Abstract
Mammalian IQGAP proteins all feature multiple ~50 amino acid sequence repeats near their N-termini and little is known about the function of these “Repeats”. We have expressed and purified the Repeats from human IQGAP1 in order to identify binding partners. We used mass spectrometry to identify 42 mouse kidney proteins that associate with the IQGAP1 Repeats including the ERM proteins ezrin, radixin and moesin. ERM proteins have an N-terminal FERM domain (four point one, ezrin, radixin, moesin) through which they bind to protein targets and phosphatidylinositol 4,5-bisphosphate (PIP2), and a C-terminal actin-binding domain, and function to link the actin cytoskeleton to distinct locations on the cell cortex. Isothermal titration calorimetry (ITC) reveals that the IQGAP1 Repeats directly bind to the ezrin FERM domain while no binding is seen for full-length “autoinhibited” ezrin or a version of full-length ezrin intended to mimic the activated protein. ITC also indicates that the ezrin FERM domain binds to the Repeats from IQGAP2 but not the Repeats from IQGAP3. We conclude that IQGAP1 and IQGAP2 are positioned at the cell cortex by ERM proteins. We propose that the IQGAP3 Repeats may likewise bind to FERM domains signaling purposes.
Keywords: IQGAP1, Repeats, ezrin, FERM domain
INTRODUCTION
IQGAPs are multiple-domain scaffold proteins involved in diverse biological processes including actin cytoskeleton reorganization, cell-cell adhesion, and several cell signaling pathways. There are three human IQGAPs and they share identical domain composition and organization, and exhibit a high degree of overall sequence identity.1 From the N-terminus to the C-terminus, IQGAPs are composed of a calponin homology domain, 5 to 6 conserved IQGAP sequence repeats, a WW domain, 4 IQ-motifs, a Ras GTPase-activating protein (GAP)-related domain and a so called RasGAP C-terminal domain. IQGAP1 appears to be ubiquitous expressed, IQGAP2 is mainly found in the liver, while IQGAP3 has been detected in brain, lung, small intestine and testis.1–4 Abnormal expression of IQGAP1 and IQGAP2 have been reported in several cancer types and it has been proposed that IQGAP1 promotes carcinogenesis whereas IQGAP2 is a tumor suppressor.5
IQGAP1 was discovered in 19943 and is to date the most studied family member. The 190 kDa protein acts as a scaffold and signaling nexus; dynamically interacting with various binding partners via its multiple domains/motifs. IQGAP1 self-associates 6, 7, and by binding to F-actin through its calponin homology domain, can crosslink F-actin 7, 8. Calmodulin (CaM) is a potent regulator of IQGAP1. CaM binds primarily to the IQ motifs with the general effect of disrupting other IQGAP1 interactions(reviewed in Briggs et al.9). Although apo-CaM is the predominant protein interactor of IQGAP1 in Ca2+-free breast epithelial cell lysate, the Ca2+-bound form of CaM binds to the IQ motifs with higher affinity.10–12 Activated forms of the Rho-family GTPases Cdc42 and Rac1 bind to IQGAP1 with the effect of promoting interactions with the microtubule -capping protein CLIP-170, and reducing the interaction between the IQGAP1 C-terminus and β-catenin.13, 14 The number of reported IQGAP1 interactors has doubled in the past six years and now exceeds 90.15, 16 With the continuing discovery of new binding partners, IQGAP1’s possible functions have expanded to include not only cytoskeletal modifications and a role in cell-cell junctions, but also intracellular trafficking and gene transcription.
Sequence repeats are commonly observed in proteins, especially those from multi-cellular eukaryotes.17 The classes of proteins that are most likely to contain sequence repeats are found associated with connective tissue, the cytoskeleton, ribonucleoproteins, muscle, brain and neuronal tissue. In an analysis of six major families of protein sequence repeats, Andrade et al. found that the most common function of sequence repeats is mediating protein-protein interactions.18 Mammalian IQGAPs contain multiple ~50 amino acid sequence repeats located just C-terminal to the calponin homology domain.1, 3 While IQGAP1 and IQGAP3 each contain 6 of these sequence repeats, IQGAP2 has just 5 repeats. Earlier, it was believed that the IQGAP1 sequence repeats (hereafter 1Repeats) comprised a coiled-coil which was responsible for the known oligomerization of IQGAP1, and it was reported that the N-terminal fragment containing residues 216–683 (within the 1Repeats) could pull down the full length IQGAP1 from bovine brain cytosol.8 However, a later study found that the IQGAP1 fragment spanning residues 2–746 and encompassing the entire 1Repeats region, failed to bind to full-length IQGAP1, but that IQGAP1 residues 763–863 abrogated self-association of full-length IQGAP1.6
In support of the notion that the 1Repeats are another protein-protein interaction determinant on IQGAP1, Smith et al. recently found that the adaptor protein ShcA directly interacts with IQGAP1 residues 401–522 (within the 1Repeats) via its PTB domain, and the interaction does not require a phosphorylated tyrosine.19 Because the 1Repeats comprise an extensive part of full-length IQGAP1 (~30%), it is likely that they mediate interactions with other proteins besides just ShcA. In order to further investigate the role of the 1Repeats, we have expressed and purified an IQGAP1 fragment encompassing residues 156–672 which contains all 6 conserved repeats, fused to an N-terminal 23 amino acid peptide containing six consecutive histidine residues. Employing nickel affinity, 2-dimensional gel electrophoresis and mass spectrometry, this fragment was used to identify binding partners from mouse kidney-cell lysate. We identified 42 mouse proteins that bound to our construct but not the control Ni2+-Sepharose resin. Surprisingly, three of these proteins were the eponymous ERM family members ezrin, radixin and moesin. To determine if binding to ezrin was direct or mediated by another protein, we used ITC and purified proteins. We discovered that the 1Repeats directly interact with the FERM domain of ezrin, but not with full-length ezrin or a phosphomimetic version of full-length ezrin thought to represent activated (phosphorylated) ezrin. We also found that the Repeats from IQGAP2 (hereafter 2Repeats) bind to the ezrin FERM domain, but the Repeats from IQGAP3 (hereafter 3Repeats) do not. The results of this study reinforce the role of IQGAP1 in cytoskeleton modulation and Rho GTPase signaling, and reveal a novel direct interactor of IQGAP1 and IQGAP2.
MATERIALS AND METHODS
DNA constructs and purification of proteins
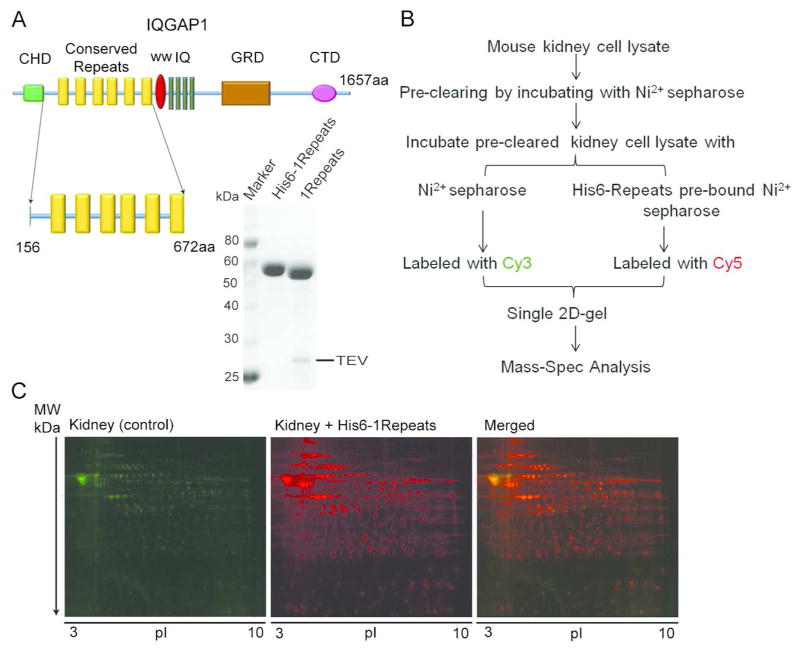
The cDNA encoding the 1Repeats (IQGAP1 residues 156–672) was generated via PCR and ligated into the pProEXHT expression vector (Invitrogen) which encodes a tobacco etch virus (TEV) protease-cleavable 6xHis tag in frame with and N-terminal to the protein of interest. PCR was also used to make expression constructs for the 2Repeats (IQGAP2 residues 211–591) and the 3Repeats (IQGAP3 residues 146–660). Both the 2Repeats and the 3Repeats expression constructs utilize an N-terminal 6xHis affinity tag that is removable using TEV protease. Recombinant proteins were expressed in RosettaTM2 (DE3) cells (Novagen) growing in LB medium. Cells containing the 1Repeats expression plasmid were grown at 37°C until the OD600 reached 0.6–1.0 and then the temperature was reduced to 27°C and 0.5 mM of isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to induce protein expression. After 3h induction, the cells were harvested by centrifugation and resuspended in 300 mM NaCl, 20 mM tris (hydroxymethyl)aminomethane(Tris)-HCL pH 8.0, 15mM imidazole, and 5% glycerol (N1 buffer). The cells were lysed at 4°C using an Emulsiflex-C5 high pressure homogenizer (Avestin) and the lysate was immediately centrifuged for 45min at 260,000g at 4°C. The supernatant was passed through a 5mL HisTrap HP column (GE Healthcare), and after extensive washing with N1 buffer, the protein was eluted with N1 buffer containing an additional 300 mM imidazole. The protein was further purified using a 26/60 Superdex 200 gel filtration column (GE Healthcare) equilibrated to S buffer (300 mM NaCl, 20 mM Tris-HCL pH 8.0, 5% glycerol and 0.5 mM Tris(2-carboxyethyl)phosphine (TCEP)) to remove high molecular weight contaminants and verify the expected monomeric state. Fractions containing the 1Repeats were identified using SDS-PAGE with Coomassie Brilliant Blue staining and pooled and concentrated to ~10mL prior to loading onto a 26/60 Superdex 75 (GE Healthcare) column equilibrated to S buffer to remove low molecular weight protein contaminants. The purity of the protein was assessed using SDS-PAGE and Coomassie staining. Preparation of all Repeats proteins to be used in Isothermal Titration Calorimetry experiments differs from the above procedure in that the eluted proteins from the His-Trap HP column were cleaved by TEV protease during overnight dialysis at 4°C against N1 buffer plus 2mM β-mercaptoethanol (BME). Dialyzed proteins were then re-loaded onto the HisTrap HP column and the flow-through was collected, concentrated and loaded onto a 26/60 Superdex 75 column equilibrated to S buffer. Superdex fractions were analyzed by SDS-PAGE with Coomassie staining, pooled, concentrated, and stored at −80°C for future use.
The cDNAs for human ezrin and human S-adenosylhomocysteine hydrolase (SAHH) were purchased from OpenBiosystems. A cDNA for human RhoGDI was a kind gift from Dr. Rafael Garcia-Mata, University of Toledo. cDNAs encoding full-length ezrin, the ezrin FERM domain (residues 1–297), the ezrin C-terminal fragment (residues 477–586), full-length SAHH and full-length RhoGDI were generated via PCR and ligated into the pET Trx vector (a kind gift from Dr. Gunter Stier, Umeå University). After ligation, this vector encodes the solubilizing carrier protein E. coli thioredoxin (Trx) N-terminal to and in-frame with a TEV protease-cleavable 6XHis affinity tag followed by the protein fragment of interest. The ezrin phosphomimetic T567D was generated via site-directed mutagenesis of the full-length ezrin construct using the QuickChange (Stratagene) methodology. D.K.W provided the expression plasmid for human Cdc42C188S which contains a cysteine-to-serine mutation at residue 188 and does not code for the terminal 3 residues (189–191) of Cdc42. Proteins were expressed as described in the previous paragraph. The supernatants were first partially purified using Ni2+-Sepharose chromatography as described previously. Proteins eluted using N1 buffer + 300mM imidazole were then cleaved by TEV protease during overnight dialysis against N1 buffer plus 2mM BME at 4°C. Dialyzed proteins were then loaded onto the 5mL HisTrap HP column equilibrated to N1 buffer, and proteins in the flow through were collected, concentrated and loaded onto a 26/60 Superdex 75 column equilibrated to S buffer. Superdex fractions were then analyzed by SDS-PAGE with Coomassie staining, pooled, concentrated, and stored at −80°C for future use.
Cell lysate preparation and pull-down assay
Two mouse kidneys (Pel-Freez Biologicals) were finely chopped and added to 10mL of binding buffer (20 mM Tris, pH 8.8, 5% glycerol, and 100 mM NaCl). This tissue mixture was lysed by sonication and then centrifuged at 20,000 × g for 20 minutes at 4°C. To reduce the non-specific interactions with Ni2+-Sepharose resin, the supernatant was pre-cleared by the addition of Ni-Sepharose resin twice for two hour incubations each time followed by centrifugation and collection of the supernatant fraction (~ 8mL). The supernatant was then divided into two equal volumes and incubated overnight at 4°C, tumbling with either Ni2+-Sepharose resin (control) or Ni2+-Sepharose resin bound to the 6xHis-tagged 1Repeats. The next day the resins were washed twice with binding buffer and then bound proteins were eluted with 1mL of CLB (20mM Tris, pH8.5, 20% glycerol, 7M Urea, 2M Thiourea, 1% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS))
Two-dimensional difference gel electrophoresis
Eluate from the control and Ni2+-Sepharose bound to IQGAP1 Repeats were labeled with the CyDye fluorochromes (GE Healthcare) Cy3 or Cy5 (50μg protein/400pmol CyDye), respectively. Before labeling, the samples were checked to affirm that the pH of the eluates were close to 8.5. After adding the CyDye, the samples were left on ice for 30min in the dark. 10 nmol of lysine was then added to terminate the labeling reaction. Next, equal amounts of the two samples were combined and electrophoresed on the same 2D-gel. The proteins were first separated by isoelectric focusing on a 24cm, 3–10NL immobiline pH gradient IPG strip (GE Healthcare) using an Ettan IPGphor II isoelectric focusing unit (GE Healthcare). The gel strip was run as follows: 300V for 2h, a 300V to 1000V gradient over 6h, a 1000V to 8000V gradient over 6h, then 8000V for 8h followed by a 200V hold. After the first dimension, the strip was incubated in equilibrium buffer (6M urea, 20% Glycerol, 2% SDS, and 375mM Tris, pH 8.8) supplemented with 20mg/mL dithiothreitol for 15 minutes at room temperature then followed by another 15 minute incubation in the same equilibrium buffer with 25mg/mL iodoacetamide. The strip was then loaded onto a manual cast 12% SDS-polyacrylamide gel. The second dimension was run using an Ettan DALT6 electrophoresis unit (GE Healthcare) using the following parameters: 5W for 30min and then 17W for 4h.
Gel-imaging and in-gel trypsin digestion
The Cy3 and Cy5 labeled proteins were imaged using excitation/emission wavelengths of 532nm (excitation) and 580nm (emission) Band Pass 30 for Cy3, and 633nm (ex) and 670nm (em) Band Pass 30 for Cy5 using a Typhoon 9400 device(GE Healthcare). The gel was then fixed in 10% methanol, 7% acetic acid for 1hr, rinsed in water three times, and then stained with SYPRO Ruby in the dark overnight. The SYPRO Ruby image was acquired on the same imager using 450nm (ex) and 610nm with a Band Pass 40 filter, and was re-imaged post-excision to ensure accurate protein excision. Protein spots were picked by the Ettan Spot Handling Workstation (GE Healthcare) and stored in a 96 well plate. After de-staining in 50mM ammonium bicarbonate and 50% methanol, 10μl of 20mg/mL sequencing grade trypsin (Promega) was added to each well and incubated overnight at 37°C followed by addition of 50% acetonitrile and 0.1% trifluoroacetic acid to extract the tryptic peptides.
Protein identification by Mass Spectrometry
Protein spots in the experimental sample were identified and the most promising, based on relative abundance, were sent to mass spec for identification after trypsinization and extraction Peptide mass was determined using a Thermo- Fisher LTQ-XL linear ion trap mass spectrometer (San Jose, CA, USA) coupled with an Eksigent nanoLC (Dublin, CA, USA). Peptide samples were loaded onto a Dionex C18 PepMap 100 trap column with dimensions of 300μm (inside diameter) × 5mm (Sunnyvale, CA) and were separated by a New Objective reversed phase C18 PicoFrit emitter with dimensions of 75μm (inside diameter) × 10cm (bed length) with 15μm tip size [part number PF7515-100-H002] (Woburn, MA). Peptides were loaded at 500 nanoliters/minute using a mobile phase of 2% acetonitrile, 0.1% formic acid and then eluted using a gradient of 5–40% acetonitrile, 0.1% formic acid over 35 minutes, with a ramp to 60% acetonitrile, 0.1% formic acid for 10 minutes, and finally a ramp to 95% acetonitrile, 0.1% formic acid for 10 minutes. A Top 5 data-dependent scan strategy was utilized. The MS1 scan range is between m/z 300–2000. The Top 5 most abundant peptides in this MS1 scan were chosen for MS/MS. The MS/MS parameters are the default; briefly, the isolation window is set to 2 Da, 35% relative collision energy (CID), dynamic exclusion is enabled with repeat count set to 1, repeat duration is 30 seconds, and an exclusion size of 100 with an exclusion duration of 75 seconds.
Raw data were analyzed by the Mascot search engine V2.2 (Matrix Science Inc, Boston, MA, USA) against the SwissProt database (August, 2010) using a significance threshold: p (probability) of <5% (confidence interval >95%). Database search was performed for b and y ion series and allowed for up to 2 missed trypsin cleavages, a peptide tolerance of 1.5 amu, a fragment ion tolerance of 1.0 amu, and possible modifications of a methionine oxidation and cysteine carbamidomethyl. Identified proteins, their number of high confidence peptides, and % sequence coverage are listed in Table 1.
Table 1.
List of murine proteins identified by MS as binding partners of the IQGAP1 Repeats.
| Protein name | Gene name | Accession number | High Confidence Peptides | Percent Sequence Coverage | Subcellular localization | Function |
|---|---|---|---|---|---|---|
| Ezrin | Ezr | P26040 | 5 | 38 | C,CM,CSK | Actin filament binding |
| Radixin | Rdx | P26043 | 4 | 24 | C,CM,CSK | Actin capping |
| Moesin | Msn | P26041 | 6 | 31 | C,CM,CSK | Cytoskeleton related (inferred) |
| F-actin-capping protein subunit α2 | Capza2 | P47754 | 1 | 10 | C,CM,CSK1 | Actin capping |
| β-centractin | Actr1b | Q8R5C5 | 5 | 28 | C,CM | ATP binding |
| Rho GDI | Arhgdia | Q99PT1 | 5 | 41 | C | GTPase activation |
| G protein subunit β1 | Gnb1 | P62874 | 2 | 13 | CM1 | Signal transducer |
| Aspartoacylase-2 | Acy3 | Q91XE4 | 6 | 39 | C,CM | Hydrolase |
| α-enolase/MBP-1 | Eno1 | P17182 | 9 | 51 | C,N | Phosphopyruvate hydratase/Mg2+ binding |
| Purine nucleoside phosphorylase | Pnp | P23492 | 9 | 37 | C | Glycosyltransferase |
| Eukaryotic initiation factor 4A-II | Eif4a2 | P10630 | 1 | 24 | C2 | Helicase/hydrolase/initiation factor |
| Elongation factor 2 | Eef2 | P58252 | 1 | 6 | C | Elongation factor |
| Adenosylhomocysteinase(SAHH) | Ahcy | P50247 | 9 | 33 | C | Hydrolase |
| 10-FTHFDH | Aldh1l1 | Q8R0Y6 | 9 | 26 | C | Oxidoreductase |
| Transaldolase | Taldo1 | Q93092 | 3 | 33 | C | Transferase |
| Aldo-keto reductase family 1 member C21 | Akr1c21 | Q91WR5 | 4 | 27 | C | Oxidoreductase |
| Homogentisicase | Hgd | O09173 | 5 | 15 | C2 | Oxidoreductase/dioxygenase |
| Peroxiredoxin-1 | Prdx1 | P35700 | 7 | 58 | C | Antioxidant/oxidoreductase/Peroxidase |
| Peroxiredoxin-2 | Prdx2 | Q61171 | 3 | 37 | C | Antioxidant/oxidoreductase/peroxidase |
| λ-crystallin homolog | Cryl1 | Q99KP3 | 4 | 28 | C | Oxidoreducatse |
| Inorganic pyrophosphatase | Ppa1 | Q9D819 | 3 | 34 | C | Hydrolase |
| ω-amidase | Nit2 | Q9JHW2 | 2 | 12 | C | Hydrolase |
| Bifunctional ATP-dependent dihydroxyacetone kinase /FAD-AMP lyase (cyclizing) | Dak | Q8VC30 | 7 | 23 | C2 | Kinase/lyase/transferase |
| Sedoheptulokinase | Shpk | Q9D5J6 | 2 | 5 | C | Kinase/transferase |
| abhydrolase domain-containing protein 14B | Abhd14b | Q8VCR7 | 4 | 40 | C,N | Hydrolase |
| Fructose-1,6-bisphosphatase1 | Fbp1 | Q9QXD6 | 9 | 50 | C1 | Hydrolase |
| 26S protease regulatory subunit 7 | Psmc2 | P46471 | 6 | 28 | C,N | ATP-binding/ATPase |
| Antiquitin-1 | Aldh7a1 | Q9DBF1 | 3 | 19 | C,M,N | Oxidoreductase |
| Sorting nexin 1 | Snx1 | Q9WV80 | 2 | 6 | C,CM,VM,G | Phosphatidylinositol binding/protein transporter |
| 78 kDa glucose-regulated protein | Hspa5 | P20029 | 2 | 11 | C,ER | ATP binding/misfolded protein binding/ribosome binding |
| Voltage-dependent anion-selective channel protein 1 | Vdac1 | Q60932 | 4 | 33 | CM,MM | Porin |
| V-type proton ATPase subunit B, brain isoform | Atp6v1b2 | P62814 | 8 | 32 | C,CM,VM,G | ATP binding/ hydrolase/transmembrane transport |
| Catalase | Cat | P24270 | 4 | 25 | P | NADP binding/catalase activity |
| Aconitate hydratase | Aco2 | Q99KI0 | 6 | 22 | M | Iron,sulfur cluster binding/hydratase |
| Cytochrome b-c1 complex subunit 1 | Uqcrc1 | Q9CZ13 | 9 | 42 | MM | Catalytic/metal ion binding |
| Peroxiredoxin-3 | Prdx3 | P20108 | 2 | 19 | M | Peroxidase |
| Pyruvate dehydrogenase E1 component subunit β | Pdhb | Q9D051 | 7 | 34 | M | Oxidoreductase |
| 2-oxoglutarate dehydrogenase | Ogdh | Q60597 | 3 | 17 | M | Oxidoreductase |
| Dimethylargininase-1 | Ddah1 | Q9CWS0 | 6 | 45 | M1 | Hydrolase |
| Heterogenous nuclear riboprotein D-like | Hnrpdl | Q9Z130 | 1 | 8 | C,N | Transcription |
| Serotransferrin | Tf | Q921I1 | 8 | 34 | S | Iron transport |
| Fibrinogen β chain | Fgb | Q8K0E8 | 2 | 15 | S | Eukaryotic cell surface binding |
1, information from neXtProt; NA: not available; C: cytoplasm; CM: cell membrane; CSK: cytoskeleton; ER: endoplasmic reticulum; G: golgi; MM: mitochondria membrane; M: mitochondrion; N: nucleus; S: secreted; P: peroxisome; VM: vacuole membranes
Confirmation of direct protein-protein interaction via isothermal titration calorimetry (ITC)
ITC was performed using either an ITC200 or a VP-ITC titration microcalorimeter (MicroCal, GE Healthcare). To eliminate possible heat changes due to buffer effects, all the proteins were dialyzed extensively against the same buffer containing 100 mM NaCl, 20 mM Tris pH 8.0, 5% glycerol and 0.5 mM TCEP. The optical absorbance at 280 nanometers of each protein sample was measured using a NanoDrop 1000 (Thermo Scientific), and the protein concentration was estimated using Beer’s law and the theoretical molar absorbance coefficients of the protein sequences provided by ProtParam at the ExPASy Bioinformatics Resource Portal. ITC200 data collection consisted of 18 2μl aliquots of titrant injected into the sample cell at time intervals of 120 seconds. Data collected using the VP-ITC, involved a preliminary 2μl injection followed by 29 × 10μl injections of titrant with 240 seconds between injections. With the exception of the ezrin FERM vs. ezrin C-terminal domain titration, which was performed at 30°C, all titration experiments were performed at 23°C. In every experiment, a control titration (titrant into sample cell containing only dialysis buffer) was subtracted from the original data to eliminate potential heats of dilution. Raw ITC data were integrated and analyzed using Origin 7.0 software (MicroCal, GE Healthcare).
RESULTS
Pull-down experiment using mouse kidneys
The 1Repeats was expressed and purified as described above (see Figure 1A). Since the human and mouse IQGAP1 proteins share 96% sequence identity (1590 residues out of 1657), and are enriched in kidney, placenta and lung tissue3, we used mouse kidney whole-cell lysate in our pull-down assay. The amount of recombinant protein used was thought to be sufficient to greatly exceed endogenous mouse IQGAP1 levels which would otherwise be expected to compete for binding partners. Numerous proteins bound to the Ni2+-Sepharose-1Repeats while only a small number of proteins were detected that bound to the control resin (see Figure 1C). 40 spots from the 2D gel were picked (Supplementary Figure S1) and from these, 42 proteins were identified (Table 1).
Figure 1. IQGAP1 domain organization and procedural overview.
(A) IQGAP1 schematic and purified 1Repeats protein. CHD, calponin homology domain; WW, tryptophan-rich polyproline-binding domain; IQ, isoleucine- and glutamine-containing motifs; GRD, RasGAP-related domain; CTD, RasGAP C-terminal domain (B) Procedure flow chart. (C) 2D-gel of proteins pulled-down by control and 6xHis-Repeats.
Functional classification of 1Repeats-associated proteins
The 42 proteins identified in our study serve a variety of enzymatic and/or structural functions. As an aid toward understanding how these proteins may be integrated into known IQGAP biology, we have provided their known or predicted subcellular locations and functions (Table 1) using for the most part, information from the UniProt Knowledge Database (UniProtKB). For eight of the identified proteins, which were missing the necessary information at UniProtKB, we used information found at either neXtProt or NCBI Gene. To facilitate understanding of the functional classification of identified proteins, the PANTHER classification tool was used to create a pie chart representation and Table (Supplementary Figure S2; Supplementary Table 1). As can be seen in these tables and figure, the majority of 1Repeats interactors are enzymes (hydrolases, hydratases, oxidoreductases, peroxidases, kinases, etc.) with the remainder performing structural/binding roles. Several of these proteins have functions consistent with known IQGAP1 biology. For instance, IQGAP1 binds F-actin and is an effector of the Rho-family small GTPases Cdc42 and Rac1 which have roles in actin cytoskeleton modulation. IQGAP1 also binds to the microtubule plus-end-tracking protein CLIP-170.13 Therefore proteins with obvious actin (ERM proteins, F-actin-capping protein subunit α2), microtubule (β-centractin), or RhoGTPase (RhoGDI) associations are perhaps more easily understood in the context of published IQGAP1 information. Since IQGAP1 has not been previously detected within mitochondria, peroxisomes, the ER, or in the extracellular environs, it is more difficult to understand how proteins from these compartments may play a role in IQGAP1 signaling. However, our study did identify 4 enzymes whose major function is the conversion of H2O2 to water and oxygen. Two of these (peroxiredoxins 1 and 2) are localized to the cytosol where IQGAP1 can be found, whereas catalase and peroxiredoxin-3 are found in peroxisomes and mitochondria, respectively, and should not be ordinarily encountered by IQGAP1. Nevertheless, on a purely functional basis catalase and peroxiredoxin-3 are consistent with the discovery of peroxiredoxins 1 and 2 and therefore, should not be easily discounted.
Ezrin binds the IQGAP1 Repeats directly through its N-terminal FERM domain
Ezrin, radixin and moesin function to link the cytoskeleton to important locations on the cell membrane (reviewed in Fehon et al.20). ERM family proteins share high sequence identities (~75%) and consist of a ~300 amino acids N-terminal FERM domain followed by a central α-helical domain and a C-terminal ERM-association domain (C-ERMAD). The FERM domain can bind directly to a number of proteins including EBP5021, CD4322, 23, CD4423–25, while the last 30 amino acids of C-ERMAD can bind to F-actin26. ERM family proteins can exist in an inactive form wherein the C-ERMAD binds to the FERM domain covering FERM residues needed for binding to other proteins. It is believed that binding to the phosphatidylinositol 4,5-bisphosphate (PIP2) and the phosphorylation of certain C-ERMAD residues reduce the interaction between the N- and C-terminus of ezrin, thereby activating the protein.27, 28
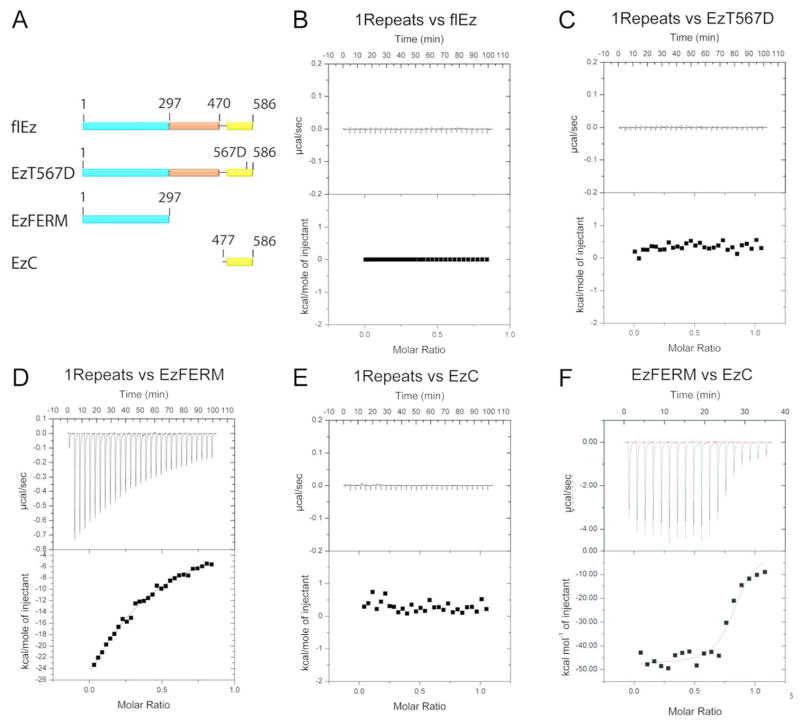
Considering the activation aspects of ezrin, several constructs were designed (Figure 2A): Full length ezrin (flEz), ezrin1–297 (EzFERM), ezrin477–586 (EzC) and a phosphomimetic mutant ezrinT567D (EzT567D). RhoGDI, SAHH and Cdc42 were also expressed and purified for ITC experiments. Whereas IQGAP1 and other effectors bind to Cdc42 and Rac1 in their active (GTP-bound) forms, RhoGDI binds to these RhoGTPases and inhibits the release of bound GDP thus stabilizing them in their inactive forms (reviewed in Garcia-Mata et al.29). Understanding the interplay between the ERM proteins, RhoGDI, RhoGTPases and IQGAP1 is a necessary step towards understanding IQGAP1’s role in cytoskeletal dynamics and signal transduction. SAHH was chosen because it was identified twice with very high Mascort scores. SAHH converts S-adenosyl-L-homocysteine to adenosine and homocysteine, and downregulation of SAHH has been linked to tumorigenesis.30, 31
Figure 2. IQGAP1 Repeats bind the Ezrin FERM domain.
(A) Schematic diagram of full length Ezrin (flEz), Ezrin T567D (EzT567D), Ezrin FERM domain (EzFERM) and Ezrin C-ERMAD (EzC) constructs. (B–D) ITC experiments of 1Repeats with flEz, EzT567D, EzFERM and EzC, respectively. The 1Repeats were the titrant and ezrin proteins were in the calorimeter cell. (F) EzFERM titrated into EzC at 30°C using an ITC200. B–F, Each panel shows the raw ITC data (top) and the integrated heats (bottom) after subtraction of any heats arising from the injection of titrant into buffer.
Proteins were expressed and purified as described in Materials and Methods, and purity was confirmed by SDS-PAGE and Coomassie Blue staining (Supplementary Figure S3). A series of exothermic peaks were observed when the 1Repeats were titrated into the calorimetry cell containing EzFERM indicating a direct protein-protein interaction (Figure 2D). No net heat was absorbed or released in titrations of the Repeats into the cell containing flEz, or EzT567D, or EzC, (Figures 2B, 2C, 2E) or SAHH or RhoGDI (Supplementary Figures S4A and S4B). Thus, in our titration conditions, the 1Repeats bind to the ezrin FERM domain but there is no detected binding of the 1Repeats to flEz, EzT567D, EzC, SAHH or RhoGDI. However, the EzFERM does bind to EzC as expected (Figure 2F) supporting the notion that both EzFERM and EzC are properly folded. Previously, RhoGDI was found to bind to a protein fragment that includes most of the mouse radixin FERM domain (residues 1–280).32 These researchers also found that a longer fragment containing the entire radixin FERM domain (residues 1–318) promoted Dbl-mediated guanine nucleotide exchange of Rho-family GTPases in the presence of RhoGDI. Similar stimulation of nucleotide exchange was achieved using residues 1–280 of ezrin and moesin. The results of these experiments led the investigators to conclude that RhoGDI binds to the FERM domains of all three ERM proteins and this interaction causes the release of RhoGDI-bound GTPases; making them available for activation by Dbl-family exchange factors. Unexpectedly, we see no interaction between RhoGDI and EzFERM in our ITC experiments (Supplementary Figure S4C). To address the possibility that RhoGDI is not properly folded, we performed an ITC experiment using RhoGDI and GDP-bound Cdc42. The ITC data show that RhoGDI can indeed interact with GDP-bound Cdc42 as expected (Supplementary Figure S4D).
The ezrin FERM domain interacts with the IQGAP2 Repeats
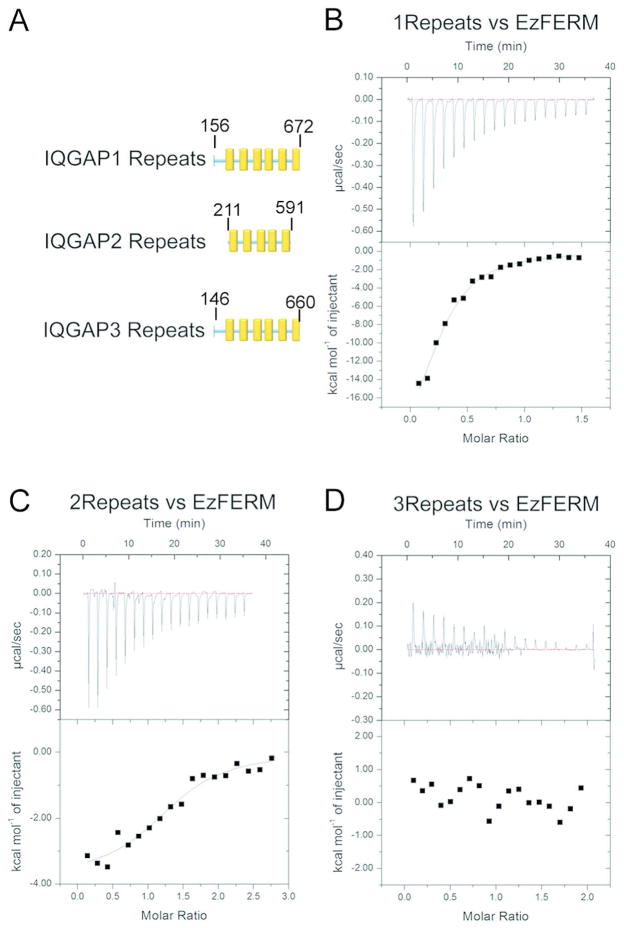
IQGAP2 and IQGAP3 are close relatives of IQGAP1 with distinct tissue distributions and some shared IQGAP1 protein interactors. To determine whether the ezrin FERM domain will bind to the Repeats of IQGAP2 and IQGAP3, we performed ITC experiments. We found that the 2Repeats do display binding to the EzFERM (Figure 3C), but surprisingly, the 3Repeats do not appear to bind to the EzFERM fragment (Figure 3D). Binding constants derived from the ITC experiments are listed in Table 2. The EzFERM/1Repeats and EzFERM/2Repeats calorimetry data were each fit to a single site model of binding since this model displayed significantly lower χ2/degrees of freedom values as compared to other models we attempted to fit (2-site, 2-site sequential, 3-site sequential). RhoGDI/Cdc42 and EzFERM/EzC were also fit to one site models. During fitting of the data, the association constant (Ka), the change in enthalpy (ΔH) and the binding stoichiometry (n) were each allowed to vary. We were not able to properly fit the data using a fixed integer value (=1) for the parameter n. Non-integer values for n are typically attributed to errors in the estimated protein concentrations and/or interaction details that are not properly modeled. Nevertheless, the 1Repeats and 2Repeats have very similar association constants; however, they show distinct thermodynamic modes of binding. While the binding of 1Repeats to EzFERM is enthalpy driven, the binding of the 2Repeats to EzFERM is governed by both enthalpic and entropic terms. Therefore, there are at least subtle differences in the details of binding between IQGAP1 and IQGAP2, but this should not be too surprising since the amino acid sequences of the respective Repeat regions are similar but not identical. Because we identified all three ERM proteins using the 1Repeats as bait, and since we verified direct binding between the ezrin FERM domain and the 1Repeats using ITC, we can posit that IQGAP1 directly binds all 3 ERM proteins via interaction between its sequence repeats and the respective FERM domains. The 2Repeats also bind to the ezrin FERM domain; however, the 2Repeats may not bind to the FERM domains of radixin and moesin.
Figure 3. Representative ITC data for IQGAP Repeats and EzFERM.
Titrations were conducted at 23°C using an ITC200. Repeats were the titrant and EzFERM was in the cell. (A) Schematic diagram of Repeats fragment constructs used in this study. (B) 1Repeats. (C) 2Repeats. (D) 3Repeats. B–D, Each panel shows the raw ITC data (top) and the integrated heats (bottom) after subtraction of any heats arising from injection of titrant into buffer alone.
Table 2.
ITC Thermodynamic Parameters.
| Ka (M−1) | ΔH (kcal mol−1) | ΔS (cal/mol/deg) | n | |
|---|---|---|---|---|
| 1RepeatsvsEzFERM | 1.71×105±3.17×104 | −49.07±15.55 | −57.1 | 0.274±0.0243 |
| 2RepeatsvsEzFERM | 1.43×105±4.58×104 | −3.98±0.39 | 10.9 | 1.33±0.0688 |
| EzFERMvsEzC | 1.13×106±3.96×105 | −47.89±1.33 | −130 | 0.815±0.0155 |
| CDC42GDPvsRhoGDI | 5.57×104±940 | −13.29±0.08 | −23.1 | 0.853±0.0003 |
CONCLUSIONS
Because the three human IQGAP proteins are quite large, we have employed a ‘divide and conquer’ approach in this study. One of the advantages of expressing well-behaved IQGAP fragments is that these fragments can be used in binding studies to identify cellular proteins that interact with a specific domain/motif. We have used this rationale and ITC to discover that the 1Repeats bind to the ezrin FERM domain. No binding was seen to full-length ezrin or the “activated” phosphomimetic version of the full-length protein. Although EzT567D has been used before as a constitutively active form of ezrin in cells, it has been previously noted that EzrinT567D fails to bind to two known Ezrin FERM domain binding partners, EBP50 and NHERF1.33, 34 In addition, neutron scattering experiments have revealed a closed conformation of full-length wild-type Ezrin and EzrinT567D. Subsequent addition of PIP2 induced an overt conformational change in both proteins.34 It is therefore logical to assume that in our hands, both of these full-length versions of ezrin were in their closed, autoinhibited conformations and that additional phosphorylation and/or PIP2 binding is required to expose the FERM domain for IQGAP1 binding.
Because we also identified radixin and moesin in our pull-down study, by extension we conclude that the 1Repeats also bind to the FERM domains of radixin and moesin. We have found that the 2Repeats bind to the ezrin FERM domain, but the 3Repeats do not show significant binding under these conditions. Presumably, not all FERM domains are alike in that they most probably mediate protein-specific interactions. Therefore, because of our positive ITC results with the Repeats from both IQGAP1 and IQGAP2, and considering the conserved domain composition and organization of the three IQGAPs and the fact that some fifty FERM domain-containing proteins have been identified thus far35, we propose that the 3Repeats are used to bind to an as yet unidentified FERM domain.
For some time, ERM proteins have been considered as linkers that join specific structures in the plasma membrane to the actin cytoskeleton.20 In 1994, Tsukita et al.24 found that ezrin, radixin and moesin are associated with the hyaluronic acid (HA) receptor CD44 in both baby hamster kidney cells and mouse fibroblasts. In 1998, Yonemura et al. 23 used immunoprecipitation and mutagenesis studies to reveal that ezrin, radixin and moesin bound to the leukosialin (CD43), hyaluronin (CD44) and intercellular adhesion molecule-2 (ICAM-2) receptors via positive charges found in those receptor’s cytoplasmic domains and the FERM domains of the respective ERM proteins36. In 2005, Bourguignon et al. discovered that IQGAP1 associates with the CD44 receptor in HA-treated ovarian tumor cells.37 Furthermore, these investigators discovered that HA stimulated the association of phosphorylated ERK2 protein with IQGAP1 resulting in Elk-1 and estrogen receptor-α phosphorylation events and transcriptional activation. A number of structural studies involving FERM domains bound to peptides, proteins and IP3 (the headgroup of PIP2) have been performed. Very recently, the X-ray structure of a ternary complex of a FERM domain bound to the GTPase Rap1 and a peptide from a transmembrane protein has been published.38 This ternary structure supports the notion that at least in some cases, FERM domains can engage multiple targets. Since the three ERM proteins bind to CD44, and IQGAP1 binds to the ERM proteins, it seems quite possible that the ERM proteins mediate IQGAP1’s reported binding to CD44 via the 1Repeats. IQGAP1 has been previously shown to bind to B-Raf39, MEK1/240 and ERK241 (and probably ERK1) through sequences outside the Repeats region. Utilization of the 1Repeats for association with CD44 via an ERM protein would presumably still allow for MAP kinase signaling through the IQGAP1 scaffold.
Rho GTPases are prenylated at their C-termini and active forms of the GTPases are found associated with membranes. Rho guanine nucleotide dissociation inhibitors (RhoGDIs) bind to the nucleotide-dependent “switch regions” of the GTPases and provide a hydrophobic groove that accommodates the GTPases’ geranylgeranyl group. These features allow RhoGDIs to function as shuttles; delivering Rho GTPases to and removing them from membranes.29 The exact molecular details involved in the release of the GTPases from the GDI at membranes are still forthcoming, but interaction with ERM proteins may be required. Direct interaction between the FERM domain of ERMs and RhoGDI using bacterially-expressed proteins and pull-downs has been shown before32, 42, so we were surprised to detect no binding in our ITC experiment. For the ezrin FERM domain, our choice of residues to express (1–297) was based upon the construct used to solve the crystal structure43, and E. coli expressed full-length RhoGDI has been used in structural studies as well44. During protein purification we always include a gel filtration purification step to monitor the aggregation state of the proteins. Our ezrin FERM domain bound to the ezrin C-terminus as expected and our RhoGDI bound to Cdc42•GDP as expected – despite the absence of the significant contribution to the binding that would be afforded by burying the hydrophobic geranylgeranyl group (our bacterially-expressed Cdc42 has no post-translational modifications). Perhaps exact buffer composition or temperature plays a significant role in FERM domain and RhoGDI binding?
Since we did not see direct binding between RhoGDI and the 1Repeats, barring the requirement for one or more post-translational modifications for this interaction, another protein intermediary would be responsible for RhoGDI’s presence on our 2D gel. We analyzed just 40 spots from the original 96 spots picked from our 2D gel. Perhaps within the remaining 56 spots there are one or more obvious candidates for such an intermediary? For instance, if we had identified Cdc42 or Rac1 in our analysis, we might postulate that the presence of a prenylated RhoGTPase bound to RhoGDI (which would provide additional interaction surfaces not present on RhoGDI alone) is required for 1Repeats binding. In Table 1 there are no obvious candidates for such an intermediary aside from moesin and radixin. However, since we failed to see binding between our purified ezrin FERM domain and RhoGDI (and we believe those proteins are properly folded, etc.), moesin and radixin as intermediates would seem unlikely.
When considered as important for IQGAP1’s biological function, many of the proteins listed in Table 1, are quite intriguing. For instance, one of the enzymes we identified twice is α-enolase, a glycolytic enzyme. An alternate transcript from the α-enolase gene is known as Myc-binding protein 1 (MBP-1) which is localized to the nucleus and functions to suppress transcription of the c-myc gene. MBP-1 (343 amino acids) is identical in sequence to α-enolase (434 amino acids) except it is missing the first 93 amino acids of the glycolytic enzyme. Although we did identify the full-length enzyme on our 2D gel, we do not know that the 1Repeats required the presence of the first 93 amino acids for binding. Therefore, it is possible that the 1Repeats will also bind to the shorter, transcription-repression version with a resulting effect on proliferation signaling. Another intriguing protein is Sorting Nexin 1. IQGAP1 has been previously shown to be involved in intracellular trafficking and this interaction would make sense.45 Again, if IQGAP1 bound to sorting nexin 1 via the 1Repeats, the better-characterized IQGAP1 domains and motifs (Figure 1A.) would in theory be still available to engage various known binding partners. Finally, hydrogen peroxide is a main contributor to oxidative damage and through selective modification of cysteine residues on sensitive targets, H2O2 can effect downstream redox signaling. The binding of several growth factors to their receptors, including epidermal growth factor (EGF), has been shown to increase intracellular H2O2 levels leading to the modulation of the activity of various kinases and phosphatases including MAP kinase cascade components.46–48 Catalase and the peroxiredoxins catalyze the breakdown of hydrogen peroxide and so counter the effects of increased H2O2 mediated by growth factors. Since IQGAP1 is known to be a signaling scaffold that binds MAP kinase pathway participants, simultaneous binding of a regulator of redox signaling seems to make sense.
In the past it was shown that several glycolytic enzymes were associated with the actin cytoskeleton.49, 50 Much more recently, it has been shown that human de novo purine biosynthetic enzymes cluster together under low-purine conditions, and these “purinosomes” are positioned in the cytoplasm by microtubule filaments. 51 In our study, we identified several enzymes involved in diverse metabolic pathways, including glycolysis, the citric acid cycle and gluconeogenesis. Since the C-terminus of ERM family proteins can bind to F-actin, it is quite likely that in our effort to identify 1Repeats interactors, we have pulled down cytoskeleton-associated proteins that may not have direct relevance to IQGAP1 biology. Therefore, until direct protein-protein interactions can be demonstrated, Table 1 should be considered as a source of interesting leads requiring further investigation.
Supplementary Material
Supplementary Figure S1. Identified mouse kidney proteins on 2D gel. iqga1 at the upper left corner was identified as the human 1Repeats used in the pull-down assay.
Supplementary Figure S2. Examples of purified proteins used in ITC experiments.
Supplementary Figure S3. ITC data. (A) IQGAP1 Repeats was titrated into the cell containing s-adenosylhomocysteine hydrolase (SAHH). (B) RhoGDI was titrated into 1Repeats. (C) RhoGDI titrated into EzFERM. (D) Cdc42•GDP was titrated into RhoGDI; Titrations were measured at 23°C using an ITC200. A–D, Each panel shows the raw ITC data (top) and the integrated heats (bottom) after subtraction of any heats arising from injection of titrant into buffer alone.
Supplementary Figure 4. PANTHER functional classification of 1Repeats-associated proteins.
Supplementary Document. Proteins identified using a single high confidence peptide. Single peptide is highlighted in yellow.
Acknowledgments
We thank the LSU-HSC Proteomics Core personnel for performing the mass spectrometry. This work was supported by National Institutes of Health Grant NIGMS GM084072 (D.K.W.).
Footnotes
Author Contributions
J.L. conducted all ITC experiments, protein purifications, original manuscript preparation; J.J. G. performed affinity pull-down, 2D gel electrophoresis and spot picking and in-gel trypsin digests; D.K.W. conceptualized and designed the research.
ASSOCIATED CONTENTS
2-Dimensional SDS–polyacrylamide gel electrophoresis (2D SDS–PAGE) of cellular proteins interacting with the 1Repeats (Supplementary Figure S1), PANTHER GO Molecular Function pie-chart (Supplentary Figure S2), SDS-PAGE of some of the purified proteins used in ITC experiments (Supplementary Figure S3), Representative ITC data of 1Repeats titrated into SAHH (A), 1Repeats titrated into RhoGDI (B), RhoGDI titrated into EzFERM (C) and Cdc42 titrated into RhoGDI (D) (Supplementary Figure S4), Proteins identified with a single high confidence peptide (Supplemental Document).
References
- 1.Wang S, Watanabe T, Noritake J, Fukata M, Yoshimura T, Itoh N, Harada T, Nakagawa M, Matsuura Y, Arimura N, Kaibuchi K. IQGAP3, a novel effector of Rac1 and Cdc42, regulates neurite outgrowth. J Cell Sci. 2007;120(Pt 4):567–77. doi: 10.1242/jcs.03356. [DOI] [PubMed] [Google Scholar]
- 2.Brill S, Li S, Lyman CW, Church DM, Wasmuth JJ, Weissbach L, Bernards A, Snijders AJ. The Ras GTPase-activating-protein-related human protein IQGAP2 harbors a potential actin binding domain and interacts with calmodulin and Rho family GTPases. Mol Cell Biol. 1996;16(9):4869–78. doi: 10.1128/mcb.16.9.4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weissbach L, Settleman J, Kalady MF, Snijders AJ, Murthy AE, Yan YX, Bernards A. Identification of a human rasGAP-related protein containing calmodulin-binding motifs. J Biol Chem. 1994;269(32):20517–21. [PubMed] [Google Scholar]
- 4.McCallum SJ, Wu WJ, Cerione RA. Identification of a putative effector for Cdc42Hs with high sequence similarity to the RasGAP-related protein IQGAP1 and a Cdc42Hs binding partner with similarity to IQGAP2. J Biol Chem. 1996;271(36):21732–7. doi: 10.1074/jbc.271.36.21732. [DOI] [PubMed] [Google Scholar]
- 5.White CD, Brown MD, Sacks DB. IQGAPs in cancer: a family of scaffold proteins underlying tumorigenesis. FEBS Lett. 2009;583(12):1817–24. doi: 10.1016/j.febslet.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ren JG, Li Z, Crimmins DL, Sacks DB. Self-association of IQGAP1: characterization and functional sequelae. J Biol Chem. 2005;280(41):34548–57. doi: 10.1074/jbc.M507321200. [DOI] [PubMed] [Google Scholar]
- 7.Bashour AM, Fullerton AT, Hart MJ, Bloom GS. IQGAP1, a Rac- and Cdc42-binding protein, directly binds and cross-links microfilaments. J Cell Biol. 1997;137(7):1555–66. doi: 10.1083/jcb.137.7.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukata M, Kuroda S, Fujii K, Nakamura T, Shoji I, Matsuura Y, Okawa K, Iwamatsu A, Kikuchi A, Kaibuchi K. Regulation of cross-linking of actin filament by IQGAP1, a target for Cdc42. J Biol Chem. 1997;272(47):29579–83. doi: 10.1074/jbc.272.47.29579. [DOI] [PubMed] [Google Scholar]
- 9.Briggs MW, Sacks DB. IQGAP1 as signal integrator: Ca2+, calmodulin, Cdc42 and the cytoskeleton. FEBS Lett. 2003;542(1–3):7–11. doi: 10.1016/s0014-5793(03)00333-8. [DOI] [PubMed] [Google Scholar]
- 10.Ho YD, Joyal JL, Li Z, Sacks DB. IQGAP1 integrates Ca2+/calmodulin and Cdc42 signaling. J Biol Chem. 1999;274(1):464–70. doi: 10.1074/jbc.274.1.464. [DOI] [PubMed] [Google Scholar]
- 11.Joyal JL, Annan RS, Ho YD, Huddleston ME, Carr SA, Hart MJ, Sacks DB. Calmodulin modulates the interaction between IQGAP1 and Cdc42. Identification of IQGAP1 by nanoelectrospray tandem mass spectrometry. J Biol Chem. 1997;272(24):15419–25. doi: 10.1074/jbc.272.24.15419. [DOI] [PubMed] [Google Scholar]
- 12.Mateer SC, McDaniel AE, Nicolas V, Habermacher GM, Lin MJ, Cromer DA, King ME, Bloom GS. The mechanism for regulation of the F-actin binding activity of IQGAP1 by calcium/calmodulin. J Biol Chem. 2002;277(14):12324–33. doi: 10.1074/jbc.M109535200. [DOI] [PubMed] [Google Scholar]
- 13.Fukata M, Watanabe T, Noritake J, Nakagawa M, Yamaga M, Kuroda S, Matsuura Y, Iwamatsu A, Perez F, Kaibuchi K. Rac1 and Cdc42 capture microtubules through IQGAP1 and CLIP-170. Cell. 2002;109(7):873–85. doi: 10.1016/s0092-8674(02)00800-0. [DOI] [PubMed] [Google Scholar]
- 14.Fukata M, Kuroda S, Nakagawa M, Kawajiri A, Itoh N, Shoji I, Matsuura Y, Yonehara S, Fujisawa H, Kikuchi A, Kaibuchi K. Cdc42 and Rac1 regulate the interaction of IQGAP1 with beta-catenin. J Biol Chem. 1999;274(37):26044–50. doi: 10.1074/jbc.274.37.26044. [DOI] [PubMed] [Google Scholar]
- 15.Brown MD, Sacks DB. IQGAP1 in cellular signaling: bridging the GAP. Trends Cell Biol. 2006;16(5):242–9. doi: 10.1016/j.tcb.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 16.White CD, Erdemir HH, Sacks DB. IQGAP1 and its binding proteins control diverse biological functions. Cell Signal. 2012;24(4):826–34. doi: 10.1016/j.cellsig.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marcotte EM, Pellegrini M, Yeates TO, Eisenberg D. A census of protein repeats. J Mol Biol. 1999;293(1):151–60. doi: 10.1006/jmbi.1999.3136. [DOI] [PubMed] [Google Scholar]
- 18.Andrade MA, Perez-Iratxeta C, Ponting CP. Protein repeats: structures, functions, and evolution. J Struct Biol. 2001;134(2–3):117–31. doi: 10.1006/jsbi.2001.4392. [DOI] [PubMed] [Google Scholar]
- 19.Smith MJ, Hardy WR, Li GY, Goudreault M, Hersch S, Metalnikov P, Starostine A, Pawson T, Ikura M. The PTB domain of ShcA couples receptor activation to the cytoskeletal regulator IQGAP1. Embo J. 2010;29(5):884–96. doi: 10.1038/emboj.2009.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fehon RG, McClatchey AI, Bretscher A. Organizing the cell cortex: the role of ERM proteins. Nat Rev Mol Cell Biol. 2010;11(4):276–87. doi: 10.1038/nrm2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finnerty CM, Chambers D, Ingraffea J, Faber HR, Karplus PA, Bretscher A. The EBP50-moesin interaction involves a binding site regulated by direct masking on the FERM domain. J Cell Sci. 2004;117(Pt 8):1547–52. doi: 10.1242/jcs.01038. [DOI] [PubMed] [Google Scholar]
- 22.Takai Y, Kitano K, Terawaki S, Maesaki R, Hakoshima T. Structural basis of the cytoplasmic tail of adhesion molecule CD43 and its binding to ERM proteins. J Mol Biol. 2008;381(3):634–44. doi: 10.1016/j.jmb.2008.05.085. [DOI] [PubMed] [Google Scholar]
- 23.Yonemura S, Hirao M, Doi Y, Takahashi N, Kondo T, Tsukita S, Tsukita S. Ezrin/radixin/moesin (ERM) proteins bind to a positively charged amino acid cluster in the juxta-membrane cytoplasmic domain of CD44, CD43, and ICAM-2. J Cell Biol. 1998;140(4):885–95. doi: 10.1083/jcb.140.4.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsukita S, Oishi K, Sato N, Sagara J, Kawai A, Tsukita S. ERM family members as molecular linkers between the cell surface glycoprotein CD44 and actin-based cytoskeletons. J Cell Biol. 1994;126(2):391–401. doi: 10.1083/jcb.126.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mori T, Kitano K, Terawaki S, Maesaki R, Fukami Y, Hakoshima T. Structural basis for CD44 recognition by ERM proteins. J Biol Chem. 2008;283(43):29602–12. doi: 10.1074/jbc.M803606200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turunen O, Wahlstrom T, Vaheri A. Ezrin has a COOH-terminal actin-binding site that is conserved in the ezrin protein family. J Cell Biol. 1994;126(6):1445–53. doi: 10.1083/jcb.126.6.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yonemura S, Matsui T, Tsukita S, Tsukita S. Rho-dependent and -independent activation mechanisms of ezrin/radixin/moesin proteins: an essential role for polyphosphoinositides in vivo. J Cell Sci. 2002;115(Pt 12):2569–80. doi: 10.1242/jcs.115.12.2569. [DOI] [PubMed] [Google Scholar]
- 28.Fievet BT, Gautreau A, Roy C, Del Maestro L, Mangeat P, Louvard D, Arpin M. Phosphoinositide binding and phosphorylation act sequentially in the activation mechanism of ezrin. J Cell Biol. 2004;164(5):653–9. doi: 10.1083/jcb.200307032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia-Mata R, Boulter E, Burridge K. The ‘invisible hand’: regulation of RHO GTPases by RHOGDIs. Nat Rev Mol Cell Biol. 2011;12(8):493–504. doi: 10.1038/nrm3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palmer JL, Abeles RH. The mechanism of action of S-adenosylhomocysteinase. J Biol Chem. 1979;254(4):1217–26. [PubMed] [Google Scholar]
- 31.Leal JF, Ferrer I, Blanco-Aparicio C, Hernandez-Losa J, Ramon YCS, Carnero A, Lleonart ME. S-adenosylhomocysteine hydrolase downregulation contributes to tumorigenesis. Carcinogenesis. 2008;29(11):2089–95. doi: 10.1093/carcin/bgn198. [DOI] [PubMed] [Google Scholar]
- 32.Takahashi K, Sasaki T, Mammoto A, Takaishi K, Kameyama T, Tsukita S, Takai Y. Direct interaction of the Rho GDP dissociation inhibitor with ezrin/radixin/moesin initiates the activation of the Rho small G protein. J Biol Chem. 1997;272(37):23371–5. doi: 10.1074/jbc.272.37.23371. [DOI] [PubMed] [Google Scholar]
- 33.Chambers DN, Bretscher A. Ezrin mutants affecting dimerization and activation. Biochemistry. 2005;44(10):3926–32. doi: 10.1021/bi0480382. [DOI] [PubMed] [Google Scholar]
- 34.Jayasundar JJ, Ju JH, He L, Liu D, Meilleur F, Zhao J, Callaway DJ, Bu Z. Open conformation of ezrin bound to phosphatidylinositol 4,5-bisphosphate and to F-actin revealed by neutron scattering. J Biol Chem. 2012;287(44):37119–33. doi: 10.1074/jbc.M112.380972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moleirinho S, Tilston-Lunel A, Angus L, Gunn-Moore F, Reynolds PA. The expanding family of FERM proteins. Biochem J. 2013;452(2):183–93. doi: 10.1042/BJ20121642. [DOI] [PubMed] [Google Scholar]
- 36.Hamada K, Shimizu T, Yonemura S, Tsukita S, Tsukita S, Hakoshima T. Structural basis of adhesion-molecule recognition by ERM proteins revealed by the crystal structure of the radixin-ICAM-2 complex. Embo J. 2003;22(3):502–14. doi: 10.1093/emboj/cdg039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bourguignon LY, Gilad E, Rothman K, Peyrollier K. Hyaluronan-CD44 interaction with IQGAP1 promotes Cdc42 and ERK signaling, leading to actin binding, Elk-1/estrogen receptor transcriptional activation, and ovarian cancer progression. J Biol Chem. 2005;280(12):11961–72. doi: 10.1074/jbc.M411985200. [DOI] [PubMed] [Google Scholar]
- 38.Gingras AR, Puzon-McLaughlin W, Ginsberg MH. The Structure of the Ternary Complex of Krev Interaction Trapped 1 (KRIT1) bound to both the Rap1 GTPase and the Heart of Glass (HEG1) cytoplasmic tail. J Biol Chem. 2013 doi: 10.1074/jbc.M113.462911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ren JG, Li Z, Sacks DB. IQGAP1 modulates activation of B-Raf. Proc Natl Acad Sci U S A. 2007;104(25):10465–9. doi: 10.1073/pnas.0611308104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roy M, Li Z, Sacks DB. IQGAP1 is a scaffold for mitogen-activated protein kinase signaling. Mol Cell Biol. 2005;25(18):7940–52. doi: 10.1128/MCB.25.18.7940-7952.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roy M, Li Z, Sacks DB. IQGAP1 binds ERK2 and modulates its activity. J Biol Chem. 2004;279(17):17329–37. doi: 10.1074/jbc.M308405200. [DOI] [PubMed] [Google Scholar]
- 42.Maeda M, Matsui T, Imamura M, Tsukita S, Tsukita S. Expression level, subcellular distribution and rho-GDI binding affinity of merlin in comparison with Ezrin/Radixin/Moesin proteins. Oncogene. 1999;18(34):4788–97. doi: 10.1038/sj.onc.1202871. [DOI] [PubMed] [Google Scholar]
- 43.Smith WJ, Cerione RA. Crystallization and preliminary crystallographic analysis of the ezrin FERM domain. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 8):1359–61. doi: 10.1107/s0907444902010004. [DOI] [PubMed] [Google Scholar]
- 44.Hoffman GR, Nassar N, Cerione RA. Structure of the Rho family GTP-binding protein Cdc42 in complex with the multifunctional regulator RhoGDI. Cell. 2000;100(3):345–56. doi: 10.1016/s0092-8674(00)80670-4. [DOI] [PubMed] [Google Scholar]
- 45.Rittmeyer EN, Daniel S, Hsu SC, Osman MA. A dual role for IQGAP1 in regulating exocytosis. J Cell Sci. 2008;121(Pt 3):391–403. doi: 10.1242/jcs.016881. [DOI] [PubMed] [Google Scholar]
- 46.Truong TH, Carroll KS. Redox regulation of epidermal growth factor receptor signaling through cysteine oxidation. Biochemistry. 2012;51(50):9954–65. doi: 10.1021/bi301441e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Truong TH, Carroll KS. Redox regulation of protein kinases. Crit Rev Biochem Mol Biol. 2013 doi: 10.3109/10409238.2013.790873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Corcoran A, Cotter TG. Redox regulation of protein kinases. Febs J. 2013;280(9):1944–65. doi: 10.1111/febs.12224. [DOI] [PubMed] [Google Scholar]
- 49.Arnold H, Henning R, Pette D. Quantitative comparison of the binding of various glycolytic enzymes to F-actin and the interaction of aldolase with G-actin. Eur J Biochem. 1971;22(1):121–6. doi: 10.1111/j.1432-1033.1971.tb01522.x. [DOI] [PubMed] [Google Scholar]
- 50.Clarke FM, Masters CJ. On the association of glycolytic enzymes with structural proteins of skeletal muscle. Biochim Biophys Acta. 1975;381(1):37–46. doi: 10.1016/0304-4165(75)90187-7. [DOI] [PubMed] [Google Scholar]
- 51.An S, Deng Y, Tomsho JW, Kyoung M, Benkovic SJ. Microtubule-assisted mechanism for functional metabolic macromolecular complex formation. Proc Natl Acad Sci U S A. 2010;107(29):12872–6. doi: 10.1073/pnas.1008451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. Identified mouse kidney proteins on 2D gel. iqga1 at the upper left corner was identified as the human 1Repeats used in the pull-down assay.
Supplementary Figure S2. Examples of purified proteins used in ITC experiments.
Supplementary Figure S3. ITC data. (A) IQGAP1 Repeats was titrated into the cell containing s-adenosylhomocysteine hydrolase (SAHH). (B) RhoGDI was titrated into 1Repeats. (C) RhoGDI titrated into EzFERM. (D) Cdc42•GDP was titrated into RhoGDI; Titrations were measured at 23°C using an ITC200. A–D, Each panel shows the raw ITC data (top) and the integrated heats (bottom) after subtraction of any heats arising from injection of titrant into buffer alone.
Supplementary Figure 4. PANTHER functional classification of 1Repeats-associated proteins.
Supplementary Document. Proteins identified using a single high confidence peptide. Single peptide is highlighted in yellow.