Abstract
Proton magnetic resonance spectroscopy (1H-MRS) is capable of noninvasively detecting metabolic changes that occur in the brain tissue in vivo. Its clinical utility has been limited so far, however, by analytic methods that focus on independently evaluated metabolites and require prior knowledge about which metabolites to examine. Here, we applied advanced computational methodologies from the field of metabolomics, specifically partial least squares discriminant analysis and orthogonal partial least squares, to in vivo 1H-MRS from frontal lobe white matter of 27 patients with relapsing–remitting multiple sclerosis (RRMS) and 14 healthy controls. We chose RRMS, a chronic demyelinating disorder of the central nervous system, because its complex pathology and variable disease course make the need for reliable biomarkers of disease progression more pressing. We show that in vivo MRS data, when analyzed by multivariate statistical methods, can provide reliable, distinct profiles of MRS-detectable metabolites in different patient populations. Specifically, we find that brain tissue in RRMS patients deviates significantly in its metabolic profile from that of healthy controls, even though it appears normal by standard MRI techniques. We also identify, using statistical means, the metabolic signatures of certain clinical features common in RRMS, such as disability score, cognitive impairments, and response to stress. This approach to human in vivo MRS data should promote understanding of the specific metabolic changes accompanying disease pathogenesis, and could provide biomarkers of disease progression that would be useful in clinical trials.
Keywords: Magnetic resonance spectroscopy (MRS), Metabolomics, Relapsing–remitting multiple sclerosis, Multivariate statistics
Introduction
The challenge of finding biomarkers for multiple sclerosis
Multiple sclerosis (MS) is an immune-mediated demyelinating disorder affecting the central nervous system; it is one of the most frequent causes of disability in young adults (Kurtzke and Wallin, 2000). Most patients suffer a relapsing–remitting (RR) disease course that over time transitions into insidious progression. The pathogenic mechanisms that underlie the relapsing phase and lead to the transition from RR to secondary progression remain poorly understood (Frohman et al., 2006); clinically, the unpredictability and variability in symptoms complicate disease management and render prognosis particularly elusive.
Knowledge of metabolic changes, which are a reflection of the underlying biochemistry, could provide biomarkers that would greatly improve the prospects of managing MS, and provide insight into the disease process itself. Ideally, MS biomarkers would be acquired non-invasively and would reflect disease-related pathogenic processes. Such biomarkers could foster early diagnosis and perhaps distinguish between those patients who present with clinically isolated syndrome, but never develop MS, from those who will develop a RR disease. Biomarkers, or specific patterns of biomarkers, would also make it possible to quantify patient response to treatments, improving the quality and specificity of clinical trials.
The strengths and limitations of conventional MRI and MRS
The ability of conventional magnetic resonance imaging (MRI) to identify demyelinating inflammatory plaques within the white matter offers a fairly noninvasive way to track disease progression by monitoring lesion burden, though there is often only a loose correlation between changes revealed by conventional MRI and clinical status (Bakshi et al., 2008). MRI can improve diagnosis of MS by distinguishing it from disorders with a similar clinical presentation, but here again it is not foolproof: T2 lesions occur in other neurological disorders and have been documented in asymptomatic aging brains (Bakshi et al., 2008; Vernooij et al., 2008), while some patients with clinically definite MS display no MRI abnormalities (Fazekas et al., 1999).
Neuroimaging methods such as magnetic resonance spectroscopy (MRS) can reveal metabolic changes in white matter that appears healthy by conventional MRI (De Stefano and Filippi, 2007), and therefore might provide a more precise means of diagnosing and following the disease course. However, MRS has its own limitations: MRS studies typically evaluate independent changes in only a small handful of major metabolites (Poullet et al., 2008; Sajja et al., 2009) such as N-acetyl groups (mainly N-acetyl aspartate (NAA)), choline-containing compounds (Cho), creatine and phosphocreatine (Cr + PCr), and myo-inositol (mI). While changes in these specific metabolites have been reported at various stages of MS (Arnold et al., 1994; Chard et al., 2002; Narayana, 2005), such targeted analyses have failed to develop an MS-specific metabolic signature. More importantly, these targeted analyses rely on prior knowledge about the metabolite's presence to calculate and compare group means–however, they overlook a vast amount of potentially valuable information that might be contained in smaller but abundant metabolites such as lipids, lactate, aspartate (Asp), glutamine (Gln), and glutamate (Glu), which are difficult to quantify using current methods.
Metabolomic techniques can circumvent these limitations
To circumvent these limitations, we turned to techniques developed in the emerging field of metabolomics. Metabolomics uses a non-targeted approach to obtain an accurate representation of the metabolome, the collection of small molecules that reflect the processes which take place in living biological systems (Griffin, 2003; Lindon and Nicholson, 2008; Smolinska et al., 2012). In contrast to the traditional approach of interrogating a specific subset of small molecules based on a predetermined hypothesis, similar to testing for high cholesterol as an indicator of heart disease, the aim of metabolomics is to acquire a functional biochemical profile that encompasses all detectable small metabolites (specifically identified or not) and trace changes in the profile over the course of development or disease to generate new hypotheses. Whether the goal is to assess the metabolic effect of diet, a response to drug therapy or differences in populations, the question is simple: what has changed?
Metabolomic analysis can be performed on any biological matrix–blood, tears, urine, cerebrospinal fluid or biopsied tissue in vitro, or living tissue in vivo–using mass spectrometry or nuclear magnetic resonance (NMR) spectroscopy (Hassan-Smith et al., 2012; Lindon and Nicholson, 2008). The resulting high-density datasets are analyzed with multivariate statistical modeling to identify metabolites that correlate with functional changes in a given system (Griffin, 2003). Metabolomic-type analysis can overcome the sorts of signal distortions that can occur with MRS, providing previously unavailable information about living tissue, in vivo. Unlike other quantification tools, metabolomic analysis of the full resolution spectra has the advantage of not requiring a priori knowledge such as the line shapes of the metabolite resonances. Therefore, the resonances that can be identified are not limited to the user's input criteria, and changes in small resonances can be extracted.
Untargeted metabolic profiling has been proven feasible for a variety of human diseases (Griffin, 2003). One of the most significant applications has been the use of NMR-based metabolomics on sera for rapid, accurate, and noninvasive assessment of coronary artery disease (Brindle et al., 2002). Other applications include the detection of oral squamous cell carcinoma using plasma (Zhou et al., 2009), epithelial ovarian cancer with sera (Odunsi et al., 2005), the characterization of inflammatory bowel disease using urine samples (Williams et al., 2009), and distinguishing multiple sclerosis patients from controls using cerebrospinal fluid samples (Hassan-Smith et al., 2012; Rajalahti et al., 2010). In vivo, a lot of work and much success have been in the area of distinguishing brain tumor type and grade using metabolic profiling in combination with other MR measures (Galanaud et al., 2006; Preul et al., 1996).
In our study, we extend multivariate statistical analyses to in vivo MRS spectra obtained from individuals with RRMS and healthy controls. We identify a metabolomic model of RRMS that distinguishes between spectra from three tissue types in vivo: the white matter of the frontal lobe in healthy controls (CTWM); the frontal lobe in RRMS patients, which appears normal by conventional MRI (normal-appearing white matter, NAWM); and the periventricular non-enhancing lesions in RRMS (NELES). We validate this metabolomic model by predicting a set of spectra not used in the model-building procedure and achieve excellent assignment of tissue type. We also show, for the first time, that the untargeted metabolomic techniques applied to in vivo MRS data can identify metabolic perturbations that correlate with clinical features common in RRMS.
Subjects and methods
Subject selection
The study was designed to focus solely on RRMS subjects whose clinical information is summarized in Supplementary Table 1. We recruited 27 individuals (22 females, age 38.6 ± 10.1 years; age range: 23–62 years) who met diagnostic criteria for RRMS (Polman et al., 2011) through the outpatient offices of the Multiple Sclerosis Comprehensive Care Center in Stony Brook, NY. RRMS subjects had to be clinically stable, which we defined as at least two months since the last relapse, ambulatory with at most bilateral assistance, and able to tolerate neuroimaging. Participants could be on or off disease-modifying therapy, but their medications had to have been stable for at least two months prior to evaluation, and participants were imaged no sooner than four weeks after their last steroid dose. Subjects with an Extended Disability Status Scale (EDSS) (Kurtzke, 1983) greater than 6.5 were excluded from the study.
The control group consisted of 14 subjects (13 females; age 31.1 ± 9.1 years; range: 21 to 51 years) recruited from a community sample of healthy volunteers who had no history of neurological disorders. All participants gave written consent to participate in the study, which was approved by the Institutional Review Boards of Stony Brook and Princeton Universities.
Subject evaluations
Neuropsychological measures were included on a subgroup of participants who consented for cognitive testing (Table 1). Neuropsy-chological measures included: the Rey Auditory Verbal Learning Test (RAVLT), a list learning task that assesses verbal learning and memory (Nici, 2000); the Symbol Digit Modality Test (SDMT), a measure of working memory and cognitive processing speed (Parmenter et al., 2007); and the Paced Auditory Serial Addition Test (PASAT)–3 s, a timed measure of working memory (Gronwell, 1977). Some patients had time restraints and could not complete all the tests, and in some cases these measures could not be completed in entirety. Subsequent to the cognitive testing, two psychological questionnaires were added: the Holmes & Rahe Social Readjustment Rating Scale (SRRS) which measures stressful life events (Hobson et al., 1998; Holmes and Rahe, 1967); and the Fatigue Severity Scale (FSS) which assesses the impact of fatigue on daily functioning (Krupp et al., 1989).
Table 1.
Clinical and cognitive findings in RRMS and control groups.
| Control group | RRMS group | p-Value | |
|---|---|---|---|
| Age (years) | 31.1 (9.1) | 38.6 (10.1) | 0.02 |
| (n = 14) | (n = 27) | ||
| Disease duration (years) | N/A | 8.1 (8.8) (n = 27) | N/A |
| EDSS | N/A | 2.3 (1.2) | N/A |
| Global neurological impairment | (n = 27) | ||
| RAVLT | 59.0 (9.4) | 46.9 (10.6) | <0.001 |
| Verbal memory | (n = 10) | (n = 20) | |
| SDMT | 65.2 (9.9) | 51.2 (12.5) | <0.001 |
| Cognitive processing speed | (n = 10) | (n = 22) | |
| PASAT | 50.1 (6.4) | 39.6 (10.2) | <0.001 |
| Working memory | (n = 10) | (n = 21) | |
| FSS | 2.2 (0.9) | 4.2 (1.2) | <0.001 |
| Fatigue | (n = 7) | (n = 14) | |
| SRRS | 11.6 (10.7) | 19.4 (10.9) | ns |
| Stress | (n = 7) | (n = 14) |
The RRMS and control groups mean values differed significantly across all the cognitive measures but only on one of the self-report measures, the FSS. ns: not significant, all values are reported as mean (SD).
Magnetic resonance imaging and spectroscopy data acquisition
MRI and MRS were conducted on a 3 T Phillips Achieva MR Scanner (Philips Medical Systems) with an 8-channel SENSE, receive-only head coil. In MS subjects, spectra from normal appearing white matter (NAWM) and non-enhancing lesions (NELES) were obtained contralaterally and exclusively in frontal lobe. NELES with a diameter of 7 mm or larger were selected for MRS, to minimize partial volume effects. In control subjects, spectra were collected in control white matter (CTWM) from the periventricular white matter in the frontal lobe, to prevent confounds in the modeling from any region-dependent metabolic differences. Whole brain segmentation was calculated in SPM8 (Wellcome Department of Cognitive Neurology, London, UK), yielding measures of whole brain gray, white and CSF fractions, i.e. total volume/intracranial volume, for each subject.
Volume of interest (VOI)
There is a delicate balance between voxel size and placement for in vivo 1H-MRS studies, because the brain is heterogeneous; a large voxel may contain multiple tissue types while decreasing the voxel size limits the signal-to-noise ratio (SNR). A 14 × 14 × 14 mm voxel (2.744 cm3) was chosen for all regions and subjects, based primarily on the size of a typical MS lesion, allowing maximum signal to be captured from the lesion volume, with minimal signal from the surrounding tissue. Care was also taken to avoid CSF within the VOI; the high water content of CSF has been shown to impair data acquisition. Because not all subjects had lesions in the frontal lobe or lesion-free NAWM in the frontal lobe, the analysis was carried out on 23 NAWM spectra and 19 NELES spectra from the 27 MS subjects, and 14 CTWM spectra from control subjects.
MRI/MRS data acquisition parameters
1H-MRS were collected using short echo time (TE/TR = 32/2000 ms), single voxel point-resolved spectroscopy (PRESS) in the frontal lobe NAWM and NELES, if present, in RRMS subjects (Fig. 1) and the frontal lobe of control subjects. Other sequence parameters were: 2 kHz acquisition bandwidth, 1024 points, 128 NEX, and CHESS water suppression (Frahm et al., 1989). To confirm that the lesions selected were non-enhancing, 3-dimensional T1-weighted gradient echo images were taken 10 min following a single-dose gadolinium contrast injection. Gadolinium was given after the MRS was done.
Fig. 1.
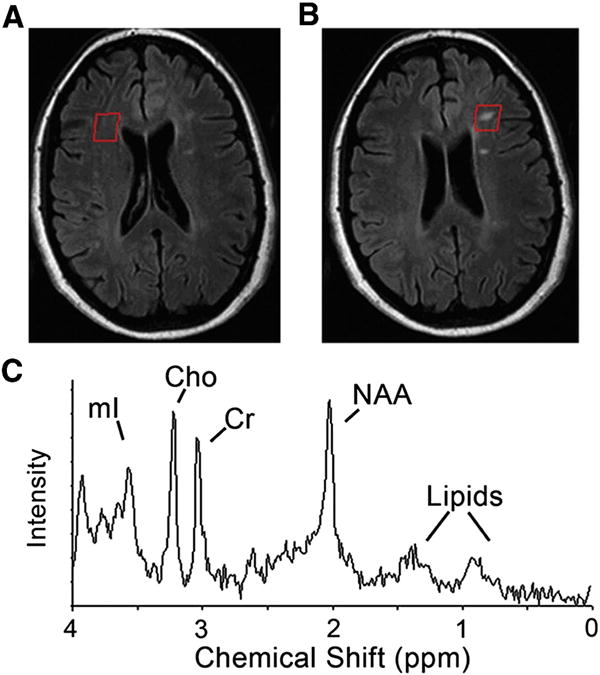
MRI and MR spectroscopy (MRS) of relapsing–remitting multiple sclerosis (RRMS) subject showing (A) normal appearing white matter and (B) non-enhancing lesion. The representative T1 weighted MRIs (upper) demonstrate the voxel placement for MRS indicated by the red square. (C) Representative MR spectra, indicating major metabolites: mI–myo-inositol; Cho–choline containing compound; Cr–creatine; NAA–N-acetyl as-partate, and lipids. Control white matter spectra were acquired from similar locations.
Magnetic resonance spectroscopy data analysis
Single voxel 1H-MRS spectra were processed using 3DiCSI software v.1.9.9 (Hatch MR Research Center, Columbia University), involving the following steps: zero filling to 2048 points, apodization with a 5 Hz Gaussian filter, and Fourier Transform. The first and zero order phase corrections were manually applied to correct any phase errors. The spectra were exported as ASCII files into MATLAB (The Mathworks, Natick, MA) where the curve-fitting toolbox was used to apply linear baseline correction. Also in MATLAB, the spectra were normalized to creatine at 3.0 ppm, the region from 4.0 to 6.0 ppm was excluded from the data, and the spectra were compiled into a matrix for subsequent statistical analysis using SIMCA-P (version 11.55, Umetric, San Jose, CA).
Metabolites were also quantified using LCModel (Provencher, 1993). The LCModel is a fully automated MRS quantification technique that uses information from a basis set of metabolite spectra in order to estimate the quantity of metabolite in a given tissue, from the MR spectrum. LCModel fits the reference spectra of the basis set of metabolites to the real data, to provide absolute and relative quantitation of individual metabolites such as NAA, Cr, Cho etc., present in the dataset.
Normalization
The absolute creatine + phosphocreatine (Cr + PCr) concentration from the LCModel and unsuppressed water signal were considered as normalization factors. Since the Cr + PCr levels were more consistent across CTWM, NAWM, and NELES than the water signal and did not vary with age, we used it as the normalization factor for the metabolomic analysis (see Supplementary Fig. 3 and Normalization of MRS data). An additional benefit of normalizing to Cr + PCr is that it allows direct comparison to many previous studies which have reported their results as ratios to Cr, although the increase in Cr in NAWM found in our study needs to be taken into account when interpreting the results.
Statistical analysis
All assessments are expressed as mean ± standard deviation. A Student's t test (unpaired, two-tailed) was applied for the comparisons of clinical assessments (RAVLT, SDMT, PASAT, FSS, and SRRS) between control and RRMS subjects.
Multivariate statistical analysis was carried out with SIMCA-P. For this analysis, the spectral points, excluding the residual water region and creatine peaks, are considered the X variables. The class information of the spectra (CTWM, NAWM, or NELES) or clinical scores are the Y variables. For more details on the metabolomic statistical methods, see the Supplemental materials. Since the multivariate methods are based on variance, i.e., they seek out the greatest variance of the data, the X variables are centered and scaled to reduce the bias it places on large variables (Craig et al., 2006). As with most analytical data, a large peak will exhibit a greater absolute variance than a small peak. To minimize this effect, the standard in metabolomics is to apply unit-variance or Pareto scaling. We chose Pareto scaling, which computes the base weight as 1/sqrt(sdj), where sdj is the standard deviation of the variable j computed around the mean, as the most suitable for minimizing the effects of the baseline and overall noise in the data.
Metabolic profiling of three tissue types (NAWM, CTWM, and NELES) was carried out using partial least squares discriminant analysis (PLS-DA). PLS-DA is a supervised chemometric tool that models the variation in the n by m data matrix X into two multiple components, where n is the number of observations and m the number of variables (in our case each spectral point is a variable), in response to an n by k dummy Y matrix containing class membership information (Barker and Rayens, 2003). The k variables are zeros and ones, where a value of one indicates class membership. This analysis results in a score plot that summarizes each spectrum as a single point as it relates to the other spectra in the dataset. The loading plot summarizes the contribution of each variable (spectral point) to the placement of the samples in the score plot.
Correlations of the metabolic features with clinical scores were modeled using orthogonal partial least squares (O-PLS). O-PLS is a chemometric tool that models the variation in data matrix X into 2 components, one linearly related to variation of the clinical variable (the first component) and one that is orthogonal and associated with uncorrelated variation (the orthogonal component) (Trygg and Wold, 2002). This extension of PLS greatly increases the interpretability of the model since the metabolic variables associated with the variation in the clinical features are condensed along one component.
To build and validate the PLS-DA model for class discrimination and prediction, the data was randomly divided into a training set (80% of the data–11 CTWM, 18 NAWM, 15 NELES) and a test set (20% of the data–3 CTWM, 5 NAWM, 4 NELES). The test set was excluded from model construction, and the model was used to predict class membership of the data in the test set.
During both O-PLS and PLS-DA model construction, an iterative method of cross-validation was used where 1/7th of the samples were randomly excluded and used for predictions by applying the loading coefficients of the model to the excluded spectra. This was repeated until each spectrum was predicted once. This results in a predicted y value for each sample (i.e. class membership in the case of PLS-DA, clinical score in the case of O-PLS) that can be compared to the actual y value to assess the model's predictability.
In addition to the metabolomic analysis, multiple ANOVA was used to determine if significant differences exist between groups for LCModel quantified metabolites. Tukey honest significant difference tests were used to determine which contrasts were significantly different while controlling for multiple comparisons. Pearson's correlation was used to determine relationships between age and MR measures.
Results
Subject evaluation
Clinically, the RRMS subjects were characterized according to their EDSS, which ranged from 0 to 4.0 (mean of 2.3 ± 1.2), and disease duration, which ranged from 0.25 to 40 years (mean of 8 ± 9 years; median 5 years) (Supplementary Table 1). Further, they had neuropsychological (see Subject evaluations and Metabolic signatures distinguish white matter of MS patients from healthy controls, and Table 1) and radiological assessments. Radiological parameters included lesion load, which ranged from 3.6 to 47.8 cm3 (mean 17.3 cm3, standard deviation 11.0 cm3) (Supplementary Table 1). Gray matter fraction significantly varied with age (Pearson's correlation r = −0.63, p < 0.001), while white matter fraction did not (p = 0.167). After correcting for age, there was a significant difference between gray matter fraction in MS (44.69% ± 2.04%) and controls (46.14% ± 2.02%, p = 0.04), as well as a difference in white matter fraction (controls 32.97% ± 2.25%, MS subjects 31.20% ± 2.51%, p = 0.03).
Effect of age on MRS data
We have analyzed frontal NAWM 1H-MRS data in subjects with established RRMS. The participants varied in age (p = 0.02), and thus we considered the influence of age on the metabolite content. Interestingly, while several studies clearly indicate metabolite differences in most regions of the brain (Inglese and Ge, 2004; Kadota et al., 2001; Leary et al., 2000; Schuff et al., 2001), the metabolites assessed in the frontal white matter have been shown to not be affected significantly by age (Chang et al., 1996, 2009; Fukuzako et al., 1997). Regardless, to evaluate if the metabolic patterns in our data had an age affect, we performed principal component analysis (PCA) which showed no trends of age in the data (Supplementary Fig. 1). Additionally, we attempted to build a PLS model with age as the Y matrix, and could not compute a validated model (data not shown). This indicates that our MRS data does not have an age dependence. To ascertain that our multivariate statistical analyses do not mask the effect of age on individual metabolites, we analyzed the effect of age on key individual metabolites estimated by LCModel (Supplementary Fig. 2). As shown, scatterplots indicate that there is no significant effect of age on NAA, Cho, mI, and macromolecule/lipid compounds. Overall, these data established that further analysis of metabolomic signatures in RRMS in all our subjects is not confounded by age.
Normalization of MRS data
We considered both the water signal from an unsuppressed spectrum and Cr + PCr as normalization factors (Supplementary Fig. 3). The water signal was found to significantly differ between all three groups (CTWM: 1.71 ± 0.21, NAWM: 1.91 ± 0.24, NELES: 2.06 ± 0.31; p-value CTWM vs. NAWM, 0.014; CTWM vs. NELES, 0.001; NAWM vs. NELES, 0.098), in addition to having a strong relationship with age (p = 0.009). The absolute concentration of Cr + PCr was significantly decreased in CTWM in comparison to NAWM, but not between CTWM and NELES or NAWM and NELES (CTWM: 4.68 ± 0.59; NAWM 5.12 ± 0.41; 4.95 ± 0.36; p-values CTWM vs. NAWM, 0.025; CTWM vs. NELES, 0.148; NAWM vs. NELES, 0.167). We therefore chose to use Cr + PCr as normalization factor since it was consistent across tissue types for our data and did not vary significantly with age.
Metabolic signatures distinguish white matter of MS patients from healthy controls
To characterize the metabolic differences between CTWM from healthy controls, and NAWM and NELES from MS patients, we performed PLS-DA. The PLS-DA score plot shows clustering amongst the spectra from each tissue type (Fig. 2A). The first component discriminates NELES from CTWM, while the second component discriminates NAWM from CTWM and NELES. The regions that are most influential in producing this discrimination are indicated by the regression coefficients derived from the PLS-DA model (Fig. 2B). A positive value along the x-axis of Fig. 2B indicates a relatively higher metabolite concentration in CTWM compared to RRMS tissue (either NAWM or NELES). A positive value along the y-axis indicates a relatively higher metabolite concentration in NAWM compared to either CTWM or NELES. On the 2-dimensional loading plot, each point represents a single point along the chemical shift scale of the MRS spectrum and is color-coded according to assigned metabolites to aid in interpretation (Fig. 2C). In general, the greatest spectral differences resulted from NAA (2.02 ppm) in CTWM; lipids (1.3 − 1.75 ppm) in NAWM, mI (3.58 ppm) in NELES; and Cho (3.22 ppm) in both RRMS tissues (i.e., NAWM and NELES). Additional metabolites which may have influenced the model include Glu (2.45 ppm) in NAWM, as well as aspartate (Asp, 2.79 ppm) and γ-aminobutyric acid (GABA, 1.89 ppm) in NELES.
Fig. 2.
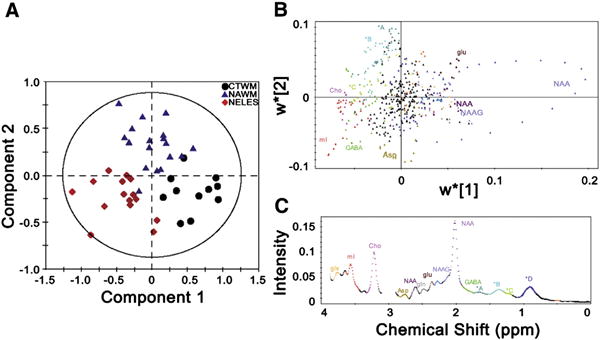
Comprehensive metabolomic model using PLS-DA distinguishes between control white matter (CTWM), normal appearing white matter (NAWM), and non-enhancing lesions (NELES) based on their metabolic profiles. (A) On the PLS-DA score plot, each single spectrum is converted to a data point that represents the relative contribution of metabolites from MRS of CTWM (black circle), NAWM (blue triangle), and NELES (red diamond) from the frontal lobe. A 2-dimensional loading plot (top panel) (B) colored by spectral features (bottom panel) (C) demonstrates the contribution each point along the spectrum makes to the component analysis in A. This aids in interpretation when there are a lot of variables, as in the case of full resolution MRS datasets. While the NAA peak did not show up in the 1-dimensional loading projection, it can be clearly seen to be the most pronounced in the CTWM. The creatine peaks (3.0 and 3.9 ppm) were left out of analysis since the spectra were normalized to creatine. Key: glx: glutamine + glutamate; mI: myo-inositol; Cho: choline; Asp: aspartate; NAA: N-acetyl aspartate; gln: glutamine; glu: glutamate; NAAG: N-acetyl aspartyl glutamate; GABA: γ-aminobutyric acid. *A: 1.65−1.75 ppm, macromolecules; *B 1.3−1.65 ppm, lipids, alanine, and lactate; *C 1.15−1.3 ppm, macromolecules, possibly lactate; *D 0.8−1.1 ppm, lipids. Metabolite assignments are based on reported chemical shifts in the in vivo MRS literature (Govindaraju et al., 2000).
Validation
The PLS-DA model successfully discriminated class membership based on spectral features. Using a class membership threshold of 0.5, the internal cross-validation shows that the model classified 38 of the 44 training spectra correctly, a rate of 86% (Table 2). Of the six spectra incorrectly classified, one CTWM was misclassified as NAWM, one NAWM was misclassified as a NELES, and two CTWM and two NAWM were not classified into any membership class based on the 0.5 criteria. There was no obvious reason for these misclassification based on the individual's clinical descriptors such as age, gender, or disease characteristics.
Table 2.
Internal cross validation of the PLS-DA model.
| CTWM | NAWM | NELES | |
|---|---|---|---|
| (n = 11) | (n = 18) | (n = 15) | |
| Correct predictions | 8 | 15 | 15 |
| Incorrect predictions | 1 | 1 | 0 |
| No prediction | 2 | 2 | 0 |
Classification from the leave one out procedure. A score of 0.5 or higher indicated class membership. CTWM, control white matter; NAWM, normal appearing white matter; NELES, non-enhancing lesion. 86% of the spectra were correctly classified.
External validation was used to exclude the possibility that PLS-DA modeling describes only the data it was built on and fails to describe or predict new data (See Supplementary materials for more detail). Compared to the internal validation, which correctly classified 86% of the spectra, the external validation successfully predicted 9 out of 12 spectra, or 75% of the data, as summarized in Table 3. However, on the score plot with the predicted scores overlaid in Fig. 3, only two spectra seem misclassified. Thus, the modeling is probably not taking into account a slight overlap between the classes when making the predictions of class membership. In either case, these results show great promise for future applications of metabolomics to in vivo assessment of neurological disorders.
Table 3.
PLS-DA model performance on the test set.
| CTWM | NAWM | NELES | |
|---|---|---|---|
| (n = 3) | (n = 5) | (n = 4) | |
| Correct predictions | 1 | 4 | 4 |
| Incorrect predictions | 1 (NAWM) | 1 (CTWM) | 0 |
| No prediction | 1 | 0 | 0 |
Classification from the test set. A score of 0.5 or higher indicated class membership. Class 1 is control white matter, Class CTWM, control white matter; NAWM, normal appearing white matter; NELES, non-enhancing lesion. 9 out of 12 of the spectra were correctly classified.
Fig. 3.
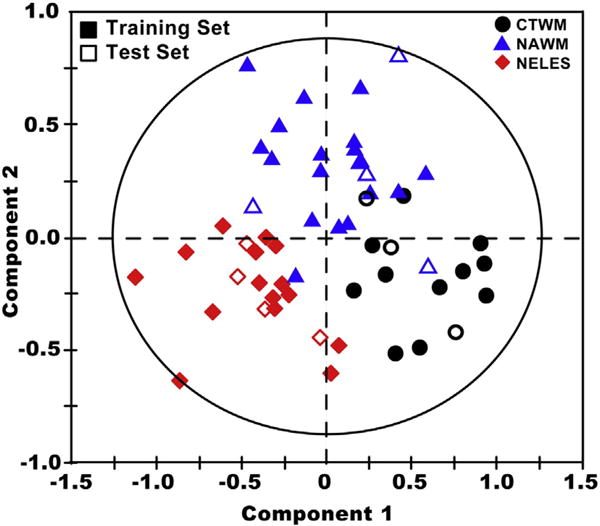
Validation of the PLS-DA model. Using the model described in Fig. 2 (training set), a second set of data (test set) was used to test the validity of the model (training set–filled symbols; test set–open symbols). The close clustering of the test set to the training set demonstrates the validity of the model described in Fig. 2 for separating out tissue types based on their metabolic properties.
Comparison to LCModel analysis
To complement the results of the PLS-DA model which used the digital resolution spectral points as the input parameters, we quantified the metabolites using LCModel, which uses a priori established reference library for a selected group of metabolites (Supplementary Table 2). If there was a trend between groups defined as α < 0.10, we performed a post hoc analysis to determine which metabolites differed between groups. Our results indicate a significant decrease in NAA + NAAG/Cr + PCr between both NAWM and NELES compared to CTWM. In addition, there was a significant increase in mI/Cr + PCr compared to NAWM, and a trend of increase in CTWM. Glu/Cr + PCr also displayed a trend of being decreased in NELES compared to CTWM and NAWM, although this did not reach significance after taking into account multiple testing. Asp/Cr + PCr displayed a significant increase in NAWM compared to CTWM and NELES, and GABA/Cr + PCr had a small but insignificant increase in NELES. The macromolecules and lipids fitted with the LCModel did not show any significant differences between groups.
The LCModel results agree with the PLS-DA analysis, as can be seen by the contributing metabolites identified by the loading plot (Fig. 2B). In the loading plot, an increase in metabolite signal is signified by the metabolite being in the same area as the tissue type cluster in the score plot and a decrease is signified by the metabolite being in the opposite area. For example, mI appears in the lower left hand corner on the loading plot, due to the fact that it is increased in NELES compared to NAWM, and it has a slight but insignificant increase compared to CTWM. While the major metabolites identified by LCModel analysis agreed with the data obtained by metabolomic model, the lipids did not. This is most likely due to the less well-defined nature of their signals. In our analysis, the lineshape of these signals is not predefined, where in the LCModel they are. Our analysis showed that macromolecule peaks at 1.3 ppm and 1.7 ppm were increased in NAWM compared to CTWM and NELES. The ANOVA for the LCModel correlates (MM14 + Lip13a + Lip13b + MM12 and MM17) did not reach significance with the threshold of α = 0.05. This could be due to the difficulty to fit these complex broad areas to the metabolites in the basis set reliably.
Clinical features common in RRMS correlate with specific metabolic signatures
To further characterize the disease process of RRMS and determine whether there is a metabolic pattern specific to certain clinical aspects of MS, we performed orthogonal partial least squares (O-PLS) with standard clinical measures of global neurological impairment, cognitive dysfunction, fatigue, and stress (see Subjects and methods). Such untargeted statistical correlations have never been established and analyzed before. The RRMS group had significantly worse scores than the control group on every test except the Social Readjustment Rating Scale (SRRS), which measures response to stressful life events (Table 1).
We built independent O-PLS models for each clinical assessment to determine if a metabolic signature correlated with these measures. Using internal cross-validation, we found three tests that correlated with certain spectral characteristics: the EDSS which measures global neurological impairment, the RAVLT measure of verbal learning, and the SRRS (Fig. 4) of perceived stress. Each O-PLS model is summarized according to a score plot colored by the clinical measure, an average MRS spectrum with the loading coefficient for the first component projected by color, and an observed versus predicted plot from the internal cross validation (Fig. 4; see Subjects and methods).
Fig. 4.
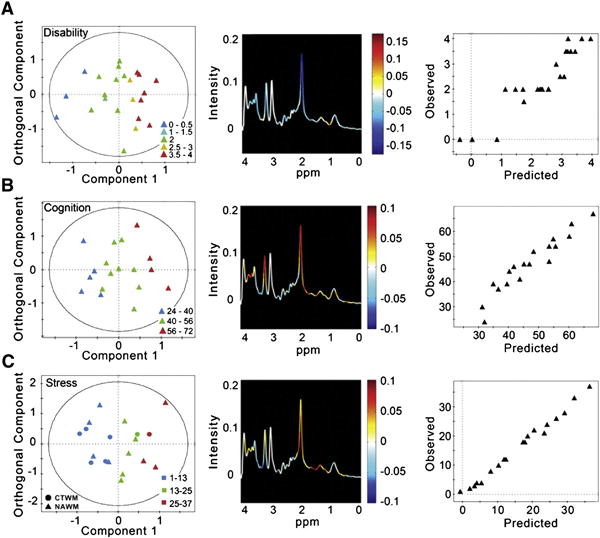
The correlation of clinical features in RRMS and metabolic profiles. A separate O-PLS model was developed for each of the following characteristics commonly seen in RRMS: global neurological impairment measured by EDSS; verbal memory measured by RAVLT; and stress measured by SRRS. (A) The first component of the score plot (left) demonstrates how the metabolic profiles for each NAWM spectrum, represented by a single point, vary with neurological impairment (EDSS, where 0 is no impairment and 4 is moderate impairment). The orthogonal component represents the variability in the dataset unrelated to neurological impairment. The spectrum (center) is color-coded by the regression coefficients driving the O-PLS model, thus summarizing the metabolites varying with neurological impairment. For example, NAA is colored blue, indicating that as NAA decreases EDSS increases. The observed versus predicted plot (right) from the internal cross validation indicates a good model with an R2 of 0.858 and RMSE of 0.466. (B) The first component of the scores plot (left) demonstrates how the metabolic profiles of NAWM vary with verbal memory measured by RAVLT, where a lower score indicates declining verbal memory. The spectrum (center) is color-coded by the O-PLS model coefficients, where red indicates metabolites increasing with increasing verbal memory (or, conversely, decreasing with memory decline), and blue metabolites are decreased with increasing memory performance. The observed vs. predicted plot (right) has an R2 of 0.909 with a RMSE of 3.55, indicating a predictive model. (C) The first component of the score plot (left) demonstrates how the metabolic profiles of CTWM and NAWM vary with stress measured by SRRS, where a higher score indicates a greater amount of stressful life events. The spectrum (center) is color-coded by the O-PLS model coefficients, where metabolites colored red are increasingand those colored blue are decreasing with increased stress. The observed vs. predicted plot (right) has an R2 of 0.991 and RMSE of 1.07, indicating a predictive model.
NAA levels correlate inversely with global neurological impairment
The EDSS scores for the RRMS subjects (n = 27) ranged from 0 to 4, with a mean of 2.3 ± 1.2. The O-PLS model estimating EDSS score from NAWM spectra shows a decrease in many of the MRS-detected metabolites and an increase in lipids around 1.3 ppm, which correlates with increased EDSS (Fig. 4A), as indicated by the loading coefficients from the first component of the model. A positive coefficient value along the first component indicates a metabolite that increases with worsening neurological impairment, whereas a negative value indicates a metabolite that decreases with worsening neurological impairment. The main contributing metabolite is NAA (2.02 ppm), which correlates negatively with the EDSS score. The internal cross-validation, summarized by the observed versus predicted plot, has a R2 of 0.858, with a root mean squared error (RMSE) of 0.466.
Decreases in NAA and Cho correlate with loss of verbal memory
Overall, the RRMS participants demonstrated significant verbal memory dysfunction compared to the control group (two-sample, two-sided t-test, p-value = 0.0065), with a mean RAVLT score of 46.85 ± 10.6 (control group average score: 59.0 ± 9.4). The O-PLS model for RAVLT was therefore built on NAWM spectra alone (n = 20), and is summarized in Fig. 4B with a score plot colored by RAVLT score, where normal memory function is defined as a RAVLT score over 50. From the internal cross validation, the R2 for the observed versus predicted plot is 0.909 with a RMSE of 3.55. The regression coefficients indicate that decreases in NAA (2.02 ppm) and Cho (3.20 ppm) are the main metabolic changes correlated with verbal memory decline.
Greater stress correlates with lower levels of lipids in both RRMS and controls
Interestingly, the controls did not differ significantly from the RRMS subjects in SRRS scores (two-sample, two-sided t-test, p-value = 0.19). Therefore, the O-PLS model for SRRS included CTWM and NAWM spectra (n = 21) (Fig. 4C). The score plot is colored by SRRS, with a low value indicating low stress and a high value indicating high stress. The R2 for the observed versus predicted plot is 0.991 and RMSE of 1.07. The loading projection shows that there is an increase in the lipids around 1.3 ppm and NAA (2.02 ppm), and a decrease in lipids resonating at 0.90 ppm and ml(3.58 ppm), which correlates with higher stress and a larger SRRS score.
Discussion
We set out to determine whether an untargeted metabolomic approach to human brain spectral data could distinguish between healthy and disease states, specifically in multiple sclerosis. We found that a multivariate statistical approach can be used to detect correlative changes of several metabolites within brain tissue, as demonstrated on CTWM, NAWM, and NELES, resulting in distinct complex metabolic signatures. We also identified metabolomic signatures that correlate with specific disease characteristics of RRMS, suggesting that this type of analysis might deepen our understanding of specific metabolic changes underlying MS pathogenesis.
We applied multivariate analysis, widely used in a variety of experimental settings (Dehmeshki et al., 2001; Weygandt et al., 2011) and in metabolomic profiling of biospecimen, but not in human brain spectroscopy, to capture all the metabolic fluctuations important for class discrimination in one analysis. This approach covers the whole spectra without prior selection of the metabolites and can identify differential behavior of small resonances that are difficult to quantify. While both the conventional LCModel analysis and PLS-DA approach lead to similar conclusions about the metabolic differences between tissue types, the PLS-DA analysis has the advantage of not testing multiple hypotheses, and therefore can be used to direct conventional analysis without producing errors due to multiple testing. Conventional quantification techniques have found significant differences between individual metabolites in CTWM, NAWM, and NELES (De Stefano and Filippi, 2007), but these differences have been neither disease-specific nor discriminatory. We propose that further studies identifying more complex metabolite signatures could yield useful disease-specific profiles of numerous neurological disorders.
Our complex metabolic signatures of NAWM, NELES, and CTWM were independent of age and are consistent with previous reports of independently measured metabolic disturbances, demonstrating the validity of our approach. The decrease of NAA we found in both NAWM and NELES has been reported (De Stefano et al., 2001; Filippi et al., 1999; Kutzelnigg et al., 2005; Ruiz-Pena et al., 2004) and is often interpreted as a sign of neurological/axonal dysfunction (De Stefano et al., 2001). The increases in mI and Cho we found to be primarily associated with NELES are also supported by previous studies: increases in mI mark glial cell proliferation in MS (Poullet et al., 2008) and an increase of Cho indicates membrane disturbances and possibly myelin breakdown (Govindaraju et al., 2000). The increase in lipids we observed in NAWM has been reported previously and interpreted to indicate demyelination and remyelination (Sajja et al., 2009). The combined increase coefficients, where metabolites colored red are increasing and those colored blue are decreasing with increased stress. The observed vs. predicted plot (right) has an R2 of 0.991 and RMSE of 1.07, indicating a predictive model. in lipids and Cho has been shown in longitudinal studies to indicate acute and severe inflammation, leading to lesion development (Narayana, 2005).
We also identified abundant small metabolites, primarily Glu, GABA, and Asp, which differ in the metabolic signatures of NAWM, CTWM, and NELES (Fig. 2). These metabolites are difficult to measure because of the small signal intensities that appear in a crowded region of the MR spectrum. Using a special MRS acquisition technique, Glu has been shown to be elevated in NAWM (Srinivasan et al., 2005), which supports our findings. The changes in GABA and Asp are an interesting discovery that merits further investigation.
We then sought to determine whether clinical characteristics of RRMS, such as global neurological impairment, cognitive dysfunction, pathological fatigue, and stressful experiences correlate with metabolic signatures of NAWM. We used O-PLS to identify metabolic profiles indicative of neurological impairment, verbal memory, and stress. Consistent with previous studies, we identified a negative correlation between NAA and global neurological impairment (as measured by EDSS) (De Stefano et al., 1998, 2001; Mainero et al., 2001; Ruiz-Pena et al., 2004). We also found that global neurological impairment negatively correlates with levels of Cho, mI, and Gln/Glu. It should be noted that the decrease in the majority of MRS-visible metabolites could indicate a change in Cr + PCr, to which the spectra were normalized. While Cr + PCr is usually considered a stable metabolite and accepted as a normalization method, alterations in Cr + PCr have been reported in numerous neurological disorders and needs to be taken into account when interpreting results (Caramanos et al., 2005). Nevertheless, a high resolution NMR spectroscopy study of post-mortem MS brains showed that Cr + PCr is unchanged in normal-appearing brain tissue of MS subjects, although the Cr + PCr concentration may be altered in the lesion tissue (Davies et al., 1995). Comparison of the absolute concentrations of Cr + PCr to the unsuppressed water signal across three tissue types indicated that Cr + PCr was more consistent in our study. Since the focus of this study was primarily on the changes and characterization of NAWM in MS, normalizing to Cr + PCr was a reasonable choice.
There have been no studies specifically addressing the relationship between metabolic fluctuations and cognitive function as measured by the RAVLT in MS, but a decrease in NAA has been associated with MS-related declines in cognition measured by other means (e.g., a cognitive dysfunction factor and a modified Brief Repeatable Battery of Neuro-psychological Tests) (Christodoulou et al., 2003; Mathiesen et al., 2006). We found that decreases in NAA, Cho and Gln/Glu, along with possible increases in scyllo-inositol, correlated with declining verbal memory. These metabolites could be important indicators of cognitive decline in RRMS.
Finally, the relationship between stress and MS disease activity is poorly understood (Mohr et al., 2000) and it is unknown whether there is a biochemical basis behind it. Our results suggest that there could be a general metabolic relationship between stressful life events and the frontal lobe white matter. In both controls and RRMS participants, increases in stress correlated with a reduction in lipid content.
Unlike the clear metabolomic correlations with neurological impairment, verbal memory, and stress response, there were no distinct metabolomic signatures associated with fatigue, cognitive processing speed, or working memory (Supplementary Fig. 5). There are three conceivable explanations: these features of MS do not have an observable metabolomic signature, our sample size was too small to detect such metabolic signatures, or metabolic changes that do correlate with these measures do not occur in the frontal lobe. Nevertheless, certain features of RRMS are clearly reflected in a distinct metabolome composition of the NAWM. Such findings could be important for practitioners, as they could indicate comorbid states in RRMS.
Conclusions
We have shown that untargeted metabolomic statistical analysis can be used for comprehensive noninvasive tissue profiling of diseased and healthy human brains in vivo. While distinguishing between CTWM, NAWM, and NELES demonstrates the utility of the technique, metabolic profiling of specific clinical aspects of disease demonstrates its potential power. In a single analysis we captured metabolic alterations that have been reported in several independent analyses. Since the metabolomic approach does not use a priori information about the metabolites in the sample, more features can be extracted from the MRS datasets if they are correlated to an aspect of disease. These techniques should be extended to a larger dataset to model disease subtype, progression, or treatment monitoring, where they may become a valuable tool not only for creating more patient-specific assessments but also for providing insight into the underlying MS pathology.
Supplementary Material
Acknowledgments
The authors wish to thank Y. Patel, V. Bhise, and D. Greenblatt for help with the subject recruitment. Wealso thank Vicky Brandt, William Rooney, and Dennis Bourdette for comments and expert revision of the paper. This work was supported by the National Library of Medicine (grant# 5-T15-LM 7088-20) (L.K.V), the Lourie Foundation Incorporated, National Multiple Sclerosis Society (grant # RG4030A2/1), National Center for Research Resources (grant # 5-MO1-RR-10710) (L.B.K.), and by the NIH Intellectual and Developmental Disabilities Research Grant (P30HD024064), Dana Foundation, and McKnight Endowment for Science (M.M.-S.).
Abbreviations
- MRS
magnetic resonance spectroscopy
- 1H-MRS
proton magnetic resonance spectroscopy
- RRMS
relapsing–remitting multiple sclerosis
- CTWM
control white matter, i.e., MRS of white matter from healthy controls
- NAWM
normal-appearing white matter, i.e., MRS from the frontal lobe in RRMS patients
- NELES
periventricular non-enhancing lesions in RRMS patients
Footnotes
Appendix A. Supplementary data: Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.neuroimage.2013.05.125.
Author contributions: L.K.V. designed the experiments, analyzed the data, and wrote the manuscript. H.J.Y. performed MRI and MRS studies. D.S. Scheduled patients for MRI studies and visits, performed cognitive evaluations, and entered data. C.C. performed neuropsychological analyses and critically reviewed the paper. M.E.W. performed MRI and MRS studies and critically reviewed the manuscript. I.P. supervised all analyses and critically reviewed the manuscript. L.B.K. clinically evaluated the subjects, designed and supervised all the experiments and wrote the manuscript. M.M.-S. designed and supervised all the experiments and wrote the manuscript.
References
- Arnold DL, Riess GT, Matthews PM, Francis GS, Collins DL, Wolfson C, Antel JP. Use of proton magnetic resonance spectroscopy for monitoring disease progression in multiple sclerosis. Ann Neurol. 1994;36:76–82. doi: 10.1002/ana.410360115. [DOI] [PubMed] [Google Scholar]
- Bakshi R, Thompson AJ, Rocca MA, Pelletier D, Dousset V, Barkhof F, Inglese M, Guttmann CRG, Horsfield MA, Filippi M. MRI in multiple sclerosis: current status and future prospects. Lancet Neurol. 2008;7:615–625. doi: 10.1016/S1474-4422(08)70137-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker M, Rayens W. Partial least squares for discrimination. J Chemom. 2003;17(3):166–173. [Google Scholar]
- Brindle JT, Antti H, Holmes E, Tranter G, Nicholson JK, Bethell HWL, Clarke S, Schofield PM, McKilligin E, Mosedale DE, et al. Rapid and noninvasive diagnosis of the presence and severity of coronary heart disease using 1H-NMR-based metabonomics. Nat Med. 2002;8(1439–1445):1439. doi: 10.1038/nm1202-802. [DOI] [PubMed] [Google Scholar]
- Caramanos Z, Narayanan S, Arnold DL. 1H-MRS quantification of tNA and tCr in patients with multiple sclerosis: a meta-analytic review. Brain. 2005;128(11):2483–2506. doi: 10.1093/brain/awh640. [DOI] [PubMed] [Google Scholar]
- Chang L, Ernst T, Poland RE, Jenden DJ. In vivo proton magnetic resonance spectroscopy of the normal aging human brain. Life Sci. 1996;58(22):2049–2056. doi: 10.1016/0024-3205(96)00197-x. [DOI] [PubMed] [Google Scholar]
- Chang L, Jiang CS, Ernst T. Effects of age and sex on brain glutamate and other metabolites. Magn Reson Imaging. 2009;27(1):142–145. doi: 10.1016/j.mri.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chard D, Griffin CM, McLean MA, Kapeller P, Kapoor R, Thompson AJ, Miller DH. Brain metabolites changes in cortical grey and normal appearing white matter in clinically early relapsing remitting multiple sclerosis. Brain. 2002;125:2342–2352. doi: 10.1093/brain/awf240. [DOI] [PubMed] [Google Scholar]
- Christodoulou C, Krupp LB, Liang Z, Huang W, Melville P, Roque C, Scherl WF, Morgan T, MacAllister WS, Li L, et al. Cognitive performance and MR markers of cerebral injury in cognitively impaired MS patients. Neurology. 2003;60(11):1793–1798. doi: 10.1212/01.wnl.0000072264.75989.b8. [DOI] [PubMed] [Google Scholar]
- Craig A, Cloarec O, Holmes E, Nicholson JK, Lindon JC. Scaling and normalization effects in NMR spectroscopic metabonomic data sets. Anal Chem. 2006;78(7):2262–2267. doi: 10.1021/ac0519312. [DOI] [PubMed] [Google Scholar]
- Davies SE, Newcombe J, Williams SR, McDonald WI, Clark JB. High resolution proton NMR spectroscopy of multiple sclerosis lesions. J Neurochem. 1995;64(2):742–748. doi: 10.1046/j.1471-4159.1995.64020742.x. [DOI] [PubMed] [Google Scholar]
- De Stefano N, Filippi M. MR spectroscopy in multiple sclerosis. J Neuroimaging. 2007;17(Suppl 1):31S–35S. doi: 10.1111/j.1552-6569.2007.00134.x. [DOI] [PubMed] [Google Scholar]
- De Stefano N, Matthews PM, Fu L, Narayanan S, Stanley J, Francis GS, Antel JP, Arnold DL. Axonal damage correlates with disability in patients with relapsing remitting multiple sclerosis. Brain. 1998;121:1469–1477. doi: 10.1093/brain/121.8.1469. [DOI] [PubMed] [Google Scholar]
- De Stefano N, Narayanan S, Francis GS, Arnaoutelis R, Tartaglia MC, Antel JP. Evidence of axonal damage in the early stages of multiple sclerosis and its relevance to disability. Arch Neurol. 2001;58:65–70. doi: 10.1001/archneur.58.1.65. [DOI] [PubMed] [Google Scholar]
- Dehmeshki J, Ruto AC, Arridge S, Silver NC, Miller DH, Tofts PS. Analysis of MTR histograms in multiple sclerosis using principal components and multiple discriminant analysis. Magn Reson Med. 2001;46(3):600–609. doi: 10.1002/mrm.1233. [DOI] [PubMed] [Google Scholar]
- Fazekas F, Barkhof F, Filippi M, Grossman RI, Li DKB, McDonald WI, McFarland HF, Paty DW, Simon JH, Wolinsky JS, et al. The contribution of magnetic resonance imaging to the diagnosis of multiple sclerosis. Neurology. 1999;53(3):448–456. doi: 10.1212/wnl.53.3.448. [DOI] [PubMed] [Google Scholar]
- Filippi M, Tortorella C, Bozzali M. Normal-appearing white matter changes in multiple sclerosis: the contribution of magnetic resonance techniques. Mult Scler. 1999;5(4):273–282. doi: 10.1177/135245859900500414. [DOI] [PubMed] [Google Scholar]
- Frahm J, Bruhn H, Gyngell ML, Merboldt KD, Hänicke W, Sauter R. Localized high-resolution proton NMR spectroscopy using stimulated echoes: initial applications to human brain in vivo. Magn Reson Med. 1989;9(1):79–93. doi: 10.1002/mrm.1910090110. [DOI] [PubMed] [Google Scholar]
- Frohman EM, Racke MK, Raine CS. Multiple sclerosis–the plaque and its pathogenesis. N Engl J Med. 2006;354(9):942–955. doi: 10.1056/NEJMra052130. [DOI] [PubMed] [Google Scholar]
- Fukuzako H, Hashiguchi T, Sakamoto Y, Okamura H, Doi W, Takenouchi K, Takigawa M. Metabolite changes with age measured by proton magnetic resonance spectroscopy in normal subjects. Psychiatry Clin Neurosci. 1997;51(4):261–263. doi: 10.1111/j.1440-1819.1997.tb02595.x. [DOI] [PubMed] [Google Scholar]
- Galanaud D, Nicoli F, Chinot O, Confort-Gouny S, Figarella-Branger D, Roche P, Fuentès S, Le Fur Y, Ranjeva J, Cozzone PJ. Noninvasive diagnostic assessment of brain tumors using combined in vivo MR imaging and spectroscopy. Magn Reson Med. 2006;55(6):1236–1245. doi: 10.1002/mrm.20886. [DOI] [PubMed] [Google Scholar]
- Govindaraju V, Young K, Maudsley AA. Proton NMR chemical shifts and coupling constants for brain metabolites. NMR Biomed. 2000;13(3):129–153. doi: 10.1002/1099-1492(200005)13:3<129::aid-nbm619>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Griffin JL. Metabonomics: NMR spectroscopy and pattern recognition analysis of body fluids and tissues for characterisation of xenobiotic toxicity and disease diagnosis. Curr Opin Chem Biol. 2003;7(5):648–654. doi: 10.1016/j.cbpa.2003.08.008. [DOI] [PubMed] [Google Scholar]
- Gronwell D. Paced auditory serial addition task: a measure of recovery from concussion. Percept Mot Skills. 1977;44:367–373. doi: 10.2466/pms.1977.44.2.367. [DOI] [PubMed] [Google Scholar]
- Hassan-Smith G, Wallace GR, Douglas MR, Sinclair AJ. The role of metabolomics in neurological disease. J Neuroimmunol. 2012;248:48–52. doi: 10.1016/j.jneuroim.2012.01.009. [DOI] [PubMed] [Google Scholar]
- Hobson CJ, Kamen J, Szostek J, Nethercut CM, Tiedmann JW, Wojnarowicz S. Stressful life events: a revision and update of the social readjustment rating scale. Int J Stress Manag. 1998;5(1):1–23. [Google Scholar]
- Holmes TH, Rahe RH. The social readjustment rating scale. J Psychosom Res. 1967;11(2):213–218. doi: 10.1016/0022-3999(67)90010-4. [DOI] [PubMed] [Google Scholar]
- Inglese M, Ge Y. Quantitative MRI: hidden age-related changes in brain tissue. Top Magn Reson Imaging. 2004;15(6):355–363. doi: 10.1097/01.rmr.0000168069.12985.15. [DOI] [PubMed] [Google Scholar]
- Kadota T, Horinouchi T, Kuroda C. Development and aging of the cerebrum: assessment with proton MR spectroscopy. Am J Neuroradiol. 2001;22(1):128–135. [PMC free article] [PubMed] [Google Scholar]
- Krupp LB, LaRocca NG, Muir-Nash J, Steinberg AD. The fatigue severity scale: application to patients with multiple sclerosis and systemic lupus erythematosus. Arch Neurol. 1989;46(10):1121–1123. doi: 10.1001/archneur.1989.00520460115022. [DOI] [PubMed] [Google Scholar]
- Kurtzke JF. Rating neurologic impairment in multiple sclerosis: an extended disability status scale (EDSS) Neurology. 1983;33:1444–1452. doi: 10.1212/wnl.33.11.1444. [DOI] [PubMed] [Google Scholar]
- Kurtzke JF, Wallin MT. Epidemiology. In: Burks JS, Johnson KP, editors. Multiple Sclerosis: Diagnosis, Medical Management, and Rehabilitation. Demos Medical Publishing, Inc.; New York: 2000. pp. 49–71. [Google Scholar]
- Kutzelnigg A, Lucchinetti CF, Stadelmann C, Brück W, Rauschka H, Bergmann M, Schmidbauer M, Parisi JE, Lassmann H. Cortical demyelination and diffuse white matter injury in multiple sclerosis. Brain. 2005;128(11):2705–2712. doi: 10.1093/brain/awh641. [DOI] [PubMed] [Google Scholar]
- Leary SM, Brex PA, MacManus DG, Parker GJM, Barker GJ, Miller DH, Thompson AJ. A 1H magnetic resonance spectroscopy study of aging in parietal white matter: implications for trials in multiple sclerosis. Magn Reson Imaging. 2000;18:455–459. doi: 10.1016/s0730-725x(00)00131-4. [DOI] [PubMed] [Google Scholar]
- Lindon JC, Nicholson JK. Spectroscopic and statistical techniques for information recovery in metabonomics and metabolomics. Annu Rev Anal Chem. 2008;1(1):45–69. doi: 10.1146/annurev.anchem.1.031207.113026. [DOI] [PubMed] [Google Scholar]
- Mainero C, De Stefano N, Iannucci G, Sormani MP, Guidi L, Federico A, Bartolozzi ML, Comi G, Filippi M. Correlates of MS disability assessed in vivo using aggregates of MR quantities. Neurology. 2001;56:1331–1334. doi: 10.1212/wnl.56.10.1331. [DOI] [PubMed] [Google Scholar]
- Mathiesen HK, Jonsson A, Tscherning T, Hanson LG, Andresen J, Blinkenberg M, Paulson OB, Sorensen PS. Correlation of global N-acetyl aspartate with cognitive impairment in multiple sclerosis. Arch Neurol. 2006;63(4):533–536. doi: 10.1001/archneur.63.4.533. [DOI] [PubMed] [Google Scholar]
- Mohr DC, Goodkin DE, Bacchetti P, Boudewyn AC, Huang L, Marrietta P, Cheuk W, Dee B. Psychological stress and the subsequent appearance of new brain MRI lesions in MS. Neurology. 2000;55(1):55–61. doi: 10.1212/wnl.55.1.55. [DOI] [PubMed] [Google Scholar]
- Narayana PA. Magnetic resonance spectroscopy in the monitoring of multiple sclerosis. J Neuroimaging. 2005;15:46S–57S. doi: 10.1177/1051228405284200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nici J. A compendium of neuropsychological tests. Arch Neurol. (2) 2000;57(2):278. [Google Scholar]
- Odunsi K, Wollman RM, Ambrosone CB, Hutson A, McCann SE, Tammela J, Geisler JP, Miller G, Sellers T, Cliby W, et al. Detection of epithelial ovarian cancer using 1H-NMR-based metabonomics. Int J Cancer. 2005;113(5):782–788. doi: 10.1002/ijc.20651. [DOI] [PubMed] [Google Scholar]
- Parmenter BA, Weinstock-Guttman B, Garg N, Munschauer F, Benedict RH. Screening for cognitive impairment in multiple sclerosis using the symbol digit modalities test. Mult Scler. 2007;13(1):52–57. doi: 10.1177/1352458506070750. [DOI] [PubMed] [Google Scholar]
- Polman CH, Reingold SC, Banwell B, Clanet M, Cohen JA, Filippi M, Fujihara K, Havrdova E, Hutchinson M, Kappos L, et al. Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann Neurol. 2011;69(2):292–302. doi: 10.1002/ana.22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poullet J, Sima DM, Van Huffel S. MRS signal quantitation: a review of time-and frequency-domain methods. J Magn Reson. 2008;195(2):134–144. doi: 10.1016/j.jmr.2008.09.005. [DOI] [PubMed] [Google Scholar]
- Preul MC, Caramanos Z, Collins DL, Villemure J, Leblanc R, Olivier A, Pokrupa R, Arnold DL. Accurate, noninvasive diagnosis of human brain tumors by using proton magnetic resonance spectroscopy. Nat Med. 1996;2(3):323–325. doi: 10.1038/nm0396-323. [DOI] [PubMed] [Google Scholar]
- Provencher SW. Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magn Reson Med. 1993;30(6):672–679. doi: 10.1002/mrm.1910300604. [DOI] [PubMed] [Google Scholar]
- Rajalahti T, Kroksveen AC, Arneberg R, Berven FS, Vedeler CA, Myhr K, Kvalheim OM. A multivariate approach to reveal biomarker signatures for disease classification: application to mass spectral profiles of cerebrospinal fluid from patients with multiple sclerosis. J Proteome Res. 2010;9(7):3608–3620. doi: 10.1021/pr100142m. [DOI] [PubMed] [Google Scholar]
- Ruiz-Pena J, Pinero P, Sellers G, Argente J, Casado A, Foronda J, Ucles A, Izquierdo G. Magnetic resonance spectroscopy of normal appearing white matter in early relapsing–remitting multiple sclerosis: correlations between disability and spectroscopy. BMC Neurol. 2004;4(1):8. doi: 10.1186/1471-2377-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajja BR, Wolinsky JS, Narayana PA. Proton magnetic resonance spectrosco-py in multiple sclerosis. Neuroimaging Clin N Am. 2009;19(1):45–58. doi: 10.1016/j.nic.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuff N, Ezekiel F, Gamst AC, Amend DL, Capizzano AA, Maudsley AA, Weiner MW. Region and tissue differences of metabolites in normally aged brain using multislice 1H magnetic resonance spectroscopic imaging. Magn Reson Med. 2001;45(5):899–907. doi: 10.1002/mrm.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolinska A, Blanchet L, Buydens LMC, Wijmenga SS. NMR and pattern recognition methods in metabolomics: from data acquisition to biomarker discovery: a review. Anal Chim Acta. 2012;750:82–97. doi: 10.1016/j.aca.2012.05.049. [DOI] [PubMed] [Google Scholar]
- Srinivasan R, Sailasuta N, Hurd R, Nelson S, Pelletier D. Evidence of elevated glutamate in multiple sclerosis using magnetic resonance spectroscopy at 3 T. Brain. 2005 May;128(5):1016–1025. doi: 10.1093/brain/awh467. [DOI] [PubMed] [Google Scholar]
- Trygg J, Wold S. Orthogonal projections to latent structures (O-PLS) J Chemom. 2002;16(3):119–128. [Google Scholar]
- Vernooij MW, de Groot M, van der Lugt A, Ikram MA, Krestin GP, Hofman A, Niessen WJ, Breteler MMB. White matter atrophy and lesion formation explain the loss of structural integrity of white matter in aging. NeuroImage. 2008;43(3):470–477. doi: 10.1016/j.neuroimage.2008.07.052. [DOI] [PubMed] [Google Scholar]
- Weygandt M, Hackmack K, Pfüller C, Bellmann-Strobl J, Paul F, Zipp F, Haynes J. MRI pattern recognition in multiple sclerosis normal-appearing brain areas. PLoS One. 2011;6(6):e21138. doi: 10.1371/journal.pone.0021138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams HRT, Cox IJ, Walker DG, North BV, Patel VM, Marshall SE, Jewell DP, Ghosh S, Thomas HJW, Teare JP, et al. Characterization of inflammatory bowel disease with urinary metabolic profiling. Am J Gastroenterol. 2009;104(6):1435–1444. doi: 10.1038/ajg.2009.175. [DOI] [PubMed] [Google Scholar]
- Zhou J, Xu B, Huang J, Jia X, Xue J, Shi X, Xiao L, Li W. 1H NMR-based metabonomic and pattern recognition analysis for detection of oral squamous cell carcinoma. Clin Chim Acta. 2009;401(1–2):8–13. doi: 10.1016/j.cca.2008.10.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


