured by RPKM (reads per kilobase of gene per million mapped reads) (Mortazavi et al. 2008) and adjusted by a scaling normalization method (Robinson and Oshlack 2010).
Expression of gsdf in different developmental stages and expression of figla in different adult gonads were quantified by conventional RT-PCR (see Supplemental Methods for details). RT-PCR analysis results of dmrt1 at different developmental stages were obtained from Chen et al. (2014) by adding the data of two additional time points (8 m and 10 m) using the 1 y sample as a control (Chen et al. 2014).
Dosage compensation analysis of Z chromosomal genes
The “male to female” or “male to pseudomale” gene expression ratios were used to measure the degree of dosage compensation for each Z-gene in female (ZW) or pseudomale (ZW) relative to normal male (ZZ), calculated as the RPKM ratio of each Z-gene between two compared samples. Only genes with RPKM greater than one in both compared samples were considered, and genes with “male to female” or “male to pseudomale” RPKM ratios between 1/1.5 and 1.5 were defined as dosage-compensated genes. In Figure 4A and Supplemental Figure S9, RPKM ratios were log2-transformed, and the running averages of 20 genes were plotted across the Z chromosome to visualize the chromosome-wide dosage compensation pattern.
To investigate whether dosage compensation of the 13.6- to15.6-Mb region of the Z chromosome in pseudomale testes was the result of down-regulating male expressions or up-regulating pseudomale expressions, we calculated the expression ratio of each Z-gene to the median of autosomal genes (Z:A) in pseudomale testes and normal male testes separately, then Z:A ratios of the Z-genes in each ∼2 Mb interval were visualized by box plot (Fig. 4B–D). Only genes with RPKM greater than one in at least one of the six transcriptome samples were used for Z:A analysis.
Data access
DNA methylome data of the five gonadal samples from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE41129.
Acknowledgments
We thank Jacobus J. Boomsma and M. Thomas P. Gilbert for their critical comments on this manuscript. This work was supported by grants from the National Nature Science Foundation of China (31130057, 41006107, and 31072202), State 863 High-Technology R&D Project of China (2012AA092203, 2012AA10A403-2), Special Fund for Agro-scientific Research in the Public Interest (200903046) of China, Taishan Scholar Project Fund of Shandong of China, JSPS RONPAKU (Dissertation PhD) Program, China National Genebank of China, and a Marie Curie International Incoming Fellowship (300837) to G.Z.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.162172.113.
References
- Angelopoulou R, Lavranos G, Manolakou P 2012. Sex determination strategies in 2012: Towards a common regulatory model? Reprod Biol Endocrinol 10: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayne RAL, Martins da Silva SJ, Anderson RA 2004. Increased expression of the FIGLA transcription factor is associated with primordial follicle formation in the human fetal ovary. Mol Hum Reprod 10: 373–381 [DOI] [PubMed] [Google Scholar]
- Beißbarth T, Speed TP 2004. GOstat: Find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics 20: 1464–1465 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I 2001. Controlling the false discovery rate in behavior genetics research. Behav Brain Res 125: 279–284 [DOI] [PubMed] [Google Scholar]
- Bonasio R, Li Q, Lian J, Mutti NS, Jin L, Zhao H, Zhang P, Wen P, Xiang H, Ding Y et al. 2012. Genome-wide and caste-specific DNA methylomes of the ants Camponotus floridanus and Harpegnathos saltator. Curr Biol 22: 1755–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull JJ. 1983. Evolution of sex determining mechanisms. Benjamin/Cummings Publishing Company, Inc., Menlo Park, CA. [Google Scholar]
- Castoria G, Lombardi M, Barone MV, Bilancio A, Di Domenico M, Bottero D, Vitale F, Migliaccio A, Auricchio F 2003. Androgen-stimulated DNA synthesis and cytoskeletal changes in fibroblasts by a nontranscriptional receptor action. J Cell Biol 161: 547–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary J, Skinner MK 1999. Basic helix-loop-helix proteins can act at the E-box within the serum response element of the c-fos promoter to influence hormone-induced promoter activation in Sertoli cells. Methods Enzymol 13: 774–786 [DOI] [PubMed] [Google Scholar]
- Chen S, Tian Y, Yang J, Shao C, Ji X, Zhai J, Liao X, Zhuang Z, Su P, Xu J, et al. 2009. Artificial gynogenesis and sex determination in half-smooth tongue sole (Cynoglossus semilaevis). Mar Biotechnol (NY) 11: 243–251 [DOI] [PubMed] [Google Scholar]
- Chen S, Ji X, Shao C, Li W, Yang J, Liang Z, Liao X, Xu G, Xu Y, Song W 2012. Induction of mitogynogenetic diploids and identification of WW super-female using sex-specific SSR markers in half-smooth tongue sole (Cynoglossus semilaevis). Mar Biotechnol (NY) 14: 120–128 [DOI] [PubMed] [Google Scholar]
- Chen S, Zhang G, Shao C, Huang Q, Liu G, Zhang P, Song W, An N, Chalopin D, Volff J, et al. 2014. Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat Genet doi: 10.1038/ng.2890 [DOI] [PubMed] [Google Scholar]
- Crews D, Bull JJ 2009. Mode and tempo in environmental sex determination in vertebrates. Semin Cell Dev Biol 20: 251–255 [DOI] [PubMed] [Google Scholar]
- Daxinger L, Whitelaw E 2010. Transgenerational epigenetic inheritance: More questions than answers. Genome Res 20: 1623–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daxinger L, Whitelaw E 2012. Understanding transgenerational epigenetic inheritance via the gametes in mammals. Nat Rev Genet 13: 153–162 [DOI] [PubMed] [Google Scholar]
- De Fazio S, Bartonicek N, Di Giacomo M, Abreu-Goodger C, Sankar A, Funaya C, Antony C, Moreira PN, Enright AJ, O’Carroll D 2011. The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature 480: 259–263 [DOI] [PubMed] [Google Scholar]
- Devlin RH, Nagahama Y 2002. Sex determination and sex differentiation in fish: An overview of genetic, physiological, and environmental influences. Aquaculture 208: 191–364 [Google Scholar]
- Feng S, Jacobsen SE, Reik W 2010. Epigenetic reprogramming in plant and animal development. Science 330: 622–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayatsu H, Tsuji K, Negishi K 2006. Does urea promote the bisulfite-mediated deamination of cytosine in DNA? Investigation aiming at speeding-up the procedure for DNA methylation analysis. Nucleic Acids Symp Ser (Oxf) 50: 69–70 [DOI] [PubMed] [Google Scholar]
- Janzen FJ 1995. Experimental evidence for the evolutionary significance of temperature dependent sex determination. Evolution 49: 864–873 [DOI] [PubMed] [Google Scholar]
- Jiang L, Zhang J, Wang J-J, Wang L, Zhang L, Li G, Yang X, Ma X, Sun X, Cai J 2013. Sperm, but not oocyte, DNA methylome is inherited by zebrafish early embryos. Cell 153: 773–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josso N, Belville C, di Clemente N, Picard JY 2005. AMH and AMH receptor defects in persistent Müllerian duct syndrome. Hum Reprod Update 11: 351–356 [DOI] [PubMed] [Google Scholar]
- Kanamori A, Toyama K, Kitagawa S, Kamehara A, Higuchi T, Kamachi Y, Kinoshita M, Hori H 2008. Comparative genomics approach to the expression of figα, one of the earliest marker genes of oocyte differentiation in medaka (Oryzias latipes). Gene 423: 180–187 [DOI] [PubMed] [Google Scholar]
- Kato Y, Kobayashi K, Watanabe H, Iguchi T 2011. Environmental sex determination in the branchiopod crustacean Daphnia magna: Deep conservation of a Doublesex gene in the sex-determining pathway. PLoS Genet 7: e1001345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattig J, Sierig R, Kruspe D, Besenbeck B, Englert C 2007. Wilms’ tumor protein Wt1 is an activator of the anti-Müllerian hormone receptor gene Amhr2. Mol Cell Biol 27: 4355–4364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kanatsu-Shinohara M, Inoue K, Ogonuki N, Miki H, Toyokuni S, Kimura T, Nakano T, Ogura A, Shinohara T 2007. Akt mediates self-renewal division of mouse spermatogonial stem cells. Development 134: 1853–1859 [DOI] [PubMed] [Google Scholar]
- Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J 2009. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics 25: 1966–1967 [DOI] [PubMed] [Google Scholar]
- Liang L, Soyal SM, Dean J 1997. FIGα, a germ cell specific transcription factor involved in the coordinate expression of the zona pellucida genes. Development 124: 4939–4947 [DOI] [PubMed] [Google Scholar]
- Macleod D, Clark VH, Bird A 1999. Absence of genome-wide changes in DNA methylation during development of the zebrafish. Nat Genet 23: 139–140 [DOI] [PubMed] [Google Scholar]
- Mank JE 2009. The W, X, Y and Z of sex-chromosome dosage compensation. Trends Genet 25: 226–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank JE 2013. Sex chromosome dosage compensation: Definitely not for everyone. Trends Genet 29: 677–683 [DOI] [PubMed] [Google Scholar]
- Manolakou P, Lavranos G, Angelopoulou R 2006. Molecular patterns of sex determination in the animal kingdom: A comparative study of the biology of reproduction. Reprod Biol Endocrinol 4: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson CK, Zarkower D 2012. Sex and the singular DM domain: Insights into sexual regulation, evolution and plasticity. Nat Rev Genet 13: 163–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda M, Nagahama Y, Shinomiya A, Sato T, Matsuda C, Kobayashi T, Morrey CE, Shibata N, Asakawa S, Shimizu N 2002. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature 417: 559–563 [DOI] [PubMed] [Google Scholar]
- Matsumoto Y, Crews D 2012. Molecular mechanisms of temperature-dependent sex determination in the context of ecological developmental biology. Mol Cell Endocrinol 354: 103–110 [DOI] [PubMed] [Google Scholar]
- Melamed E, Arnold AP 2007. Regional differences in dosage compensation on the chicken Z chromosome. Genome Biol 8: R202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell NJ, Janzen FJ 2010. Temperature-dependent sex determination and contemporary climate change. Sex Dev 4: 129–140 [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Munger SC, Capel B 2012. Sex and the circuitry: Progress toward a systems-level understanding of vertebrate sex determination. Wiley Interdiscip Rev Syst Biol Med 4: 401–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myosho T, Otake H, Masuyama H, Matsuda M, Kuroki Y, Fujiyama A, Naruse K, Hamaguchi S, Sakaizumi M 2012. Tracing the emergence of a novel sex-determining gene in medaka, Oryzias luzonensis. Genetics 191: 163–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita S, Kageyama D, Nomura M, Fukatsu T 2007. Unexpected mechanism of symbiont-induced reversal of insect sex: Feminizing Wolbachia continuously acts on the butterfly Eurema hecabe during larval development. Appl Environ Microbiol 73: 4332–4341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Martín L, Viñas J, Ribas L, Díaz N, Gutiérrez A, Di Croce L, Piferrer F 2011. DNA methylation of the gonadal aromatase (cyp19a) promoter is involved in temperature-dependent sex ratio shifts in the European sea bass. PLoS Genet 7: e1002447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ospina-Alvarez N, Piferrer F 2008. Temperature-dependent sex determination in fish revisited: Prevalence, a single sex ratio response pattern, and possible effects of climate change. PLoS ONE 3: e2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potok ME, Nix DA, Parnell TJ, Cairns BR 2013. Reprogramming the maternal zebrafish genome after fertilization to match the paternal methylation pattern. Cell 153: 759–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn AE, Georges A, Sarre SD, Guarino F, Ezaz T, Graves JA 2007. Temperature sex reversal implies sex gene dosage in a reptile. Science 316: 411. [DOI] [PubMed] [Google Scholar]
- Raymond CS, Murphy MW, O’Sullivan MG, Bardwell VJ, Zarkower D 2000. Dmrt1, a gene related to worm and fly sexual regulators, is required for mammalian testis differentiation. Genes Dev 14: 2587–2595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M, Chuma S, Tanaka T, Franz T, Stark A, Pillai RS 2009. Loss of the Mili-interacting Tudor domain-containing protein-1 activates transposons and alters the Mili-associated small RNA profile. Nat Struct Mol Biol 16: 639–646 [DOI] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A 2010. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Paul-Prasanth B, Suzuki A, Usami T, Nakamoto M, Matsuda M, Nagahama Y 2010. Expression of gonadal soma derived factor (GSDF) is spatially and temporally correlated with early testicular differentiation in medaka. Gene Expr Patterns 10: 283–289 [DOI] [PubMed] [Google Scholar]
- Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R, Oberdoerffer S 2011. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature 479: 74–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyal SM, Amleh A, Dean J 2000. FIGα, a germ cell-specific transcription factor required for ovarian follicle formation. Development 127: 4645–4654 [DOI] [PubMed] [Google Scholar]
- Stelkens RB, Wedekind C 2010. Environmental sex reversal, Trojan sex genes, and sex ratio adjustment: Conditions and population consequences. Mol Ecol 19: 627–646 [DOI] [PubMed] [Google Scholar]
- Sun M, Yang L, Feldman RI, Sun X, Bhalla KN, Jove R, Nicosia SV, Cheng JQ 2003. Activation of phosphatidylinositol 3-kinase/Akt pathway by androgen through interaction of p85α, androgen receptor, and Src. J Biol Chem 278: 42992–43000 [DOI] [PubMed] [Google Scholar]
- Teranishi M, Shimada Y, Hori T, Nakabayashi O, Kikuchi T, Macleod T, Pym R, Sheldon B, Solovei I, Macgregor H 2001. Transcripts of the MHM region on the chicken Z chromosome accumulate as non-coding RNA in the nucleus of female cells adjacent to the DMRT1 locus. Chromosome Res 9: 147–165 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL 2009. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance SA 1996. Morphological and behavioural sex reversal in mermithid-infected mayflies. Proc R Soc Lond B Biol Sci 263: 907–912 [Google Scholar]
- Wallace H, Badawy GM, Wallace BM 1999. Amphibian sex determination and sex reversal. Cell Mol Life Sci 55: 901–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Saxe JP, Tanaka T, Chuma S, Lin H 2009. Mili interacts with tudor domain-containing protein 1 in regulating spermatogenesis. Curr Biol 19: 640–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner DA, Shine R 2005. The adaptive significance of temperature-dependent sex determination: Experimental tests with a short-lived lizard. Evolution 59: 2209–2221 [PubMed] [Google Scholar]
- Warner DA, Shine R 2008. The adaptive significance of temperature-dependent sex determination in a reptile. Nature 451: 566–568 [DOI] [PubMed] [Google Scholar]
- Wu GC, Chiu PC, Lyu YS, Chang CF 2010. The expression of amh and amhr2 is associated with the development of gonadal tissue and sex change in the protandrous black porgy, Acanthopagrus schlegeli. Biol Reprod 83: 443–453 [DOI] [PubMed] [Google Scholar]
- Xiang H, Zhu J, Chen Q, Dai F, Li X, Li M, Zhang H, Zhang G, Li D, Dong Y, et al. 2010. Single base-resolution methylome of the silkworm reveals a sparse epigenomic map. Nat Biotechnol 28: 516–520 [DOI] [PubMed] [Google Scholar]
- Youngson NA, Whitelaw E 2008. Transgenerational epigenetic effects. Annu Rev Genomics Hum Genet 9: 233–257 [DOI] [PubMed] [Google Scholar]
- Zhuang Z, Wu D, Zhang S, Pang Q, Wang C, Wan R 2006. G-banding patterns of the chromosomes of tonguefish Cynoglossus semilaevis Günther, 1873. J Appl Ichthyology 22: 437–440 [Google Scholar]