Abstract
The major vectors of malaria in sub-Saharan Africa belong to subgenus Cellia. Yet, phylogenetic relationships and temporal diversification among African mosquito species have not been unambiguously determined. Knowledge about vector evolutionary history is crucial for correct interpretation of genetic changes identified through comparative genomics analyses. In this study, we estimated a molecular phylogeny using 49 gene sequences for the African malaria vectors An. gambiae, An. funestus, An. nili, the Asian malaria mosquito An. stephensi, and the outgroup species Culex quinquefasciatus and Aedes aegypti. To infer the phylogeny, we identified orthologous sequences uniformly distributed approximately every 5 Mb in the five chromosomal arms. The sequences were aligned and the phylogenetic trees were inferred using maximum likelihood and neighbor-joining methods. Bayesian molecular dating using a relaxed log normal model was used to infer divergence times. Trees from individual genes agreed with each other, placing An. nili as a basal clade that diversified from the studied malaria mosquito species 47.6 million years ago (mya). Other African malaria vectors originated more recently, and independently acquired traits related to vectorial capacity. The lineage leading to An. gambiae diverged 30.4 mya, while the African vector An. funestus and the Asian vector An. stephensi were the most closely related sister taxa that split 20.8 mya. These results were supported by consistently high bootstrap values in concatenated phylogenetic trees generated individually for each chromosomal arm. Genome-wide multigene phylogenetic analysis is a useful approach for discerning historic relationships among malaria vectors, providing a framework for the correct interpretation of genomic changes across species, and comprehending the evolutionary origins of this ubiquitous and deadly insect-borne disease.
Introduction
Malaria vectors belong to taxonomically diverse groups of anopheline mosquitoes. The genus Anopheles is divided into six subgenera including Anopheles, Cellia, Kerteszia, Lophopodomyia, Nyssorhynchus and Stethomyia [1]. Although the six Anopheles subgenera are monophyletic in origin, major malaria vectors do not form a monophyletic groupare polyphyletic [2]. Most malaria vectors are members of species complexes, which include both vectors and nonvectors [3], [4]. All major malaria vectors in Sub-saharan Africa belong to the subgenus Cellia, which consist of six series: Cellia, Neocellia, Myzomyia, Pyretophorus, Paramyzomyia, and Neomyzomyia. Previous analyses of the rDNA, and combined rDNA plus mtDNA data have supported the monophyly of a clade that includes Pyretophorus, Myzomyia, Neocellia and Neomyzomyia series [5]. Anopheles gambiae and An. arabiensis are major vectors of malaria in Africa and are members of the An. gambiae complex, which belongs to the series Pyretophorus. Anopheles gambiae consists of two molecular forms: the S form is widely distributed and the M form is restricted to West and Central Africa [6], [7]. A recent study has proposed to elevate the taxonomic status of the M form to a formal species level with the new name Anopheles coluzzii [8]. The An. gambiae complex also includes minor vectors (An. bwambae, An. merus, An. melas) and nonvectors (An. quadriannulatus and An. amharicus) [3], [8]. Anopheles funestus belongs to the Funestus subgroup, which is classified under the series Myzomyia and is divided in to five subgroups: Aconitus, Culicifacies, Funestus, Minimus, and Rivulorum [1]. Anopheles moucheti also belongs to the series Myzomyia. Anopheles nili is a member of the An. nili group that belongs to the series Neomyzomiya [1]. The Asian malaria mosquito An. stephensi, which is often used for phylogenetic comparisons with African mosquito species [5], [9]–[12], belongs to the series Neocellia within the subgenus Cellia [1].
Malaria in tropical humid savannas of Africa is quite stable with entomological inoculation rates (EIR: number of infective bites per person per year) varying between 50 and 300 [13]. Anopheles gambiae (M and S molecular forms), An. arabiensis, An. funestus, and An. nili are responsible for the majority of malaria cases in these areas [13]. These species, together with An. moucheti, a major vector in the equatorial forest of Central Africa, are responsible for >95% of the total malaria transmission on the African continent [14]. The habitat of these species varies considerably, and their contact with human habitation is of critical importance for addressing malarial transmission. Anopheles gambiae, An. arabiensis, and An. funestus breed in temporal or permanent freshwater pools. Anopheles gambiae is found mostly in humid savannas, An. arabiensis occupies arid savannas and steppes [15], while An. funestus has a continent-wide distribution in a broad variety of habitats [16]. Anopheles nili is as widely distributed as An. gambiae and is spread across most of West, Central, and East Africa, mainly in humid savannah and degraded rainforest areas [17], [18]. However, unlike the other major vectors, An. nili breeds in fast-moving streams and large lotic rivers exposed to light and containing vegetation or debris [13], [18]. A study of the ecological niche profiles of major malaria vectors in Cameroon demonstrated that the habitats of An. gambiae, An. arabiensis, and An. funestus have more overlap with each other than with the habitat of An. nili [17]. These results indicate a much more unusual geographic distribution of An. nili, and a different setting in which the species comes into contact with humans, thus revealing its pivotal role in malaria transmission in degraded, open-canopy forests in this region of Africa, and potentially elsewhere in the continent [18].
Knowledge about the phylogenetic relationships among the major African malaria vectors is essential for understanding the species-specific immune system responses from Plasmodium falciparum infection, and the accompanying evolutionary changes in the genomes of the mosquito species. However, it is still unknown if a particular lineage originated a long time ago or has emerged only recently. The diverse taxonomic positions of the major malaria vectors suggest that vectorial capacity evolved independently in all of these species. Each species group or complex, to which the major vectors belong, also includes nonvectors [1]. A phylogenetic reconstruction using mtDNA and rDNA has demonstrated a polyphyletic relationship among An. arabiensis, An. gambiae, An. funestus, An. moucheti, and An. nili [9]. However, stem-group relationships among the major African malaria vectors have not been unambiguously identified. For example, a Bayesian phylogenetic analysis using combined rDNA (18S and 28S) sequences, places the An. gambiae complex as sister to the remaining species of the subgenus Cellia, including An. stephensi, An. funestus and An. nili. However, a bootstrap consensus tree estimated with mtDNA sequences has been unable to provide strong support for these relationships due to a very low phylogenetic signal [9]. In contrast, other phylogenetic trees based on combined rDNA and mtDNA sequences have placed Anopheles dirus and Anopheles farauti (species from the same series as An. nili) as sister to the remaining species of the subgenus Cellia, including An. gambiae, An. funestus and An. stephensi [5].
An alternative approach to inferring phylogenetic relationships among species is a multigene phylogenetic analysis, which with the availability of many novel genetic markers and next-generation sequencing, has been successfully performed in many organisms. For example, 78 protein-coding genes have been effectively used to reconstruct a multigene phylogenetic tree of Choanozoa (unicellular protozoan phylum) and their evolutionary relationships to animals and fungi [19]. In another study, a phylogenetic tree based on 22 gene segments in Mustelidae, carnivorous mammals in the weasel family, has been inferred, and provides good support for both deep and shallow nodes in the group [20]. Importantly, multigene phylogenetics based on numerous concatenated gene sequences provides greater resolution and support, and is able to accurately reconstruct even ancient divergences, e.g. between animals and fungi [19]-[21]. According to a phylogenetic study of 106 orthologous genes from eight yeast species, the sufficient number of concatenated genes that are required to achieve the mean bootstrap value of 70% is three; while a minimum of 20 genes is required to recover >95% bootstrap values for each branch of the species tree [22]. Another study demonstrated an efficient phylogenetic approach by sampling and assembling transcriptomes of 10 mosquito species into data matrices containing hundreds of thousands of orthologous nucleotides from hundreds of genes [10]. However, that study did not include the major African malaria vectors except 3 species from the An. gambiae complex.
In our study, we investigated the phylogenetic relationships among major African malaria vectors as well as an Asian vector, An. stephensi. We selected 49 genes from the An. gambiae PEST strain genome [23], [24] distributed throughout 5 chromosomal arms. We identified orthologous sequences in the genomes of An. nili [25], An. stephensi [12], [26], C. quinquefasciatus [27], and A. aegypti [28], and in the transcriptome of An. funestus [29]. Phylogenetic trees were generated using a maximum likelihood (ML) and neighbor-joining (NJ) method from all genes individually, and with genes concatenated according to chromosomal arms. Results from different chromosomal arms were consistent with each other, and (1) placed An. nili as sister to the other African anopheline species, (2) indicated that the An. gambiae lineage split from the An. funestus and An. stephensi clade, and (3) African An. funestus and Asian An. stephensi were most closely related and most recently diverged taxa.
Materials and Methods
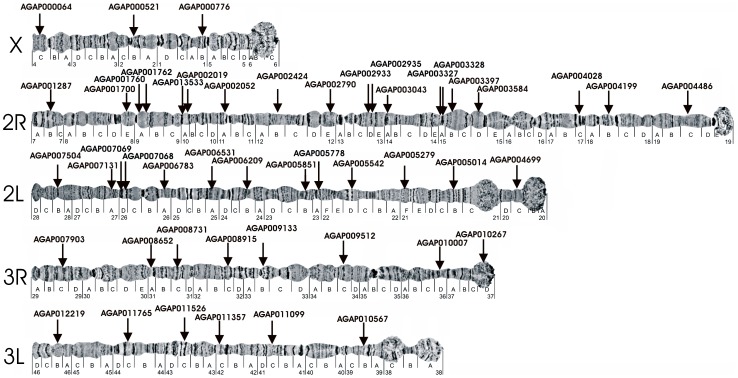
Genome-wide selection of genetic markers
To ensure that gene trees provide multiple independent estimates of the species tree, it is important that genetic markers are distributed as uniformly as possible across the genome rather than clustered in a particular genomic region/single linkage group [30]–[32]. To resolve the molecular phylogeny of African malaria vectors, we selected 49 genes as molecular markers, widely distributed throughout the genome in all five chromosomal arms of the An. gambiae cytogenetic map [33] (Figure 1). In order to select genes evenly distributed throughout the genome, the AgamP3 genome assembly of the An. gambiae PEST strain (https://www.vectorbase.org/organisms/anopheles-gambiae/pest/agamp3) [24] was divided into 5 Mb segments. Genes were randomly selected within the 5 Mb segments of the An. gambiae genome with exon lengths between 364 and 1400 bp. Selected exons were transferred to Geneious 5.1.5 software (www.geneious.com) and used for finding homologous sequences in the An. nili genome assembly (DDBJ/EMBL/GenBank accession ATLZ00000000) [25] using Basic Local Alignment Search Tool (BLAST). The AnilD1 genome assembly of the An. nili Dinderesso strain is also available at VectorBase for BLAST and download (https://www.vectorbase.org/downloadinfo/anopheles-nili-dinderessocontigsanild1fagz). If no BLAST hits were present, another exon or gene was then selected. Appropriate exons from the An. gambiae PEST genome that had significant e-values (<1e-10) in BLAST searches against the An. nili genome were the candidate genes for further BLAST analysis. These exons were used to BLAST against the An. gambiae M5 (M form, Mali strain) (https://www.vectorbase.org/organisms/anopheles-gambiae/mali-nih-m-form/m5) and G4 (S form, Pimperena strain) (https://www.vectorbase.org/organisms/anopheles-gambiae/pimperena-s-form/g4) genome assemblies [34], the An. stephensi AsteI1 (DDBJ/EMBL/GenBank accession ALPR00000000, VectorBase: https://www.vectorbase.org/downloadinfo/anopheles-stephensi-indianscaffoldsastei1fagz) [12], [26], the C. quinquefasciatus (https://www.vectorbase.org/organisms/culex-quinquefasciatus/johannesburg/cpipj1) [27], and A. aegypti (https://www.vectorbase.org/organisms/aedes-aegypti/liverpool-lvp/aaegl1) [28] genome assemblies, as well as An. funestus transcriptome sequences (http://funcgen.vectorbase.org/annotated-transcriptome/Crawford_et_al_Anopheles_funestus) [29].
Figure 1. Distribution of genic phylogenetic markers in five chromosomal arms of An. gambiae.
Names of the arms are placed near telomeres.
Orthology detection
Two genes are orthologs if they diverged in a speciation event and are related by common ancestry [35]. To find orthologous genes, a Reciprocal Best Hits (RBH) method with an e-value threshold of at least 1e-10 was used [36]. In this method, orthologous pairs should have best reciprocal BLAST hits. In order to detect the RBH, An. nili, An. stephensi, An. funestus, C. quinquefasciatus and A. aegypti sequences were used to BLAST against the An. gambiae PEST strain, which has a mixture of sequences from M and S forms. Orthology was confirmed if reciprocal BLAST then finds the originally selected sequences in the An. gambiae PEST genome as the best hits.
Gene alignment and phylogenetic analyses
Orthologous sequences with significant e-values in the BLAST search were then transferred to Molecular Evolutionary Genetics Analysis (MEGA 5.05) program [37]. The sequences were aligned using ClustalW alignment option in MEGA. Alignments were performed by adding the presumed most closely related species followed by the outgroup species. The sequence alignments are available upon request. Phylogenetic trees for each gene were constructed using a NJ method [38] in MEGA and a ML based method in RAxML version 7.5.3 [39]. A general time reversible (GTR) model with gamma distributed rate heterogeneity of nucleotide substitution (GTR + Γ) was used in the ML analysis. Support values for each clade were generated in RAxML by 1000 rapid bootstrap replicates. The 49 genes in 5 chromosomal arms were concatenated into a dataset of 42,300 bp and similarly analyzed using a RAxML tree search, while treating each gene as a separate unlinked partition. PhyloBayes version 3.3 was then used to estimate node divergence times under a relaxed clock log normal model [40], [41]. We used the best RAxML tree as a starting topology for PhyloBayes tree searches. Three fossils were used to calibrate the phylogeny: Anopheles dominicanus, 34 million years ago (mya) [42], Culex winchesteri, 34 mya [43], and Paleoculicis minutus, 70 mya, [44]. We assigned calibration fossils to the following nodes in the analysis: (1) An. dominicanus to the most recent node shared by An. gambiae and An. nili, (2) C. winchesteri to the most recent node shared by C. quinquefasciatus, and Ae. aegypti, and P. minutus to the most recent node shared by An. nili and Ae. aegypti. Placement of fossil calibrations were based on the global similarity or intuitive method that searches for the extant taxon that best corresponds to the fossil based on morphological similarity [45]. The best ML tree from the concatenated dataset was used as a rooted starting topology in the PhyloBayes analysis. Upon completion of the analysis, one fifth of the total chain length was discarded as burnin, and the posterior distribution of likelihoods, branch lengths, and rates were averaged and divergence dates summarized by the command: “readdiv”. Individual trees were visualized in FigTree version 1.4.0 [46] and consensus trees (those with a full complement of the 8 genomes) in DensiTree 2.0 [47].
Results and Discussion
DNA fragment length, alignment and matrix assembly
Based on the data obtained from VectorBase [48], the length of each chromosomal arm (in base pairs) is the following: X, 24393108; 2R, 61545105; 2L, 49364325; 3R, 53200684 and 3L, 41963435 (Table 1). In proportion to the length of each arm: 3, 19, 13, 8 and 6 genes were selected from X (Table S1), 2R (Table S2), 2L (Table S3), 3R (Table S4) and 3L (Table S5), respectively. Our sequencing data consisted of ≥364 bp-long gene fragments resulting in a total alignment of 41,124 bp based on the An. gambiae AgamP3 genome assembly. We obtained orthologous sequences of the selected genes from 8 genome assemblies representing 6 species. Support values for each clade were generated by 1000 bootstrap replicates and the reliability of nodes in the phylogenetic trees were assessed based on 70% as the cut-off value [49].
Table 1. Genome-wide distribution of genes used in the phylogenetic study.
| Chromosome arm | Length (Mb) | Number of genes | Genes per 5 Mb |
| X | 24.4 | 3 | 0.6 |
| 2R | 61.5 | 19 | 1.5 |
| 2L | 49.4 | 13 | 1.3 |
| 3R | 53.2 | 8 | 0.8 |
| 3L | 42.0 | 6 | 0.7 |
| Total | 230.5 | 49 | 1.0 |
Phylogenetic relationships among African malaria vectors
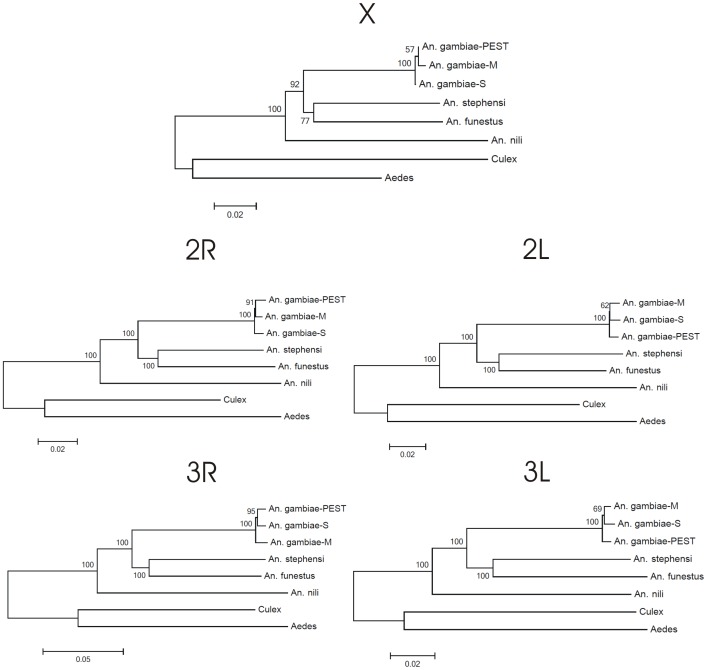
Trees obtained using ML and NJ methods from individual genes agreed with each other, with a few exceptions. The most frequent branching pattern is: ((Culex, Aedes), (An. nili, ((An. funestus, An. stephensi), An. gambiae))). The phylogenies based on three X chromosome genes placed An. nili nested within other Anopheles vectors. Only in one of these trees with a high bootstrap value (>70%), was An. nili sister to the remaining malarial vectors and consistent with the dominant branching pattern (Figure S1, Figure S2). For the X chromosome trees, bootstrap values were generally low and branching pattern variable. Yet, the AGAP000064 gene tree recovers the common branching pattern mentioned above. For the 2R arm, 11 out of 19 in NJ phylogenetic trees and 8 out of 19 in ML phylogenetic trees recovered An. nili as sister with a high bootstrap value (Figure S3, Figure S2). For 2L arm, 8 NJ and 6 ML phylogenetic trees out of 13 recovered An. nili as sister to the other species of Anopheles (Figure S4, Figure S2). Eight phylogenetic trees were constructed for the 3R arm, of which 5 NJ and 3 ML trees had a high bootstrap value placing An. nili as sister to the ingroup species (Figure S5, Figure S2). Finally, 6 phylogenetic trees were constructed based on the genes located on the 3L chromosomal arm, of which 4 NJ and 4 ML trees had a high bootstrap value supporting the sister group placement of An. nili relative to other African vectors (Figure S6, Figure S2).
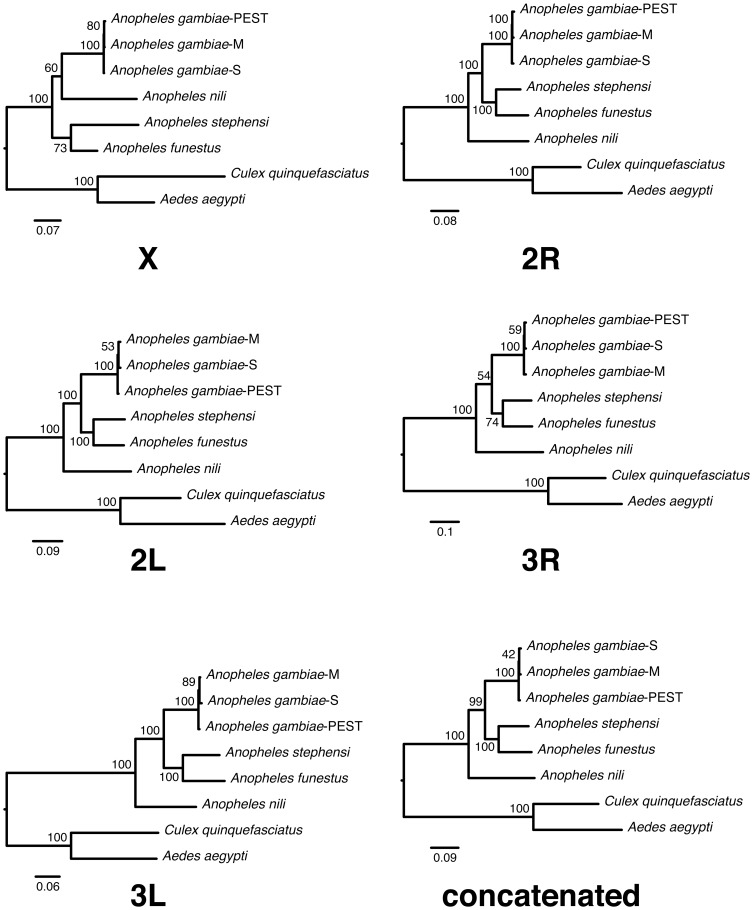
Previous studies have shown that analysis of data sets of concatenated genes is useful for resolving species phylogenetic trees [22]. This is especially the case when independent gene trees converge on a dominant topology, as is shown in our data. We created trees using sequences for all genes concatenated according to the five chromosomal arms with NJ and ML methods (Figures 2, 3). In all trees, A. aegypti and C. quinquefasciatus were clustered separately as outgroup species. The NJ and ML trees inferred with concatenated genes provided consistent support that An. nili is sister to the rest of Anopheles species, except for the X chromosome ML tree where An. nili is swapped with An. funestus + An. stephensi (Figure 3). There is also consistent support for An. funestus clustered together with An. stephensi. The Anopheles gambiae branch, which itself comprises two rapidly diverged molecular forms (M and S) as well as the PEST reference strain, is consistently recovered as sister in relation to the An. funestus + An. stephensi clade.
Figure 2. Phylogenetic NJ trees build from concatenated sequences located in 5 chromosomal arms.
Bootstrap values are shown on branches of phylogenetic trees as percentages.
Figure 3. Phylogenetic ML trees build from concatenated sequences located in 5 chromosomal arms and the partitioned ML tree for all arms combined.
Bootstrap values are shown on branches of phylogenetic trees as percentages.
We attempted to compare our phylogenetic approach to other previously used methods. The only other phylogenetic study that included An. gambiae, An. funestus, An. nili, and An. stephensi used mtDNA and nuclear rDNA [9]. A parsimony bootstrap consensus tree based on the mtDNA sequence had a very low phylogenetic signal (resulting from highly variable COI and COII sequences) and, therefore, could not resolve the phylogeny with confidence. A Bayesian phylogenetic analysis using rDNA sequences placed the An. gambiae complex sister to the remaining species of Cellia, including An. stephensi, An. funestus and An. nili [9]. However, Marshall et al. 2005 included seven Cellia species not represented in our study. Although potentially related to taxon sampling and/or a smaller dataset, the trees—even with the seven additional taxa pruned—are in contradiction with our concatenated trees, which consistently show the branching pattern: (An. nili, ((An. funestus, An. stephensi), An. gambiae)). This dominant pattern was also recovered by using the consensus-based hierarchical clustering method in DensiTree (Figure 4). Interestingly, another phylogenetic study using combined rDNA and mtDNA sequences has demonstrated a sister group position of An. dirus and An. farauti, in relation to other species of the subgenus [5]. The Asian mosquitoes An. dirus and An. farauti together with the African mosquito An. nili all belong to the series Neomyzomiya, consistently suggesting that this series is a phylogenetically separate sister clade from the remaining series: Pyretophorus (An. gambiae, An. arabiensis), Myzomyia (An. funestus, An. moucheti) and Neocellia (An. stephensi) [1].
Figure 4. A consensus cladogram of the 49 gene trees obtained with the hierarchical clustering method implemented in DensiTree.
Hypothesized evolutionary history of African malaria vectors
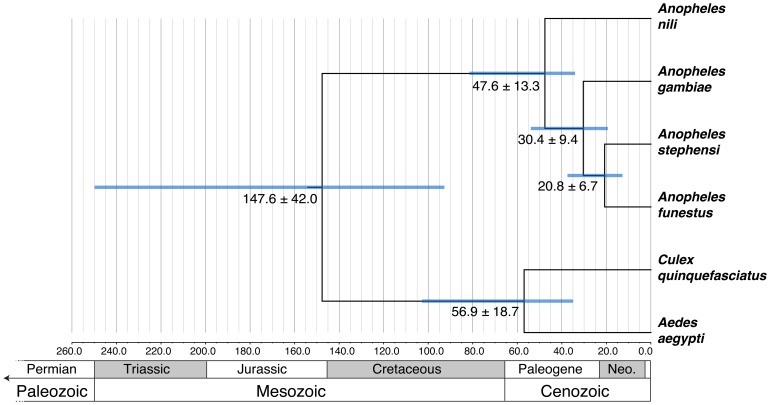
Our phylogenetic analysis of multiple genes indicates that the An. nili lineage split from the other African Anopheles species 47.6±13.3 mya (Figure 5). This date is congruent with the estimate of 43.1 mya of divergence between An. gambiae and Anopheles atroparvus in a prior study investigating divergence times in Culicidae [50]. A basal phylogenetic position of An. nili with respect to the other major malaria vectors indicates that traits relevant to increased vectorial capacity evolved in a convergent pattern. African vectors appear to have originated less frequently in the lineage to which An. nili belongs. (However, this appears to be the pattern for contemporary species as we do not yet know species extinction rates in the lineage, or differential losses and gains of vectorial capacity.) Four species had been described within the An. nili group based on morphological and genetic (isoenzymes and ribosomal DNA second internal transcribed spacer (rDNA ITS2) and D3 28S regions) differences: An. nili s.s., Anopheles somalicus, Anopheles carnevalei, and Anopheles ovengensis [51], [52]. These species, spare An. nili, show decreased vectorial capacity. A comprehensive study in Cameroon confirmed that An. nili is the only major malaria vector of the group and in contrast emphasized the exophagic behavior of An. ovengensis and An. carnevalei [53], [54]. Anopheles nili is mainly present in degraded forests, while An. ovengensis and An. carnevalei are found in the deep forests of Cameroon [18]. Anopheles ovengensis is a secondary vector, An. carnevalei is an inefficient vector, and An. somalicus is a nonvector because of its strong zoophilic and exophilic habits [54], [55]. In contrast, the clade that is sister to the An. nili lineage has produced several efficient African malaria vectors: An. funestus, An. arabiensis, An. gambiae, and, possibly, An. moucheti.
Figure 5. Time-calibrated tree and divergence dates estimated with PhyloBayes. Nodes are at mean divergence dates (in millions of years with standard errors).
Blue bars indicate a minimum/maximum 95% confidence interval estimated from the post burnin parameter distribution. Geologic time scale derived from the Geological Society of America: http://www.geosociety.org/science/timescale/.
A recent study on the genetic structure of species of the An. nili group using a combination of microsatellites, rDNA and mitochondrial DNA markers, demonstrated unexpectedly high genetic divergence among new cryptic members of the An. nili group in Cameroon [56]. Also, a comparative cytogenetic analysis of polytene chromosomes revealed significant differences in banding pattern and structure of heterochromatin between An. nili and An. ovengensis [57]. This high genetic and chromosomal divergence within the An. nili group in central Africa suggests that the lineage originated and diversified in the region corresponding to the present day equatorial forest. At 55.0 mya, the Paleocene-Eocene Thermal Maximum was underway. At this time, there was extensive forest proliferation and simultaneous rapid diversification and increase in the abundance of mammals, including primates, even-toed ungulates, and horses [58]. The divergence of the An. nili group 47.6 mya from its sister group—i.e. the remaining African malaria vectors—corresponds in timing to this event (Figure 5). In contrast, the species and population diversity in the equatorial forest is very low for the An. gambiae complex and the An. funestus group.
Regardless of the possible region of origin in Africa, each of the major African malaria vectors seems to have a sister taxon among Asian malaria mosquito species. For example, a Bayesian phylogenetic analysis using rDNA sequences indicated that An. nili is a sister taxon with Asian An. dirus and An. farauti [9]. According to our phylogenetic tree, An. funestus and An. stephensi are closely related and more recently diversified (20.8±6.7 mya) (Figures 4, 5). Another study based on morphological characteristics as well as rDNA and mtDNA sequences considered Afrotropical Funestus and Afro-Oriental Minimus groups as sister taxa [9], [59]. However, the African continent was surrounded by water during the Neogene period (from 23.03±0.05 to 2.588 mya) making mosquito migrations between Africa and Asia unlikely. In our analysis, the An. gambiae lineage diverged 30.4±9.4 mya, but the members nested within the An. gambiae diversified much more recently (Figures 4, 5). Consistent with this Asian-African sister group trend, a clade composed of An. gambiae and An. arabiensis is a sister taxon with a clade composed of the Asian Anopheles species subpictus and sundaicus in a combined phylogenetic analysis of mtDNA and rDNA [9]. Moreover, the fixed 2La chromosomal inversion typical to An. arabiensis and An. merus was also found in two species from the Middle Eastern An. subpictus complex [60]. Anopheles merus was not sampled in our phylogeny, or in the phylogeny of Marshall et al. 2005 [9], but it was a sister to other members of the An. gambiae complex in the chromosomal phylogeny [12]. Therefore, the An. gambiae complex may be closely related to other Asian malaria vectors. Overall, these data suggest that mosquitoes from divergent lineages of the subgenus Cellia (An. gambiae, An. arabiensis, An. funestus, and An. nili) experienced potentially repeated migrations between Africa and Asia spanning the Cenozoic Era (from 66 mya to the present).
Conclusion
Comparative genomic analyses of epidemiologically important traits will be more informative if performed within an accurate phylogenetic framework. Inferring the evolutionary history of African malaria vectors is crucial for establishing the association between evolutionary genomic changes with key features like the origin and loss of human blood choice, ecological and behavioral adaptations, and association with human habitats. A recent reconstruction of chromosomal phylogeny in the An. gambiae complex strongly suggests a repeated origin of increased vectorial capacity during evolution of African mosquitoes [12]. New genome assemblies for 16 species of Anopheles mosquitoes is a valuable resource for phylogenetic studies and comparative genomic analyses [61]. Although, the 16 species cluster includes vectors from different regions of the world, it lacks some of the major African malaria vectors, such as An. nili. Spanning diverse parts of the phylogenetic tree, African malaria vectors of subgenus Cellia represent a unique system for studying the evolution of vectorial capacity. Our study concludes that An. nili belongs to a basal lineage probably originating 47.6±13.3 mya. Other African malaria vectors originated more recently, and independently acquired traits related to vectorial capacity. This phylogeny will affect the interpretation of results from comparative genomics studies of malaria mosquito species. For example, genetic variation shared with An. nili might be considered ancestral polymorphism. We found strong agreement between gene trees reconstructed using multiple unlinked genes from distinct chromosomes indicating that next-generation sequence data are highly valuable for accurately inferring phylogenetic relationships among mosquito species, and providing an informative evolutionary context to understand the origins and maintenance of this pervasive and debilitating human disease.
Supporting Information
Phylogenetic NJ trees build for AGAP000064, AGAP000521, and AGAP000776 gene sequences located in the X chromosome. Bootstrap values are shown on branches of phylogenetic trees as percentages.
(TIF)
Phylogenetic ML trees build from sequences of 49 genes located in 5 chromosomal arms. Bootstrap values are shown on branches of phylogenetic trees as percentages.
(PDF)
Phylogenetic NJ trees build for AGAP001287, AGAP001700, AGAP002019, AGAP002252, AGAP002424, AGAP002790, AGAP003043, AGAP003397, AGAP003584, AGAP004028, AGAP004199, AGAP004486, AGAP001760, AGAP001762, AGAP002933, AGAP002935, AGAP013533, AGAP003327, and AGAP003328 gene sequences located in the 2R chromosomal arm. Bootstrap values are shown on branches of phylogenetic trees as percentages.
(TIF)
Phylogenetic NJ trees build for AGAP004699, AGAP005014, AGAP005279, AGAP005542, AGAP005851, AGAP006209, AGAP006531, AGAP006783, AGAP007131, AGAP007504, AGAP005778, AGAP007068, and AGAP007069 gene sequences located in the 2L chromosomal arm. Bootstrap values are shown on branches of phylogenetic trees as percentages.
(TIF)
Phylogenetic NJ trees build for AGAP007903, AGAP008652, AGAP008731, AGAP008915, AGAP009133, AGAP009512, AGAP010007, and AGAP010267 gene sequences located in the 3R chromosomal arm. Bootstrap values are shown on branches of phylogenetic trees as percentages.
(TIF)
Phylogenetic NJ trees build for AGAP010567, AGAP011099, AGAP011357, AGAP011526, AGAP011765, and AGAP012219 gene sequences located in the 3L chromosomal arm. Bootstrap values are shown on branches of phylogenetic trees as percentages.
(TIF)
Selected genes from X chromosome and length of orthologous sequences in 6 species.
(DOCX)
Selected genes from 2R chromosome and length of orthologous sequences in 6 species.
(DOCX)
Selected genes from 2L chromosome and length of orthologous sequences in 6 species.
(DOCX)
Selected genes from 3R chromosome and length of orthologous sequences in 6 species.
(DOCX)
Selected genes from 3L chromosome and length of orthologous sequences in 6 species.
(DOCX)
Funding Statement
This work was supported by National Institutes of Health (http://www.nih.gov/) grants 5R21AI079350 and 5R21AI094289 (to I. V. S). C. A.-N. was supported by a Wellcome Trust (http://www.wellcome.ac.uk/) Intermediate Fellowship in Public Health and Tropical medicine (WTO86423MA). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Harbach R (2004) The classification of genus Anopheles (Diptera: Culicidae): a working hypothesis of phylogenetic relationships. Bulletin of Entomological Research 94: 537–554. [DOI] [PubMed] [Google Scholar]
- 2. Krzywinski J, Besansky NJ (2003) Molecular systematics of Anopheles: from subgenera to subpopulations. Annu Rev Entomol 48: 111–139. [DOI] [PubMed] [Google Scholar]
- 3. Coluzzi M, Sabatini A, della Torre A, Di Deco MA, Petrarca V (2002) A polytene chromosome analysis of the Anopheles gambiae species complex. Science 298: 1415–1418. [DOI] [PubMed] [Google Scholar]
- 4. Harbach RE (2004) The classification of genus Anopheles (Diptera: Culicidae): a working hypothesis of phylogenetic relationships. Bull Entomol Res 94: 537–553. [DOI] [PubMed] [Google Scholar]
- 5. Sallum MAM, Schultz TR, Foster PG, Aronstein K, Wirtz RA, et al. (2002) Phylogeny of Anophelinae (Diptera: Culicidae) based on nuclear ribosomal and mitochondrial DNA sequences. Systematic Entomology 27: 361–382. [Google Scholar]
- 6. Favia G, Della Torre A, Bagayoko M, Lanfrancotti A, Sagnon NF, et al. (1997) Molecular identification of sympatric chromosomal forms of Anopheles gambiae and further evidence of their reproductive isolation. Insect Molecular Biology 6: 377–383. [DOI] [PubMed] [Google Scholar]
- 7. della Torre A, Tu Z, Petrarca V (2005) On the distribution and genetic differentiation of Anopheles gambiae ss molecular forms. Insect Biochemistry and Molecular Biology 35: 755–769. [DOI] [PubMed] [Google Scholar]
- 8. Coetzee M, Hunt RH, Wilkerson R, Della Torre A, Coulibaly MB, et al. (2013) Anopheles coluzzii and Anopheles amharicus, new members of the Anopheles gambiae complex. Zootaxa 3619: 246–274. [PubMed] [Google Scholar]
- 9. Marshall JC, Powell JR, Caccone A (2005) Short report: Phylogenetic relationships of the anthropophilic Plasmodium falciparum malaria vectors in Africa. Am J Trop Med Hyg 73: 749–752. [PubMed] [Google Scholar]
- 10. Hittinger CT, Johnston M, Tossberg JT, Rokas A (2010) Leveraging skewed transcript abundance by RNA-Seq to increase the genomic depth of the tree of life. Proc Natl Acad Sci U S A 107: 1476–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sharakhova MV, Antonio-Nkondjio C, Xia A, Ndo C, Awono-Ambene P, et al. (2011) Cytogenetic map for Anopheles nili: Application for population genetics and comparative physical mapping. Infect Genet Evol 11: 746–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kamali M, Xia A, Tu Z, Sharakhov IV (2012) A new chromosomal phylogeny supports the repeated origin of vectorial capacity in malaria mosquitoes of the Anopheles gambiae complex. PLoS Pathog 8(10): e1002960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fontenille D, Simard F (2004) Unravelling complexities in human malaria transmission dynamics in Africa through a comprehensive knowledge of vector populations. Comp Immunol Microbiol Infect Dis 27: 357–375. [DOI] [PubMed] [Google Scholar]
- 14.Mouchet J, Carnevale P, Coosemans M, Julvez J, Manguin S, et al. (2004) Biodiversité du paludisme dans le monde Editions. John Libbey Eurotext Montrouge, France.
- 15. Coluzzi M, Sabatini A, Petrarca V, Di Deco M (1979) Chromosomal differentiation and adaptation to human environments in the Anopheles gambiae complex. Transactions of the Royal Society of Tropical Medicine and Hygiene 73: 483–497. [DOI] [PubMed] [Google Scholar]
- 16. Hay SI, Guerra CA, Gething PW, Patil AP, Tatem AJ, et al. (2009) A world malaria map: Plasmodium falciparum endemicity in 2007. PLoS Medicine 6: e1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ayala D, Costantini C, Ose K, Kamdem GC, Antonio-Nkondjio C, et al. (2009) Habitat suitability and ecological niche profile of major malaria vectors in Cameroon. Malar J 8: 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Antonio-Nkondjio C, Ndo C, Costantini C, Awono-Ambene P, Fontenille D, et al. (2009) Distribution and larval habitat characterization of Anopheles moucheti, Anopheles nili, and other malaria vectors in river networks of southern Cameroon. Acta Trop 112: 270–276. [DOI] [PubMed] [Google Scholar]
- 19. Shalchian-Tabrizi K, Minge MA, Espelund M, Orr R, Ruden T, et al. (2008) Multigene phylogeny of choanozoa and the origin of animals. PLOS ONE 3: e2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Koepfli K-P, Deere K, Slater G, Begg C, Begg K, et al. (2008) Multigene phylogeny of the Mustelidae: resolving relationships, tempo and biogeographic history of a mammalian adaptive radiation. BMC Biology 6: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gao F, Katz LA, Song W (2013) Multigene-based analyses on evolutionary phylogeny of two controversial ciliate orders: Pleuronematida and Loxocephalida (Protista, Ciliophora, Oligohymenophorea). Molecular Phylogenetics and Evolution 68: 55–63. [DOI] [PubMed] [Google Scholar]
- 22. Rokas A, Williams BL, King N, Carroll SB (2003) Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature 425: 798–804. [DOI] [PubMed] [Google Scholar]
- 23. Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, et al. (2002) The genome sequence of the malaria mosquito Anopheles gambiae . Science 298: 129–149. [DOI] [PubMed] [Google Scholar]
- 24. Sharakhova MV, Hammond MP, Lobo NF, Krzywinski J, Unger MF, et al. (2007) Update of the Anopheles gambiae PEST genome assembly. Genome Biol 8: R5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Peery A, Sharakhova MV, Antonio-Nkondjio C, Ndo C, Weill M, et al. (2011) Improving the population genetics toolbox for the study of the African malaria vector Anopheles nili: microsatellite mapping to chromosomes. Parasites and Vectors 4: 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kamali M, Sharakhova M, Baricheva E, Karagodin D, Tu Z, et al. (2011) An integrated chromosome map of microsatellite markers and inversion breakpoints for an Asian malaria mosquito, Anopheles stephensi. . Journal of Heredity 102: 719–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Arensburger P, Megy K, Waterhouse RM, Abrudan J, Amedeo P, et al. (2010) Sequencing of Culex quinquefasciatus establishes a platform for mosquito comparative genomics. Science 330: 86–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nene V, Wortman JR, Lawson D, Haas B, Kodira C, et al. (2007) Genome sequence of Aedes aegypti, a major arbovirus vector. Science 316: 1718–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Crawford JE, Guelbeogo WM, Sanou A, Traore A, Vernick KD, et al. (2010) De novo transcriptome sequencing in Anopheles funestus using Illumina RNA-seq technology. PLoS ONE 5: e14202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Michel AP, Guelbeogo WM, Grushko O, Schemerhorn BJ, Kern M, et al. (2005) Molecular differentiation between chromosomally defined incipient species of Anopheles funestus . Insect Mol Biol 14: 375–387. [DOI] [PubMed] [Google Scholar]
- 31. Neafsey DE, Lawniczak MK, Park DJ, Redmond SN, Coulibaly MB, et al. (2010) SNP genotyping defines complex gene-flow boundaries among African malaria vector mosquitoes. Science 330: 514–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Besansky NJ, Krzywinski J, Lehmann T, Simard F, Kern M, et al. (2003) Semipermeable species boundaries between Anopheles gambiae and Anopheles arabiensis: evidence from multilocus DNA sequence variation. Proc Natl Acad Sci U S A 100: 10818–10823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. George P, Sharakhova MV, Sharakhov IV (2010) High-resolution cytogenetic map for the African malaria vector Anopheles gambiae . Insect Mol Biol 19: 675–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lawniczak MK, Emrich SJ, Holloway AK, Regier AP, Olson M, et al. (2010) Widespread divergence between incipient Anopheles gambiae species revealed by whole genome sequences. Science 330: 512–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fitch WM (2000) Homology: a personal view on some of the problems. Trends in genetics 16: 227–231. [DOI] [PubMed] [Google Scholar]
- 36. Moreno-Hagelsieb G, Latimer K (2008) Choosing BLAST options for better detection of orthologs as reciprocal best hits. Bioinformatics 24: 319–324. [DOI] [PubMed] [Google Scholar]
- 37. Kumar S, Nei M, Dudley J, Tamura K (2008) MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Briefings in bioinformatics 9: 299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular biology and evolution 4: 406–425. [DOI] [PubMed] [Google Scholar]
- 39. Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- 40. Lartillot N, Lepage T, Blanquart S (2009) PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25: 2286–2288. [DOI] [PubMed] [Google Scholar]
- 41. Thorne JL, Kishino H, Painter IS (1998) Estimating the rate of evolution of the rate of molecular evolution. Mol Biol Evol 15: 1647–1657. [DOI] [PubMed] [Google Scholar]
- 42. Zavortink TJ, Poinar GO (2000) Anopheles (Nyssorhynchus) dominicanus sp. n. (Diptera: Culicidae) from Dominican amber. Annals of the Entomological Society of America 93: 1230–1235. [Google Scholar]
- 43. Cockerell TDA (1919) The oldest mosquitoes. Nature 103: 44. [Google Scholar]
- 44. Poinar GO, Zavortink TJ, Pike T, Johnson PA (2000) Paleoculicis minutus (Diptera: Culicidae) n. gen., n. sp., from Cretaceous Canadian amber, with a summary of described fossil mosquitoes. Acta Geologica Hispanica 35: 119–130. [Google Scholar]
- 45. Sauquet H, Ho SY, Gandolfo MA, Jordan GJ, Wilf P, et al. (2012) Testing the impact of calibration on molecular divergence times using a fossil-rich group: the case of Nothofagus (Fagales). Syst Biol 61: 289–313. [DOI] [PubMed] [Google Scholar]
- 46.Rambaut A (2012) FigTreeApplication.java. Available: http://figtree.googlecode.com/svn-history/r209/trunk/src/figtree/application/FigTreeApplication.java. Accessed 2014 March 14.
- 47. Bouckaert RR (2010) DensiTree: making sense of sets of phylogenetic trees. Bioinformatics 26: 1372–1373. [DOI] [PubMed] [Google Scholar]
- 48. Megy K, Emrich SJ, Lawson D, Campbell D, Dialynas E, et al. (2012) VectorBase: improvements to a bioinformatics resource for invertebrate vector genomics. Nucleic Acids Res 40: D729–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Maes P, Matthijnssens J, Rahman M, Van Ranst M (2009) RotaC: a web-based tool for the complete genome classification of group A rotaviruses. BMC microbiology 9: 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Reidenbach KR, Cook S, Bertone MA, Harbach RE, Wiegmann BM, et al. (2009) Phylogenetic analysis and temporal diversification of mosquitoes (Diptera: Culicidae) based on nuclear genes and morphology. BMC Evol Biol 9: 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Awono-Ambene HP, Simard F, Antonio-Nkondjio C, Cohuet A, Kengne P, et al. (2006) Multilocus enzyme electrophoresis supports speciation within the Anopheles nili group of malaria vectors in Cameroon. Am J Trop Med Hyg 75: 656–658. [PubMed] [Google Scholar]
- 52. Kengne P, Awono-Ambene P, Antonio-Nkondjio C, Simard F, Fontenille D (2003) Molecular identification of the Anopheles nili group of African malaria vectors. Med Vet Entomol 17: 67–74. [DOI] [PubMed] [Google Scholar]
- 53. Antonio-Nkondjio C, Kerah CH, Simard F, Awono-Ambene P, Chouaibou M, et al. (2006) Complexity of the malaria vectorial system in Cameroon: contribution of secondary vectors to malaria transmission. J Med Entomol 43: 1215–1221. [DOI] [PubMed] [Google Scholar]
- 54. Awono-Ambene HP, Antonio-Nkondjio C, Toto JC, Ndo C, Etang J, et al. (2009) Epidemiological importance of the Anopheles nili group of malaria vectors in equatorial villages of Cameroon, Central Africa. Africa Sci Med Afr 1: 13–20. [Google Scholar]
- 55. Awono-Ambene HP, Kengne P, Simard F, Antonio-Nkondjio C, Fontenille D (2004) Description and bionomics of Anopheles (Cellia) ovengensis (Diptera: Culicidae), a new malaria vector species of the Anopheles nili group from south Cameroon. J Med Entomol 41: 561–568. [DOI] [PubMed] [Google Scholar]
- 56. Ndo C, Simard F, Kengne P, Awono-Ambene P, Morlais I, et al. (2013) Cryptic Genetic Diversity within the Anopheles nili group of Malaria Vectors in the Equatorial Forest Area of Cameroon (Central Africa). PLOS ONE 8: e58862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sharakhova MV, Peery A, Antonio-Nkondjio C, Xia A, Ndo C, et al. (2013) Cytogenetic analysis of Anopheles ovengensis revealed high structural divergence of chromosomes in the Anopheles nili group. Infection, Genetics and Evolution 16: 341–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gingerich PD (2003) Mammalian responses to climate change at the Paleocene-Eocene boundary: Polecat Bench record in the northern Bighorn Basin, Wyoming. 463–478 p.
- 59. Garros C, Harbach RE, Manguin S (2005) Morphological assessment and molecular phylogenetics of the Funestus and Minimus Groups of Anopheles (Cellia). Journal of medical entomology 42: 522–536. [DOI] [PubMed] [Google Scholar]
- 60. Ayala FJ, Coluzzi M (2005) Chromosome speciation: humans, Drosophila, and mosquitoes. Proc Natl Acad Sci U S A 102 Suppl 1: 6535–6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Neafsey DE, Christophides GK, Collins FH, Emrich SJ, Fontaine MC, et al. (2013) The evolution of the Anopheles 16 genomes project. G3 (Bethesda) 3: 1191–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic NJ trees build for AGAP000064, AGAP000521, and AGAP000776 gene sequences located in the X chromosome. Bootstrap values are shown on branches of phylogenetic trees as percentages.
(TIF)
Phylogenetic ML trees build from sequences of 49 genes located in 5 chromosomal arms. Bootstrap values are shown on branches of phylogenetic trees as percentages.
(PDF)
Phylogenetic NJ trees build for AGAP001287, AGAP001700, AGAP002019, AGAP002252, AGAP002424, AGAP002790, AGAP003043, AGAP003397, AGAP003584, AGAP004028, AGAP004199, AGAP004486, AGAP001760, AGAP001762, AGAP002933, AGAP002935, AGAP013533, AGAP003327, and AGAP003328 gene sequences located in the 2R chromosomal arm. Bootstrap values are shown on branches of phylogenetic trees as percentages.
(TIF)
Phylogenetic NJ trees build for AGAP004699, AGAP005014, AGAP005279, AGAP005542, AGAP005851, AGAP006209, AGAP006531, AGAP006783, AGAP007131, AGAP007504, AGAP005778, AGAP007068, and AGAP007069 gene sequences located in the 2L chromosomal arm. Bootstrap values are shown on branches of phylogenetic trees as percentages.
(TIF)
Phylogenetic NJ trees build for AGAP007903, AGAP008652, AGAP008731, AGAP008915, AGAP009133, AGAP009512, AGAP010007, and AGAP010267 gene sequences located in the 3R chromosomal arm. Bootstrap values are shown on branches of phylogenetic trees as percentages.
(TIF)
Phylogenetic NJ trees build for AGAP010567, AGAP011099, AGAP011357, AGAP011526, AGAP011765, and AGAP012219 gene sequences located in the 3L chromosomal arm. Bootstrap values are shown on branches of phylogenetic trees as percentages.
(TIF)
Selected genes from X chromosome and length of orthologous sequences in 6 species.
(DOCX)
Selected genes from 2R chromosome and length of orthologous sequences in 6 species.
(DOCX)
Selected genes from 2L chromosome and length of orthologous sequences in 6 species.
(DOCX)
Selected genes from 3R chromosome and length of orthologous sequences in 6 species.
(DOCX)
Selected genes from 3L chromosome and length of orthologous sequences in 6 species.
(DOCX)