Abstract
Up to now, the direct ligation of two DNA fragments with opposite directions to obtain 3′-3′ or 5′-5′ phosphate ester bonds is still challenging. The only way to obtain DNA oligonucleotides containing a 3′-3′ or 5′-5′ inversion of polarity sites is based on professional DNA chemical synthesis. Herein, we demonstrate a convenient template-directed chemical ligation that enables 3′-3′ and 5′-5′ linkages of two DNA oligonucleotides. This method is based on the assembly of two oligonucleotides on a template in opposite directions through forming antiparallel and parallel duplexes simultaneously, followed by coupling with N-Cyanoimidazole under mild condition. Moreover, on the basis of DNA oligonucleotides with 5′-5′ linkage obtained through our template-directed chemical ligation, we developed a new cDNA display technique for in vitro selection of functional polypeptides.
DNA is one of the most important biopolymers constructed from nucleotides, which are made up of a sugar molecule (called deoxyribose), a phosphate group and nitrogen-containing base. DNA strands have directionality, and the different ends of a single strand are called the 3′ (three-prime) end and the 5′ (five-prime) end. These terms refer to the carbon atom in deoxyribose to which the next phosphate in the chain is attached. In duplex DNA, the two strands of DNA are antiparallel, which means they run in opposite directions to each other. Directionality has consequences in DNA synthesis. For example, a DNA strand can be extended in a 5′ to 3′ direction by either DNA polymerase on a DNA template or reverse transcriptase on a RNA template with adding dNTP (deoxyribonucleotide triphosphates). DNA ligase could link two separated singled stranded DNA fragments which have the same direction on a DNA splint together through catalyzing the formation of a phosphodiester bond between adjacent 5′-phosphate and 3′-hydroxyl termini. At the beginning of life on earth, due to the lack of proteins availability, reactions catalyzed by ligases would have been achieved in a different fashion, for instance, some small molecules or so called non-proteinic compounds are speculated as precursors of ligases1,2,3. Though, in most cases, all nucleotides in a DNA strand have the same direction, DNA ligases could quantitatively convert 5′-phosphoryl DNA donor into 5′,5′-Adenylyl pyrophosphoryl (5′-pp-5′) DNA by adding one ATP molecule. The capping enzyme is one kind of guanylyl transferase enzyme which can catalyze the methylation and addition of a nucleoside-5′-monophosphate to the 5′-end of the messenger RNA molecule to form 5′-ppp-5′ inversion of polarity sites at the beginning of in vivo gene expression4. Up to now, the enzyme that could catalyze the ligation of two DNA fragments with opposite directions to obtain 3′-3′ or 5′-5′ phosphate ester bonds (3′-P-3′ and 5′-P-5′) has not been discovered in nature. However, DNA oligonucleotides containing a 3′-3′ or 5′-5′ inversion of polarity sites have been broadly applied in studying of DNA structure including parallel stranded DNA5,6, Holliday junction7, Triplex-Helix8,9,10 and G-quadruplex11. Moreover, cyclicons containing two oligonucleotides linked to each other through 3′-3′ or 5′-5′ have been used as probes for real-time PCR12,13. The only way to get these modified oligonucleotides is based on chemical DNA synthesis, in which regular CPG solid support (3′-attached nucleoside), 5′-monomer-attached CPG, 3′ phosphoramidites and 5′ phosphoramidites must be rationally combined and programmed on DNA synthesizer. Therefore, such synthesis technology is too complicated to carry out in the normal laboratory setup, and it is seldom available commercially. Herein, we report a novel template-directed chemical ligation of two DNA oligonucleotides with different orientations to obtain 3′-3′ and 5′-5′ linkages conveniently. Based on this method, we developed a new cDNA display strategy by using specially modified DNA oligonucleotides that contains 5′-5′ inversion of polarity site and one 3′-end puromycin modifier.
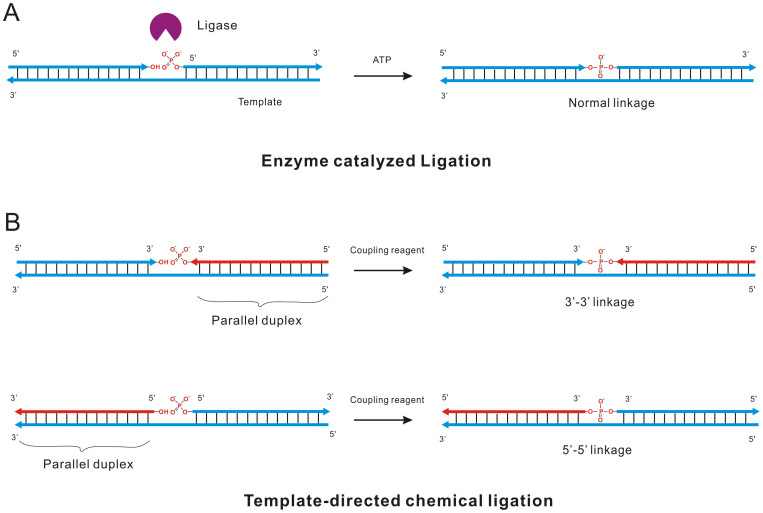
The principle of our method is illustrated in Figure 1. Unlike the ligation catalyzed by DNA ligase that could link the adjacent 5′-phosphate and 3′-hydroxyl together (Figure 1A), the template directed chemical ligation is based on the assembly of two DNA oligonucleotides with opposite directions through forming antiparallel and parallel duplexes on a template simultaneously (Figure 1B). DNA containing only A-T base pairs can form parallel stranded duplex structure, which is stabilized by Hoogsteen base-pairing6,14. Therefore, two DNA oligonucleotides with different directions could assemble on one template to obtain adjacent 3′-phosphate+3′-hydroxyl or 5′-phosphate+5′-hydroxyl termini (Figure 1B). Subsequently, the chemical reagents can be applied to activate the phosphate group, promoting the coupling reaction to obtain new phosphodiester bond.
Figure 1. Ligation reactions catalyzed by different mechanisms.
(A) Normal 5′-3′ ligation catalyzed by DNA ligase. At the presence of template, ATP and T4 DNA ligase, two oligonucleotides were ligated together and a 5′-3′ phosphodiester bonds was formed. (B) Template-directed chemical ligation of 3′-3′ and 5′-5′ oligonucleotides activated by the coupling reagent N-Cyanoimidazole. Arrows in red represent the parallel oligonucleotide with template.
Results and discussion
Optimization of ligation reaction conditions
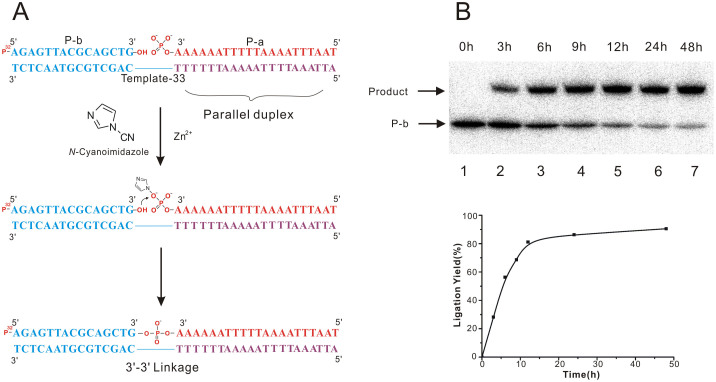
The design of template is very crucial to realize this strategy because it ought to hybridize with two DNA oligonucleotides to form duplexes with different directions. To test the feasibility of our idea, Template-33 was applied in the pilot experiments for 3′-3′ ligation (Figure 2A). The purple domain of Template-33 contained only T and A nucleotides (Figure 2A), which could form parallel duplex through Hoogsteen base-pairing pattern with the DNA oligonucleotides P-a that is complementary to the purple domain in the same direction (Figure 2A). P-a was 3′-phosphorylated in advance for the following ligation reaction. Meanwhile, the blue part of Template-33 contained all four kinds of nucleotides, which could align with P-b in antiparallel fashion via Watson-Crick base-pairing interaction (Figure 2A). P-b was 5′-32P labeled for the following denaturing PAGE analysis after ligation reaction (Figure 2A). According to previous report15, EDCI was used as coupling reagent to promote ligation reaction at the beginning of our research, but no ligation product was detected (data not shown). A chemical compound named N-Cyanoimidazole that can be obtained by the reaction of imidazole and hydrogen bromide was applied as a coupling agent for the ligation reaction between two oligonucleotides in the presence of a DNA template16,17,18,19,20. However, the reaction with N-Cyanoimidazole was not good enough and the product was very low (less than 10%) at beginning. It has been proved that divalent metal ions could promote the coupling reactions. Therefore, different kinds of divalent metal ions were added into the ligation reaction with N-Cyanoimidazole9,16, and it was found that ZnCl2 could significantly improve the yield of ligation. The optimum concentrations of ZnCl2 and N-Cyanoimidazole were found to be 1 mM in reaction solution (Supplementary Figure S1-S3). And the best temperature for ligation reaction was 20°C. Under optimal conditions, more than 80% of 3′-3′ ligation products were obtained in 12 h, and further increasing incubation time could not help to increase the yield significantly (Figure 2B). Meanwhile, the oligonucleotides P-a without 3′-phosphoryl group was not able to be ligated with P-b in the presence of Template-33 under the same reaction condition (data not shown).
Figure 2. Time-dependent 3′-3′ chemical ligation.
(A) Proposed mechanism for the chemical ligation promoted by N-Cyanoimidazole in the presence of template and Zn2+ when incubated at 20°C. (B) PAGE analysis of 3′-3′ ligation with different reaction time. Lane 1-7: Ligation reaction for 0, 3, 6, 9, 12, 24 and 48 h. Graph was determined by phosphor imager calculation of gel. 32P in red denotes a labeled phosphate. Full-length blot images are available in Supplementary Figure S10.
Verification of the formation of our ligation products
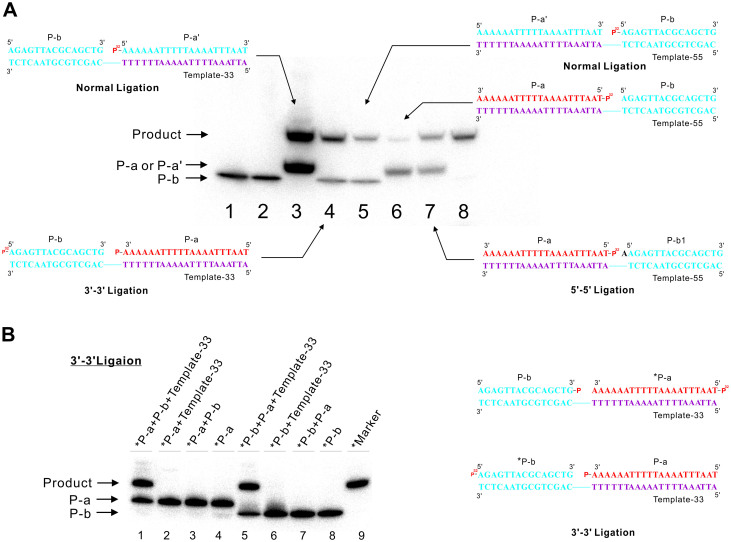
The 5′-5′ ligation was carried out on the Template-55 in which the parallel (purple) and antiparallel (blue) domains were exchanged compared to Template-33 (Figure 3A). However, under the same reaction conditions, the 5′-5′ ligation of duplex DNA did not afforded the similar yield of ligation product as that of 3′-3′ ligation (lane 4 and 6, Figure 3A). We speculate that the phosphate and hydroxyl group on the 5′-5′ inversion of polarity site is not closed enough for the coupling reaction. Therefore, an additional adenine base was added to the sequence of P-b without changing the sequence of the template. In the presence of Template-55, the yield of 5′-5′ ligation of two oligonucleotides P-a and P-b1 increased remarkably and was comparable to that of 3′-3′ ligation (lane 7, Figure 3A). Though both 3′-3′ and 5′-5′ ligation products have the predicted length as compared with the products of normal ligation catalyzed by ligase (Lane 3 and Lane 5, Figure 3A), we further carried out experiments to verify that the product obtained were same as our expectation. Firstly, no ligation product could be detected in both 3′-3′ and 5′-5′ ligation reactions containing no template or another probe, which exclude the possibility of self ligation (Lane 3 and Lane 7 in both Figure 3B and Supplementary Figure S4). Secondly, we exchange the phosphate group at the 3′-3′ and 5′-5′ inversion of polarity sites as well as the 32P labeling sites (Figure 3B and Supplementary Figure S4). As we expected, the same products with similar yield were obtained, which indicated that the DNA template-directed coupling reaction could tolerate the position exchange of reactive groups (Lane 1 and Lane 5, Figure 3B and Supplementary Figure S4). Thirdly, DNA melting experiments by thermal denaturation were carried out to investigate the difference between the products of 5′-5′ and 3′-3′ ligation with normal ligation products. Results showed that the Tm values of duplex products with 3′-3′ and 5′-5′ linkage were lower than their corresponding normal ligation products by 3.5 and 6.7°C (Supplementary Figure S5), which could be caused by the relatively less stability of parallel domain in 5′-5′ and 3′-3′ ligation products comparing to normal ligation productions. Fourthly, to verify the existence of abnormal 3′-3′ and 5′-5′ linkage, a new ligation reaction was designed in the recognition site of restriction endonuclease Xba I. The ligated duplex products containing 3′-3′ and 5′-5′ linkage were not recognized and digested by the endonuclease, but in the positive control reaction with normal ligation product, the normal duplex were digested efficiently (Supplementary, Figure S6). All those result reveal that the template-directed chemical ligation could couple two DNA oligonucleotides with opposite directions to obtain new phosphodiester bond, which was further validate by mass spectrometry analysis of ligation products (Supplementary Figure S7 and Figure S8).
Figure 3. Ligation reactions by using N-Cyanoimidazole.
(A) Lane 1: Marker of 5′-P32 labeled P-b; Lane 2: 3′-3′ ligation without N-Cyanoimidazole; Lane 3: 5′-3′ ligation that is similar as in 3′-3′ ligation in length. P-a' has the same sequence with that of P-a in opposite direction; Lane 4: 3′-3′ ligation under standard reaction condition; Lane 5: 5′-3′ ligation which similar as in 5′-5′ ligation in length; Lane 6: 5′-5′ ligation under standard reaction condition; Lane 7: same as lane 6, 5′-5′ ligation by using P-b1 that has one more A than P-b at 5-end. Lane 8: marker of 5′-P32 labeled Template-33. (B) Verification experiments of 3′-3′ chemical ligation. Lane 1: Ligation reaction containing 5′-P32 labeled P-a ligated to 3′-end phosphorylated oligonucleotides P-b in the presence of Template-33; Lane 2: the same reaction as lane 1 but without P-b; Lane 3: the same reaction as lane 1 but without Template-33; Lane 4: Marker of 5′-P32 labeled P-a; Lane 5: Ligation reaction containing 5′-P32 P-b with 3′-phosphorylated oligonucleotides P-a in the presence of Template-33; Lane 6: the same reaction as lane 5 but without P-a; Lane 7: the same reaction as lane 5 but without Template-33; Lane 8: Marker of 5′-P32 labeled P-b; Lane 9: marker. P32 in red indicated the isotope labeling position. The asterisk denotes isotope labeling. Full-length blot images are available in Supplementary Figure S10.
DNA display using 5′-5′ ligaiton product
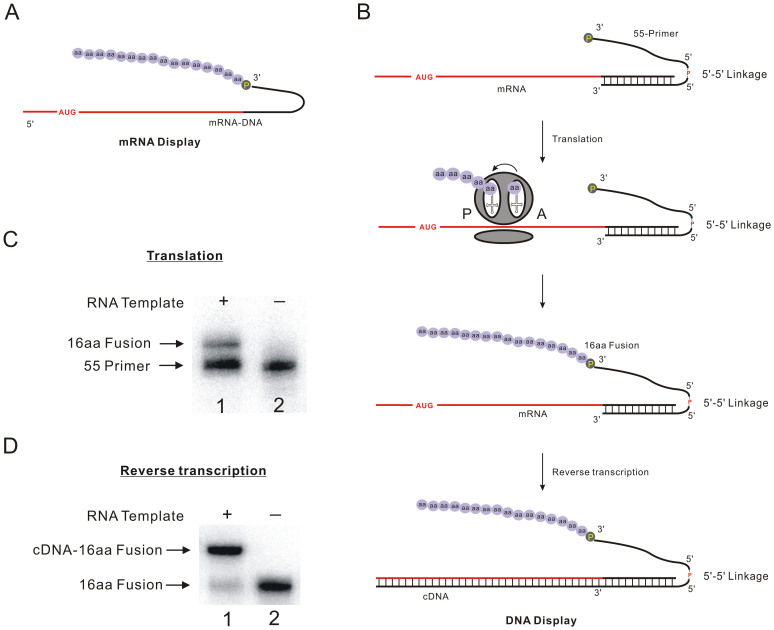
mRNA display is a powerful in vitro selection technique that can be used to identify peptide with desired properties from both natural proteome and random libraries21,22,23. Peptides were covalently connected to their mRNA through puromycin which was tethered on the 3′-end of mRNA-DNA oligonucleotides to capture the nascent peptides after translation by the ribosome (Figure 4A). However, the use of RNA as the library-encoding nucleic acid is less convenient compared to the use of DNA due to its susceptibility towards RNase degradation. Recently, a lot of effort was devoted to DNA display, in which peptides are directly linked to their cDNA. To realize this purpose, a oligonucleotide with two 3′-ends is required. Therefore, branched DNA primers tethered with puromycin have been developed by different groups24,25. And we approached this problem by using the reverse DNA synthesis based on new puromycin modifier26. However, all those methods need to be done in a laboratory with capacity of chemical DNA synthesis. Herein, based on our template-directed chemical ligation of two commercial available DNA sequence, DNA oligonucleotides could be obtained with 5′-5′ linkage to realize DNA display conveniently. As shown in Figure 4B, a DNA oligonucleotide 55-Primer with 5′-5′ linkage have two 3′-ends, in which one is puromycin-modified and another 3′-end is complementary to the sequence of mRNA that encode a polypeptide containing 16 amino acids (16aa). During the in vitro translation of mRNA, when the ribosome reaches the duplex domain, puromycin that is similar to the 3′-end of aminoacyl tRNA can enter the ribosomal A-site and form a stable amide linkage with the nascent polypeptide. Afterwards, DNA oligonucleotide 55-Primer could be retranscripted to obtain the cDNA-peptide complex. According to PAGE analysis, around 20% of 55-Primer was tethered with polypeptide after in vitro translation (Figure 4C), which was verified by the peptides degradation experiments with Proteinase K to indicate the formation of peptide-DNA fusion (Supplementary Figure S9). The peptide-tethered 55-Primer could be extended by reverse transcriptase to obtain the cDNA with high yield, which encoded the nascent ploypeptide (Figure 4D). Based on this strategy, 1012-1013 of peptide-DNA fusion could be successfully constructed in one 1.5 ml eppendorf tube in one day.
Figure 4. The application of oligonucleotides with 5′-5′ linkage in DNA display.
(A) mRNA-peptide fusion obtained from mRNA diplay. (B) The mechanism of DNA display strategy: 55-Primer containing 5′-5′ linkage with hybridized with mRNA that encode a polypeptide contains 16 amino acids (16aa). During the in vitro translation of mRNA, the puromycin will enter the A site of ribosome to capture the nascent peptide when the ribosome comes to the junction of single stranded RNA and the duplex region. After reverse transcription, the cDNA-peptide fusion is obtained. P in yellow represents puromycin. (C) Result of in vitro translation. RNA Template and puromycin-tethered, 5′-5′ structure-included DNA sequence 55-primer. Lane 1: In vitro translation mRNA in presence of P32 labeled 55-primer. The upper band represents the DNA-peptide fusion (16aa Fusion); Lane 2: In vitro translation containing P32 labeled 55-primer without RNA template as negative control, (D) Reverse transcription of 16aa Fusion on mRNA template. Lane 1: Reverse transcription containing P32 labeled 16aa Fusion and mRNA. The upper band represents the reverse transcription product cDNA-peptide fusion; Lane 2: reverse transcription without RNA template as negative control. Full-length blot images are available in Supplementary Figure S10.
Conclusions
In this study, we have demonstrated a convenient template-directed chemical ligation that enables 3′-3′ and 5′-5′ linkages of two DNA oligonucleotides. This method is based on the assembly of the two oligonucleotides in opposite directions on a template and the coupling with N-Cyanoimidazole. The conditions are mild and have been shown to be very efficient. The formation of 3′-3′ and 5′-5′ phosphodiester bond in ligation products were identified through rigorous biochemical experiments and mass spectrometry. Unlike the complicated DNA synthesis carried out in professional laboratory to obtain oligonucleotides with inversion of polarity sites, the procedure of template-directed chemical ligation is easy to operate, and all oligonucleotides and chemical reagents of our method are commercially available for ordinary labs. Moreover, based on DNA oligonucleotides with 5′-5′ linkage obtained through our template-directed chemical ligation, we developed a new DNA display technology for in vitro selection of functional polypeptide. We hope that our work will attract great attention to the research of DNA oligonucleotides with 3′-3′ and 5′-5′ inversion of polarity sites and broaden the scope of their application in biotechnology.
Methods
General information
T4 polynucleotide kinase (PNK), T4 DNA ligase, T7 RNA Polymerase and Bsm DNA Polymerase were purchased from Thermo Scientific. Taq DNA Polymerase and M-MLV reverse transcriptase were purchased from Transgen (Beijing, China). ATP, NTP, dNTP and all of the DNA sequences used in our research were purchased from Sangon Biotech (Shanghai, China) Co., Ltd. [γ-32P]ATP were purchased from Perkin Elmer. Restriction endonuclease was purchased from TaKaRa Biotechnology (Dalian, China) Co., Ltd. Rabbit Reticulocyte Lysate a translation system was purchased from Promega. The molecular weight of the ligation products was measured by the high resolution mass spectrometry (HRMS). The melting temperatures were detected by HITACHI U-1900 spectrophotometer. N-Cyanoimidazole was synthesized by the reaction between cyanogen bromide and imidazole and dissolved in chloroform after purification by filtration and rotary evaporation. Radioactive isotope γ-32P was detected by Cyclone Plus Phosphor Imager (Perkin Elmer).
Labeling Reaction
A reaction mixture containing oligonucleotides with 50 mM Tris-HCl (pH 7.8 at 23°C), 40 mM NaCl, 10 mM MgCl2, 1 mg/mL BSA, 10 μCi [γ-32P]ATP and 10 units of T4 polynucleotide kinase was incubated for 1 h at 37°C for DNA phosphorylation. The labeled product was purified by 10% denaturing PAGE (Polyacrylamide Gel Electrophoresis).
Chemical ligation reaction
3′-3′ template-directed chemical ligation reactions were performed in 20 μL volumes incubated for 12 h at 20°C containing 20 μM 3′-phosphorylated P-a, 20 μM 5′-phosphorylated P-b, 20 μM template-33, 1 mM N-Cyanoimidazole and 1 mM ZnCl2. 5′-5′ template-directed chemical ligations were performed in the same system as 3′-3′ ligation except ligation substrates and splint were replaced with 20 μM 5′-phosphorylated P-a, 20 μM P-b1 and 20 μM template-55. The stock solution of 1 M ZnCl2 was adjusted to pH = 4.9 with 1 M HCl and diluted to a final concentration of 10 mM with double distilled water before adding into the reaction. Ligation results were analyzed by 10% PAGE.
Author Contributions
H.C., Y.Z. and Z.T. conceived and designed the research. H.C., F.S., G.C. and F.D. performed ligation and DNA-display experiments. H.C., A.Y. and Z.T. analyzed the data and wrote the manuscript.
Supplementary Material
Supporting Information
Acknowledgments
This study was supported by Chinese Academy of Sciences (Hundreds of Talents Program) and National Sciences Foundation of China (Grant No. 21172215 and 21322208) and Innovation Program of the Chinese Academy of Sciences (Grant No. KSCX2-EW-J-22).
References
- Li T. & Nicolaou K. C. Chemical self-replication of palindromic duplex DNA. Nature 369, 218–221 (1994). [DOI] [PubMed] [Google Scholar]
- Vonkiedrowski G. A self-replicating hexadeoxynucleotide. Angew. Chem. Int. Ed. Engl. 25, 932–935 (1986). [Google Scholar]
- Xu L. J. & Liu D. S. Functional evolution on the assembled DNA template. Chem. Soc. Rev. 39, 150–155 (2010). [DOI] [PubMed] [Google Scholar]
- Hakansson K. & Wigley D. B. Structure of a complex between a cap analogue and mRNA guanylyl transferase demonstrates the structural chemistry of RNA capping. Proc. Natl. Acad. Sci. U S A 95, 1505–1510 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesande J. H. et al. Parallel stranded DNA. Science 241, 551–557 (1988). [DOI] [PubMed] [Google Scholar]
- Ramsing N. B. & Jovin T. M. Parallel stranded duplex DNA. Nucleic Acids Res. 16, 6659–6676 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns F. W. The holliday junction on its thirtieth anniversary. Genetics 186, 767–773 (2010).21062962 [Google Scholar]
- Horne D. A. & Dervan P. B. Recognition of mixed-sequence duplex DNA by alternate-strand triple-helix formation. J. Am. Chem. Soc. 112, 2435–2437 (1990). [Google Scholar]
- Luebke K. J. & Dervan P. B. Nonenzymatic ligation of double-helical DNA by alternate-strand triple helix formation. Nucleic Acids Res. 20, 3005–3009 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandimalla E. R. & Agrawal S. Hoogsteen DNA duplexes of 3′-3′ and 5′-5′-linked oligonucleotides and triplex formation with RNA and DNA pyrimidine single strands: Experimental and molecular modeling studies. Biochemistry 35, 15332–15339 (1996). [DOI] [PubMed] [Google Scholar]
- Zhou T. Y. et al. Synthesis of unimolecularly circular G-quadruplexes as prospective molecular probes. Nucleic Acids Res. 32, e1731–e1739 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandimalla E. R. & Agrawal S. ‘Cyclicons' as hybridization-based fluorescent primer-probes: Synthesis, properties and application in real-time PCR. Bioorg. Med. Chem. 8, 1911–1916 (2000). [DOI] [PubMed] [Google Scholar]
- Jiang Z. W. et al. Pseudo-cyclic oligonucleotides: In vitro and in vivo properties. Bioorg. Med. Chem. 7, 2727–2735 (1999). [DOI] [PubMed] [Google Scholar]
- Liu K. L., Miles H. T., Frazier J. & Sasisekharan V. A novel DNA duplex - a parallel-stranded DNA helix with hoogsteen base-pairing. Biochemistry 32, 11802–11809 (1993). [DOI] [PubMed] [Google Scholar]
- Rohatgi R., Bartel D. P. & Szostak J. W. Nonenzymatic, template-directed ligation of oligoribonucleotides is highly regioselective for the formation of 3′-5′ phosphodiester bonds. J. Am. Chem. Soc. 118, 3340–3344 (1996). [DOI] [PubMed] [Google Scholar]
- Kanaya E. & Yanagawa H. Template-directed polymerization of oligoadenylates using cyanogen-bromide. Biochemistry 25, 7423–7430 (1986). [DOI] [PubMed] [Google Scholar]
- Ferris J. P., Huang C. H. & Hagan W. J. N-Cyanoimidazole and diimidazole imine: water-soluble condensing agents for the formation of the phosphodiester bond. Nucleosides & Nucleotides 8, 407–414 (1989). [DOI] [PubMed] [Google Scholar]
- McCallum P. B. W., Weavers R. T., Grimmett M. R. & Blackman A. G. Reaction of imidazoles with cyanogen bromide: Cyanation at N1 or bromination at C2? Aust. J. Chem. 52, 159–165 (1999). [Google Scholar]
- Li T. H. et al. Construction of circular oligodeoxyribonucleotides on the new structural basis of i-motif. J. Am. Chem. Soc. 123, 12901–12902 (2001). [DOI] [PubMed] [Google Scholar]
- Liu D. S. et al. Small circular oligodeoxynucleotides achieved from self-assembling entities. Angew. Chem. Int. Ed. Engl. 42, 797–799 (2003). [DOI] [PubMed] [Google Scholar]
- Roberts R. W. & Szostak J. W. RNA-peptide fusions for the in vitro selection of peptides and proteins. Proc. Natl. Acad. Sci. U S A 94, 12297–12302 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keefe A. D. & Szostak J. W. Functional proteins from a random-sequence library. Nature 410, 715–718 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto N., MiyamotoSato E., Husimi Y. & Yanagawa H. In vitro virus: Bonding of mRNA bearing puromycin at the 3′-terminal end to the C-terminal end of its encoded protein on the ribosome in vitro. Febs Lett. 414, 405–408 (1997). [DOI] [PubMed] [Google Scholar]
- Tabuchi I., Soramoto S., Nemoto N. & Husimi Y. An in vitro DNA virus for in vitro protein evolution. Febs Lett. 508, 309–312 (2001). [DOI] [PubMed] [Google Scholar]
- Kurz M., Gu K., Al-Gawari A. & Lohse P. A. cDNA - Protein fusions: Covalent protein-gene conjugates for the in vitro selection of peptides and proteins. Chembiochem 2, 666–672 (2001). [DOI] [PubMed] [Google Scholar]
- Chen H. et al. DNA display for drug discovery. RSC Advances 3, 16251–16254 (2013). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information