Abstract
BACKGROUND
The treatment of prostate cancer has been impeded by the lack of both clinically relevant disease models and metabolic markers that track tumor progression. Hyperpolarized (HP) 13C MR spectroscopy has emerged as a new technology to investigate the metabolic shifts in prostate cancer. In this study, we investigate the glucose reprogramming using HP 13C pyruvate MR in a patient-derived prostate tissue slice culture (TSC) model.
METHODS
The steady-state metabolite concentrations in freshly excised human prostate TSCs were assessed and compared to those from snap-frozen biopsy samples. The TSCs were then applied to a perfused cell (bioreactor) platform, and the bioenergetics and the dynamic pyruvate flux of the TSCs were investigated by 31P and HP 13C MR, respectively.
RESULTS
The prostate TSCs demonstrated steady-state glycolytic and phospholipid metabolism, and bioenergetics that recapitulate features of prostate cancer in vivo. 13C spectra following injection of HP 13C pyruvate showed significantly increased pyruvate to lactate flux in malignant as compared to the benign prostate TSCs. This increased flux in the malignant prostate TSCs correlated with both increased expression of monocarboxylate transporters (MCT) and activity of lactate dehydrogenase (LDH).
CONCLUSIONS
We provide the first mechanistic evidence for HP 13C lactate as a prostate cancer biomarker in living human tissues, critical for the interpretation of in vivo studies. More broadly, the clinically relevant metabolic model system in combination with HP MR can facilitate the identification of clinically translatable biomarkers of prostate cancer presence, aggressiveness, and treatment response.
Keywords: translational biomarkers; metabolism, metabolic flux; magnetic resonance spectroscopy
INTRODUCTION
Prostate cancer is the second most common cancer occurring among Americans, with 241,740 men diagnosed in the United States in 2012 [1]. Increased screening using serum prostate-specific antigen (PSA) and extended-template transrectal ultrasound (TRUS)-guided biopsies have led to the earlier detection and staging of patients [2], but clinical management remains difficult. This difficulty is due to the biological diversity in prostate cancers, resulting in varied treatment approaches, including active surveillance and, if aggressive tumor is suspected, surgical removal, radiation and focal therapies [3]. Additionally, many prostate cancers may remain indolent without metastasizing, proving to have no effect on the quality of life of the individual [4]. Unfortunately, the natural progression of tumors as well as their response to therapy cannot be confidently predicted with current clinical or imaging biomarkers [5].
Development of biomarkers of both prostate cancer aggressiveness and response to therapy requires a robust model that accurately represents human disease. Therapeutics tested using currently available models of prostate cancer have resulted in poor translation to effective clinical treatments [6]. In many recent reviews, the lack of clinically relevant models has been cited as one of the limiting factors in the development of robust cancer drugs [7,8]. Typically, several models are used to evaluate pre-clinical activity of experimental compounds including monolayer primary and immortalized cell cultures, co-cultures of stromal and epithelial cells, three-dimensional cultures in matrices, and murine models. However, these models each have deficiencies [9]. It is nearly impossible to recapitulate complex cell–cell and cell–matrix interactions in traditional monolayer and three-dimensional cell cultures of prostate cancer [10,11]. Such interactions are critical for studying in vivo prostate cancer metabolism and the metabolic reprogramming that occurs (Fig. 1a) with tumor progression. Although transgenic mouse models, such as TRAMP [12] and PTEN knockouts [13], demonstrate several key aspects of human prostate cancer, there are a number of important differences between these animal models and human disease. First, the rodent gland is anatomically different from the human gland in its multiple free-lobe configuration, resulting in significantly less stroma about the epithelial acini and ducts than seen in the human prostate. Second, transgenic mouse models of prostate cancer, due to their genetic construction, initiate and progress at a faster rate than the human disease [14]. The transgenic components driving oncogenesis are continuously “on” throughout the epithelium of the rodent gland, a feature not seen in human prostate cancer [9]. Additionally, these models have difficulty recapitulating Gleason grades (particularly Gleason 3), since they rapidly progress to a poorly differentiated state [15]. A final significant difference is that the rodent models have a low incidence of bony metastases at the advanced stage, while human prostate cancer is marked by widely disseminated bony disease. These differences suggest that there are significant pathophysiological differences between artificial animal and cell models of prostate cancer and the human disease.
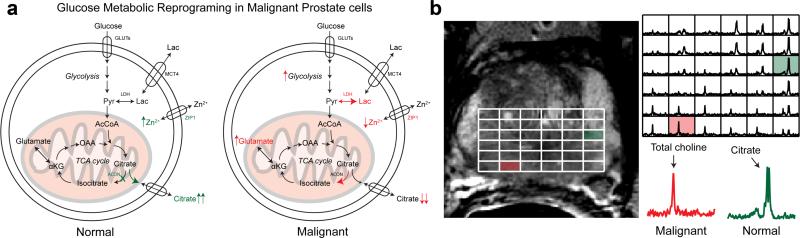
Fig. 1.
a: Change in glucose metabolism when comparing normal prostate glandular epithelial and malignant prostate cells. b: Representative in vivo T2-weighted image and corresponding 3D 1H MRSI array of a patient with Gleason 3 + 4 cancer in the right base of the prostate gland. Inlaid spectra corresponding to a normal and malignant voxel demonstrate the observed differences in metabolism for these regions. GLUTs, glucose transporters; MCT1,4, monocarboxylate transporters; ZIP1, zinc transporter, ACON, aconitase; OAA, oxaloacetate; αKG, α-ketogluterate; Pyr, pyruvate; Lac, lactate; LDH, lactate dehydrogenase.
Preliminary work in primary human tissue slice culture (TSC) suggests that the applications of prostate TSCs could provide a model that is closer to the human condition [16]. These slices are precision-cut from fresh tissues obtained during surgery and cultured on a rotary apparatus that takes the TSCs in and out of gaseous and liquid phases. This process provides optimal nutrient and gas exchange, and maintains the tissue viability. TSCs provide a preserved 3D architecture, thereby maintaining essential cell–cell interactions in the extracellular matrix of prostatic tissue [16–18]. It is possible to culture these primary tissue slices for the study of genomics, proteomics, and metabolomics prior to and following experimental therapeutics [18,19]. Moreover, TSCs of multiple types can be easily selected for analysis, including primary tumors of varying Gleason grade, metastases, and the normal prostate gland. This feature is especially important, since low grade (Gleason 3) lesions may be distinguished from high grade (Gleason 4/5) cancer, a major goal of biomarker discovery. Therefore TSCs may serve as an ideal model for the development of clinically translatable biomarkers of prostate cancer presence, aggressiveness, and response to therapy.
Hyperpolarized (HP) 13C MR is an emerging technique that allows for the rapid and non-invasive study of metabolism [20]. Typically a 13C-labeled probe is polarized using the dynamic nuclear polarization (DNP) method, and injected into a living system where the substrate is metabolized and its products imaged in real time [21,22]. Hyperpolarization provides the necessary enhancement (>10,000-fold increase in signal to noise) to observe metabolites that are crucial for understanding and monitoring cancer metabolism, but are beyond the detection threshold of traditional MR. HP probes can be used to investigate dynamic metabolic processes as well as disease phenotypes [23]. This technology lends itself to not only individual endogenous probes, but also their combination for the evaluation of multiple processes simultaneously [24]. HP 13C MR has been successfully applied to cell culture bioreactors, perfused organs, and animal models. More recently, HP 13C pyruvate studies have been performed in a Phase 1 safety trial in prostate cancer patients [25].
Although this trial demonstrated the safety of HP 13C pyruvate, and confirmed that HP 13C lactate signals can be observed in the human prostate, additional mechanistic studies are needed to interpret these in vivo results with direct correlations to tissue pathology, histochemistry, and genetic analyses. Freshly excised human tissue slices (TSCs) interfaced with HP MR provide an ideal platform to perform these mechanistic studies, both for 13C pyruvate and other clinically translatable probes. In this study, we first characterized the steady-state metabolism of prostate TSCs and compared to that of immortal cell-lines, primary prostate cancer cells, and patient-derived prostate biopsy samples. We then investigated the steady-state bioenergetics and dynamic metabolic flux in human prostate tissues using HP 13C pyruvate in an MR-compatible bioreactor. We showed, for the first time, that prostate TSCs recapitulate features of prostate cancer in vivo, and that the glucose metabolic reprogramming in human prostate cancer tissues can be rapidly visualized using clinically translatable HP 13C pyruvate.
MATERIALS AND METHODS
See Supplementary methods for full descriptions of the abbreviated methods below.
Cell Culture: PC-3, VCaP, Primary Prostate Cancer Cells
PC-3 cells, an androgen insensitive cell line derived from a prostate cancer vertebral metastasis [26] were obtained from American Type Culture Collection (ATCC, VA; obtained June 2010; authentication performed at ATCC was via Short Tandem Repeat (STR) Profiling). VCaP cells, which express the androgen receptor, were also originally isolated from a vertebral metastasis [26] and the line was obtained from ATCC (obtained June 2009; authentication performed at ATCC was via STR Profiling). PC-3 and VCaP cells were grown to 80% confluency in Dulbecco's Modified Eagle's medium (DMEM) with high glucose and supplemented analogous to the PFMR-4A previously described [27], which included 10 nM R1881. Primary prostate cancer cells were isolated from radical prostatectomy patients and cultured as previously described [28] and samples from both patients of Gleason grade 4 were used. For the labeling studies, medium was aspirated from the culture flask and replaced with DMEM containing 5 mM [3-13C] pyruvate (Isotec, Miamisburg, OH) and 1 mM unlabeled glucose for 2 hr.
Acquisition and Preparation of Human Prostate Biopsies (BYs)
This study was approved by the UCSF Institutional Review Board (IRB), and informed consent was obtained from all patients. TRUS-guided prostate biopsies were acquired from previously untreated patients (mean age = 64 ± 10 years [range: 1.6–115.9 μg/L]). Biopsies were immediately placed in individual cryovials and snap-frozen on dry ice (≤15 sec), and then stored at –80°C and analyzed within 2 weeks of harvesting, as previously described [29].
Prostate Tissue Slice (TSC) Acquisition and Culture
Fresh tissue cores (8-mm diameter) from radical prostatectomy specimens were embedded in agarose, mounted in a Krumdieck tissue slicer (Alabama Research and Development, Munford, AL) and rapidly sectioned (250–300 μm thickness) while immersed in chilled physiologic fluid. The tissue slices were then cultured in the same PFMR-4A medium as cell studies on a rotator inside of a standard cell culture incubator as previously described [17,18,27]. Following culture, the slices were either used in the bioreactor for hyperpolarized carbon MR or used for HR-MAS experiments.
High-Resolution Magic Angle Spinning (HR-MAS) NMR
1H HR-MAS spectroscopy was performed at 11.7 T (500 MHz for 1H), 1°C, and a 2,250 Hz spin rate using a Varian INOVA NMR spectrometer equipped with a 4-mm gHX nanoprobe (Varian Inc., Palo Alto, CA). Fully relaxed pulse-acquire spectra were acquired with a 2-sec presaturation delay, 2-sec acquisition time, 40,000 points, 20,000-Hz spectral width, 128 transients, and 4 steady-state pulses. T1 relaxation parameters were measured using an inversion recovery scheme and similar parameters as previously described [30,31]. Additionally, Carr-Purcell-Meiboom-Gill (CPMG), 1H–1H TOCSY, and 1H–13C HSQC spectra were acquired as previously described [28,29,32].
3DMR-Compatible Bioreactors
Tissue slices were then transferred to either an MR-compatible bioreactor or processed for pre-culture histopathological analysis. The custom designed bio-reactor is a completely contained 3D culture system with a continuous flow of 37°C media [33] (perfused with the same medium as was used for cell culture).
Hyperpolarized MR
Hyperpolarization was conducted by the DNP method using the HyperSense (Oxford Instruments, Oxford, UK) to an average polarization of 20% as previously published [34,35]. The compound polarized was a mixture of [1-13C]pyruvic acid (14.2 M) and the trityl radical OX063 (15 mM, GE Health, Menlo Park, CA). Samples were dissolved in a dissolution solution containing NaPO4 (50 mM)/ethylenediaminetetraacetic acid (EDTA) (0.3 mM) to bring the sample to 10 mM and average pH of 7.5. A solution of 1 ml of the HP pyruvate was injected into the bioreactor system with continuous flow [33].
Histopathology
Immortal cell lines (PC-3 and VCaP), primary prostate cancer cells derived from patients, human biopsies and TSCs were fixed in formalin, embedded in paraffin, sectioned at 5-mm. For each sample, adjacent sections were stained with hematoxylin and eosin (H&E) and Ki-67 nuclear antigen staining. For Ki-67, the slides were incubated for 60 min with a monoclonal mouse Ki-67 antibody, clone MIB-1 (M7240, Dako, Copenhagen), diluted 1:100 at room temperature. Secondary antibody was peroxidase labeled HRP polymer (Dako) and incubated for 60 min. The antigen localization was achieved by the DAB-chromogen (Dako). Nuclei were considered Ki67-positive if any nuclear staining was present, regardless of staining intensity.
The pathology slides were reviewed by two trained pathologists on two separate days. They determined the percentage of benign epithelium, benign stroma, the percentage and grade of prostate cancer, and the percentage of the cancer that stained positive for Ki-67. Samples containing more than 5% of other confounding pathologies, including chronic inflammation (prostatitis), high-grade prostatic intraepithelial neoplasia (HGPIN), corpora amylacea (inspissated secretions), mucin, and atrophy were excluded from the analysis. For malignant biopsies and TSCs, tissue that contained >35% cancer cells were included in this study (average % cancer in biopsies—73 ± 6% and average % cancer in TSCs—51 ± 10%, both groups 50% Gleason grade 3 and 4). A Ki-67 labeling index was defined as the percentage of positively staining cells for each cell type, as determined by counting approximately 1,000 cells of that type. Pathology readings and Ki-67 labeling index were then recorded into a database and averaged for both readers and days. For TSC post-bioreactor experiments, the two pathologists used a five-point scale (1 = excellent, 5 = poor) to quantify the quality of the pathology after perfusion as previously described [36].
Biochemical Assays
Lactate dehydrogenase (LDH) activity was assayed using standard methods. qRT-PCR for the expression of the monocarboxylate transporters (MCT1 and 4) as well as LDHA utilized primers purchased from Applied Biosystems (Foster City, CA). Expanded methods are listed in Supplementary Methods.
Data Analysis and Statistics
All NMR data was processed using a combination of ACD Lab 1D and 2D NMR processor (version 9; ACD/Labs, Toronto, Canada) and jMRUI. Peak areas or volumes were integrated and used to derive the necessary concentrations. All statistics were calculated using JMP software (SAS Corporation, Cary, NC). Significance was reported using a two-sided Student's t-test for all comparisons and a P-value < 0.05.
RESULTS AND DISCUSSION
Relaxation Parameters and Histopathology Confirm Preserved Glandular Architecturein Prostate TSCs
After the prostate was surgically removed, cores were taken from benign and malignant areas and TSCs incubated in serum-free medium. To characterize the structural state of this tissue, both spin–lattice (T1) and spin–spin (T2) relaxation constants were determined for the relevant metabolic intermediates in TSCs after 2 hr in culture. These parameters would be expected to change if the culture process impacted the tissue architecture, due to changes in the nuclear environment. For example, if the glandular structure were disrupted, the citrate T1 and T2 would change due to the loss of ductal structure. As anticipated, both T1 and T2 relaxation rates of various metabolites in the TSCs were not significantly different from those previously published for prostate tissues (Table S1) [30]. This result implies that both the ductal morphology of the benign glandular prostatic tissue as well as the malignant architecture are preserved in these TSCs. This result is further validated in formalin-fixed and paraffin-embedded TSCs (Fig. 2c), in which H&E staining demonstrates preserved cellular architecture post ex vivo culture.
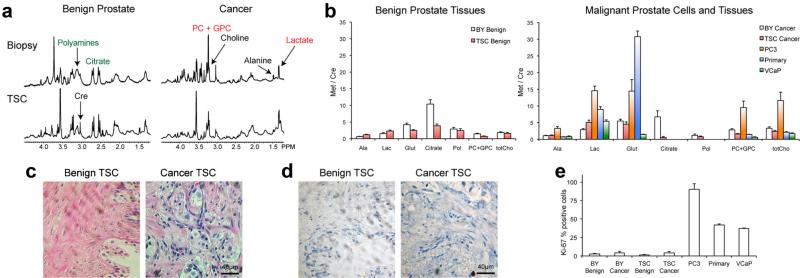
Fig. 2.
a: Representative 1H HR-MAS spectra from benign and malignant snap-frozen human biopsies and TSCs post-2 hr of culture. b: Quantified metabolite ratios for benign biopsies and TSCs (N = 16 and 14, respectively) as well as malignant BY and TSC(N = 12 and 9, respectively) compared to PC-3, primary prostate cancer cells and VCaP cells (all N = 4). c: Representative hematoxylin- and eosin- (H&E) staining and (d) Ki-67 immunohistochemistry of TSCs. e: Average percent positive Ki-67 staining of TSCs as compared to the other prostate tissues and cells. Ala, alanine; Lac, lactate; Glut, glutamate; Cre, creatine; Pol, polyamines; PC + GPC, combined phosphocholine and glycero-phosphocholine; totCho, total choline. All ratios reported as mean ± SE.
1HHR-MAS Studies Indicate That Prostate TSCs Approximate Snap-Frozen Biopsy Samples With Respect to Steady-State Citrate, Lactate, Glutamate, Choline And Polyamine Concentrations
Metabolism is dynamic and fluctuates depending on the interaction of genes, proteins, and the environment. To better understand the HP pyruvate metabolism, we first evaluated steady-state metabolite concentrations in prostate TSCs and compared them to those found in immortalized prostate cancer cells (PC-3 and VCaP cell lines), primary human prostate cancer cells, and snap-frozen, patient-derived prostate biopsy samples (BYs). The snap-frozen, patient-derived prostate biopsy samples have been shown to reflect metabolism of the in vivo prostate in patients [29,36]. High levels of citrate (derived from tricarboxylic acid (TCA) cycle metabolism) and polyamines are hallmarks of normal prostate metabolism and are typically observed in vivo (Fig. 1) [37]. Figure 2a demonstrates that metabolite concentrations seen in snap-frozen prostate biopsies and TSCs cultured for 2 hr are similar as measured by 1H HR-MAS. The cit-rate and polyamine metabolism of the normal prostate was recapitulated in TSCs from benign tissues, and is similar to that of prostate biopsies. Both citrate and polyamines were significantly higher in benign compared to malignant TSCs (Fig. 2b, P < 0.001 for citrate and 0.03 for polyamines). Although levels of citrate were lower in both malignant and benign TSCs as compared to biopsies, polyamine levels were not significantly different between TSCs and prostate biopsy samples. In contrast to the prostate TSCs and biopsies, citrate and polyamines were not detected in immortal cell lines (PC-3 and VCaP) or primary prostate cancer cells in culture.
Levels of lactate for both benign and malignant TSCs were on the same order as those in their biopsy counterparts. Also, lactate levels were lower in prostate TSCs than in the immortal PC-3 cells and primary prostate cancer cells (Fig. 2b and Fig. S1). These differences in the lactate levels highlight the differences in glycolytic metabolism between primary tissues and isolated cells in culture. Although increased aerobic glycolysis [38] is a feature of both malignant prostate TSCs and prostate cancer cells, the production of lactate in either benign or malignant human tissues is not on the same order as that observed in cells in monolayer culture. It is important to note that increased glycolysis with lactate production in prostate cancer, although significant, is difficult to visualize in conventional in vivo 1H MRSI of the prostate, since lactate signals overlap the nearly insuppressible periprostatic lipids [39].
Steady-state concentrations of glutamate in prostate TSCs were similar to those of biopsies, while dramatically different from both immortal and primary prostate cancer cells (Fig. 2b). Both PC-3 and primary prostate cancer cells demonstrated extremely elevated levels of glutamate, in contrast to the low concentration in VCaP cells. Low steady-state levels of glutamate in VCaP cells, which are derived from a vertebral metastatic lesion [26], were most likely the result of its cataplerotic use as a building block in other synthetic pathways [40]. In normal prostate metabolism, citrate is produced at high levels and secreted into the prostatic ducts. This citrate is generated via shunting of the tricarboxylic acid (TCA) cycle at aconitase due to the accumulation of high levels of mitochondrial zinc and the subsequent transport of citrate out of the cell (Fig. 1a) [41]. It has been postulated that in prostate cancer, the zinc transporters hZIP1 and hZIP3 are down regulated, resulting in a decreased intracellular concentration of zinc, increased aconitase activity and thus increased incorporation of carbons into TCA [42]. Shifting of these carbons back into TCA cycle leads to increased aketoglutarate, which is in equilibrium with glutamate. Accordingly, we observed a 31% increase in steady-state glutamate in both malignant prostate biopsies and TSCs as compared to their benign counterparts (Fig. 2b). Moreover, with increased carbon flux into the TCA cycle, the concentrations of several TCA intermediates are increased, yielding essential building blocks for cataplerotic use in rapid cell division, including nucleotides and fatty acids. Additionally, the increase in glutamate could be coupled to an increase in glutaminolysis [43], further increasing glutamate concentrations. This metabolic switch in response to zinc levels is difficult to modulate in both cell culture and animal models, though it is readily observed in TSCs, highlighting the relevance of TSCs as a realistic model system to recapitulate the in vivo human condition.
The Kennedy cycle is responsible for the metabolism of phospholipids, and changes in this pathway have been implicated in most cancers including breast, prostate, and brain [44]. To meet the demands of increased cellular proliferation, flux through the Kennedy cycle is increased, resulting in increased concentrations of phosphorylated intermediates such as phosphocholine and glycerophosphocholine. These intermediates have been studied in the context of therapies that inhibit Kennedy cycle flux and cell division. Typically, immortal cell lines demonstrate extremely high levels of Kennedy cycle intermediates, and PC-3 cells are no exception. The total choline levels of PC-3 cells, when normalized to creatine, were 3.5-fold higher than those of malignant prostate biopsies and 4.9-fold higher than those of malignant TSCs (Fig. 2b). Conversely, VCaP cells demonstrated low levels of total choline, below those of benign biopsies and TSCs. Total phosphorylated choline was also increased in malignant TSCs and biopsies relative to their benign counterparts, but both were significantly lower than that of PC-3 cells (Fig. S1). Interestingly, both VCaP and primary prostate cancer cells demonstrated lower levels of both phosphorylated and total choline as compared to malignant biopsies and TSCs. These phospholipid differences were coupled with differences in the cellular proliferation index, as measured by the nuclear antigen Ki-67 staining [32]. PC-3 cells exhibited the highest percentage of Ki-67 positive cells (90%), with VCaP and primary prostate cancer cells on the order of 40% positive. On the other hand, prostate biopsies and TSCs demonstrated significantly lower proliferation index, on the order of 5– 10% (P < 0.001, Fig. 2e).
It is important to note that differences in metabolism between TSCs and cell cultures are in part due to the variety of cell types present in tissue, consistent with the human situation, rather than a pure population of cancer cells. Additionally, this micro-environmental difference results in stromal–epithelial interactions, which are absent in cell cultures. Stromal–epithelial interactions are known to be extremely important in normal prostate biology, the development and progression of prostate cancer [45,46], and response to therapy [47]. The lack of tumor micro-environmental factors in prostate cancer cell cultures also contributes to their unnaturally rapid mitotic rate relative to the human situation [32] that also further impacts the observed metabolic profile. The purpose of cell culture and TSC metabolic profile comparisons was to show the wide range of metabolic profiles and their deviation from the in situ patient situation (snap-frozen biopsy metabolic profiles). This has important implications for metabolic biomarker development. Taken together, these data suggest that isolated cells in culture are not an appropriate surrogate for human prostate cancer, due to their high levels of phospholipid intermediates, and high proliferation rates. In contrast, TSCs provide a model that more closely approximates the in vivo human condition.
Intracellular [3-13C] Pyruvate Labelingin Culture Demonstrates Increased Fractional Enrichment of [3-13C] Lactate and Decreased Fractional Enrichmentof [4-13C] Citratein Malignant TSCs
Pyruvate is at the entry point of several important energy and biosynthesis pathways, and has a central role in cellular metabolism. In order to understand the changes in HP pyruvate metabolism in TSCs and in vivo, we evaluated the intracellular [3-13C] pyruvate labeling of prostate TSCs in 2D cultures. 2D 1H–13C HSQC NMR spectra of TSCs demonstrated visual differences in the labeling of intracellular metabolites (Fig. 3a). Malignant TSCs produced higher [3-13C] lactate than benign TSCs when normalized to creatine (0.99 ± 0.38 vs. 0.18 ± 0.04, P = 0.07), though these levels were much lower than those of primary prostate cancer cells [28]. The increased labeling also resulted in an increased fractional enrichment (FE) of intracellular lactate (Fig. 3b). The differences in the labeled lactate between TSCs and isolated cell cultures again suggest that the metabolism of isolated cells in culture departs from clinically relevant conditions, potentially as a result of lost extracellular matrix, cell–cell interactions, and changes in oxygenation [9].
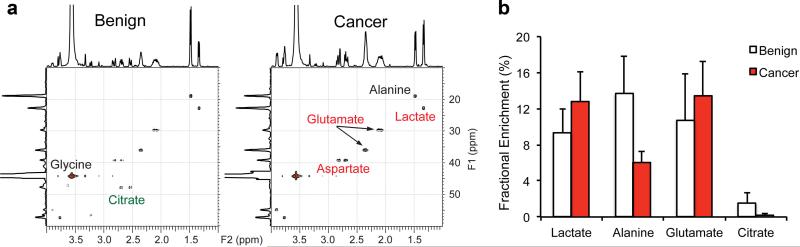
Fig. 3.
a: Representative 2D 1H–13C HSQC HR-MAS NMR spectra of benign (N = 7) and malignant (N = 4) prostate TSCs post-incubation with 4 mM [3-13C] pyruvate for 2 hr. b: Fractional Enrichment of each metabolite after 2 hr of labeling in TSCs. All ratios reported as mean ± SE.
[3-13C] alanine labeling was similar in both primary prostate cancer cells and TSCs after incubation with pyruvate (Fig. 3a). Malignant and benign TSCs produced similar concentrations of labeled alanine (0.13 ± 0.05 vs. 0.15 ± 0.03, P = 0.4), though the fractional enrichment was significantly lower in malignant TSCs (6 ± 1% vs. 14 ± 4%, P < 0.01). This difference between FE and alanine concentration implies that the metabolically active pool size of alanine in the malignant TSCs is larger. Additionally it suggests that the kinetics of alanine aminotransferase (ALT) in both the primary prostate cancer cells and TSCs are unchanged.
Of greater importance for the use of prostate TSCs in metabolic research is the recapitulation of citrate catabolism. When labeling with [3-13C] pyruvate, malignant TSCs produce higher levels of labeled [4-13C] glutamate relative to benign TSCs (0.7 ± 0.22 vs. 0.21 ± 0.06, P = 0.06) and significantly lower levels of [4-13C] citrate relative to benign TSCs (0.01 ± 0.005 vs. 0.08 ± 0.03, P = 0.03). The FE of glutamate did not change significantly, indicating that the increase of labeled glutamate scales with the pool size. In contrast, the citrate FE decreased significantly (P = 0.04, Fig. 3b). The 3.2-fold increase in 13C glutamate reflects both increases in steady-state glutamate as well as the shift of carbons from citrate back into the TCA cycle. Supporting this is the 10-fold decrease in 13C labeled citrate in malignant prostate TSCs compared to benign. The visible citrate metabolism in prostate TSCs is indicative of the preservation of the specialized metabolism of the prostate.
31P NMR Bioreactor Studies Confirm Preserved Bioenergeticsin Prostate TSCs
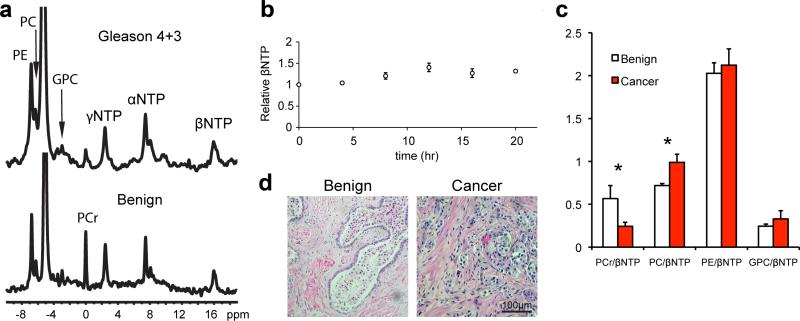
Since these initial studies demonstrated that prostate TSCs are a more realistic model of the human disease than cell cultures, we then set out to study the TSCs further in an MR-compatible bioreactor, which provides a controlled and physiological setting for real time metabolic evaluations. We first assessed the viability of TSCs in the bioreactor. TSCs were perfused in gas-equilibrated medium in order to maintain viability. Histology was compared between slices that had been perfused in the bioreactor and those that had been cultured using the rotary apparatus (Fig. 4). We also monitored the β-NTP resonance of the TSCs in the bioreactor in order to assess tissue bioenergetics. The β-NTP arises from the β phosphate group of the nucleotide triphosphates and is chemically shifted from both the nucleomono and nucleodiphosphates, allowing for the assessment of the triphopshates independently and viability assessment [48]. The viability of TSCs in the bioreactor was unchanged over the course of 24 hr (Fig. 4b). This stability ensures that the HP measurements made in the magnet are both reproducible in time and robust for comparison. Representative 31P spectra from both benign and malignant prostate TSCs in the bioreactor (Fig. 4a) demonstrate profiles similar to previously published in vivo prostate spectra [49]. In concordance with previous in vivo studies, the ratio of phosphocreatine to ATP decreased by 57% in malignant TSCs compared to benign (0.56 ± 0.15 vs. 0.24 ± 0.05, P = 0.04), and the phosphocholine to ATP ratios increased by 37% in the malignant TSCs (0.72 ± 0.02 vs. 0.99 ± 0.09, P = 0.02; Fig. 4c). Additionally, histology of TSCs following 24 hr of perfusion in the bioreactor demonstrated preservation of in vivo tissue structure (Fig. 4d), with good pathologic scores (3.3 ± 0.2) [36], reaffirming that the TSCs assessed non-invasively in the bioreactor are analogous to those in rotary culture. The preserved bioenergetics as assessed by 31P MR were indicative of not only maintained tissue viability, but also preserved in vivo metabolism in these ex vivo MR-compatible bioreactor studies.
Fig. 4.
a: 31P spectra from living malignant and benign TSCs perfused in the MR-compatible bioreactor. b: Timecourse of βNTP demonstrating preservation of ATP in the bioreactor. c: Ratios of metabolites to βNTP in benign and malignant TSCs (N = 4). d: Representative H&E staining of TSCs post-perfusion in the bioreactor. PCr, phosphocreatine; PC, phosphocholine; PE, phosphoethanolamine; GPC, glycerophosphocholine; α, β, γ-NTPs, nucleotide triphosphates. All ratios reported as mean ± SE. *P < 0.05.
HP [1-13C] Pyruvate to Lactate Flux Is Increasedin Malignant TSCs and Correlated With LDH and MCT Expression
Given the central role of pyruvate in glycolytic metabolism and the recent HP MR trial in patients, the metabolism of [1-13C] pyruvate in TSCs was investigated using the bioreactor platform. It has been postulated that increased HP lactate, the metabolic product of HP pyruvate, correlates with a number of pathologies, including both the presence and aggressiveness of prostate cancer in transgenic models [23]. Moreover, in the first phase 1 clinical trial of hyperpolarized 13C MR, HP lactate was visualized in prostate cancer in patients [25]. Accordingly, we observed increased HP lactate label generated after injection of HP pyruvate in malignant as compared to benign prostate TSCs (Fig. 5a).
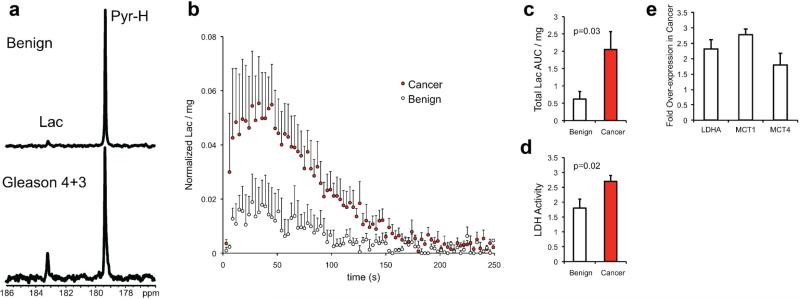
Fig. 5.
a: A single spectrum taken at 90 sec post-injection of HP [1-13C] pyruvate in the bioreactor with benign (N = 4) or malignant TSCs (N = 4). b: Average HP Lactate dynamics for TSCs. c: Total normalized lactate area under the curve (AUC) and (d) LDH activity for be nign and malignant TSCs. e: mRNA expression of lactate dehydrogenase A (LDHA), monocarboxylate transporters1 (MCT1) and 4 (MCT4). All plots are represented as mean ± SE.
Average dynamic kinetic data demonstrate a visually distinguishable difference in the total production of HP lactate over time between malignant and benign TSCs, though the time to maximum lactate peak was similar between the two (Fig. 5b). These lactate kinetics are similar to those obtained from the first-inman HP pyruvate studies in prostate cancer patients, and support the relevance of this ex vivo model to the in vivo condition. The total lactate area under the curve (AUC) was significantly elevated in malignant compared to benign TSCs (2.05 ± 0.51 vs. 0.62 ± 0.22, P = 0.03, Fig. 5c). In concordance with the HP pyruvate data, there was a 50% increase in the LDH activity in malignant relative to benign TSCs (2.7 ± 0.2 vs. 1.8 ± 0.3, P = 0.02, Fig. 5d). mRNA expression of LDHA was also elevated twofold in malignant relative to benign TSCs (Fig. 5e). LDHA is responsible for encoding the M subunit of LDH and favors the reaction of LDH toward lactate, making this enzyme a cancer target [50,51]. Since the measurement of HP lactate signal generation is in non-steady state dynamics, increased LDH activity would contribute to an increase in HP lactate signal. Moreover, increases in LDH activity have been implicated in prostate cancer, and this was demonstrated in TSCs.
Additionally, significant changes in the mRNA expression of monocarboxylate transporters MCT1 and MCT4 were observed between the benign and malignant prostate TSCs (Fig. 5e). These transporters are responsible for both the uptake of HP pyruvate (predominantly MCT1) and export of lactate (MCT4). Export of lactate out of the cells by MCT4 and corresponding acidification of the extracellular space is one method of promoting high metabolic flux through the LDH enzymatic reaction [52], and can promote cancer aggressiveness. MCT4 in particular has also been shown to be necessary for the survival of prostate cancer cells [53], and could be an important drug target for future studies. It is possible that HP MR evaluation of the dynamics of lactate export via MCT4 may provide improved biomarkers of prostate cancer presence and aggressiveness, and facilitate drug design that targets glycolysis and lactate export.
CONCLUSION
In this study we have characterized the metabolism of patient-derived prostate tissue slices in cultures, a novel model for study of prostate cancer. In contrast to immortal and primary prostate cancer cells in culture, the prostate TSCs exhibit structure, function, and metabolism that recapitulate the in vivo situation. Utilizing this model, we interrogated the glucose reprogramming in human prostate cancer tissues using HP 13C pyruvate MR, and provided mechanistic evidence for HP 13C lactate as a prostate cancer bio-marker. More broadly, this clinically relevant in vitro metabolic model system in combination with HP MR can facilitate the identification of clinically translatable biomarkers of prostate cancer presence and aggressiveness, which are important for treatment selection. Moreover, this approach addresses one of the most pressing issues in prostate cancer research, the development of treatments and multimodality companion metabolic biomarkers of response that can be easily translated into the clinic. In future studies this research will be extended to smaller cultures (i.e., single human biopsies), the analysis of the metabolism of prostate cancer samples of increasing pathologic grade, the investigation of hyperpolarized probes other than [1-13C] pyruvate, the use of other molecular imaging approaches including PET, and the use of novel therapeutics.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the funding support from National Institute of Health grants P41 EB013598 (J.K. and D.B.V.), K99 EB014328 (K.R.K.), R01 CA166766 (D.M.W.), Department of Defense W81XWH-10-1-0334 (J.K. and D.M.P.), W81XWH-12-1-0328 (R.S.), and Peer Reviewed Cancer Research Concept Award (Z.J.W.). We would also like to acknowledge assistance from Dr. Mark Albers, David Joun and Rosalie Nolley in the preliminary studies as well Drs. Robert Bok, Romelyn DelosSantos and Z. Laura Tabatabai for their help with histopathology analysis.
Grant sponsor: Department of Defense Synergistic Idea Development Award; Grant number: W81XWH-10-1-0334; Grant sponsor: Postdoctoral Fellowship; Grant number: W81XWH-12-1-0328; Grant sponsor: Peer Reviewed Cancer Research Concept Award; Grant sponsor: National Institute of Health; Grant number: P41 EB013598, K99 EB014328, R01 CA166766.
REFERENCES
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Han M, Partin AW, Piantadosi S, Epstein JI, Walsh PC. Era specific biochemical recurrence-free survival following radical prostatectomy for clinically localized prostate cancer. J Urol. 2001;166:416–419. [PubMed] [Google Scholar]
- 3.McNeal JE, Bostwick DG, Kindrachuk RA, Redwine EA, Freiha FS, Stamey TA. Patterns of progression in prostate cancer. Lancet. 1986;1:60–63. doi: 10.1016/s0140-6736(86)90715-4. [DOI] [PubMed] [Google Scholar]
- 4.Wei JT, Dunn RL, Sandler HM, McLaughlin PW, Montie JE, Litwin MS, Nyquist L, Sanda MG. Comprehensive comparison of health-related quality of life after contemporary therapies for localized prostate cancer. J Clin Oncol. 2002;20:557–566. doi: 10.1200/JCO.2002.20.2.557. [DOI] [PubMed] [Google Scholar]
- 5.Bill-Axelson A, Holmberg L, Ruutu M, Haggman M, Andersson SO, Bratell S, Spangberg A, Busch C, Nordling S, Garmo H, Palmgren J, Adami HO, Norlen BJ, Johansson JE. Radical prostatectomy versus watchful waiting in early prostate cancer. N Engl J Med. 2005;352:1977–1984. doi: 10.1056/NEJMoa043739. [DOI] [PubMed] [Google Scholar]
- 6.Ocana A, Pandiella A, Siu LL, Tannock IF. Preclinical development of molecular-targeted agents for cancer. Nat Rev Clin Oncol. 2011;8:200–209. doi: 10.1038/nrclinonc.2010.194. [DOI] [PubMed] [Google Scholar]
- 7.Hutchinson L, Kirk R. High drug attrition rates—Where are we going wrong? Nat Rev Clin Oncol. 2011;8:189–190. doi: 10.1038/nrclinonc.2011.34. [DOI] [PubMed] [Google Scholar]
- 8.Begley CG, Ellis LM. Drug development: Raise standards for preclinical cancer research. Nature. 2012;483:531–533. doi: 10.1038/483531a. [DOI] [PubMed] [Google Scholar]
- 9.Peehl DM. Primary cell cultures as models of prostate cancer development. Endocr Relat Cancer. 2005;12:19–47. doi: 10.1677/erc.1.00795. [DOI] [PubMed] [Google Scholar]
- 10.Romero D, O'Neill C, Terzic A, Contois L, Young K, Conley BA, Bergan RC, Brooks PC, Vary CP. Endoglin regulates cancer–stromal cell interactions in prostate tumors. Cancer Res. 2011;71:3482–3493. doi: 10.1158/0008-5472.CAN-10-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peehl DM. Are primary cultures realistic models of prostate cancer? J Cell Biochem. 2004;91:185–195. doi: 10.1002/jcb.10691. [DOI] [PubMed] [Google Scholar]
- 12.Gingrich JR, Greenberg NM. A transgenic mouse prostate cancer model. Toxicol Pathol. 1996;24:502–504. doi: 10.1177/019262339602400414. [DOI] [PubMed] [Google Scholar]
- 13.Wang S, Gao J, Lei Q, Rozengurt N, Pritchard C, Jiao J, Thomas GV, Li G, Roy-Burman P, Nelson PS, Liu X, Wu H. Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell. 2003;4:209–221. doi: 10.1016/s1535-6108(03)00215-0. [DOI] [PubMed] [Google Scholar]
- 14.Pienta KJ, Abate-Shen C, Agus DB, Attar RM, Chung LW, Greenberg NM, Hahn WC, Isaacs JT, Navone NM, Peehl DM, Simons JW, Solit DB, Soule HR, VanDyke TA, Weber MJ, Wu L, Vessella RL. The current state of preclinical prostate cancer animal models. Prostate. 2008;68:629–639. doi: 10.1002/pros.20726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hensley PJ, Kyprianou N. Modeling prostate cancer in mice: Limitations and opportunities. J Androl. 2012;33:133–144. doi: 10.2164/jandrol.111.013987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blauer M, Tammela TL, Ylikomi T. A novel tissue-slice culture model for non-malignant human prostate. Cell Tissue Res. 2008;332:489–498. doi: 10.1007/s00441-008-0602-z. [DOI] [PubMed] [Google Scholar]
- 17.Zhao H, Nolley R, Chen Z, Peehl DM. Tissue slice grafts: An in vivo model of human prostate androgen signaling. Am J Pathol. 2010;177:229–239. doi: 10.2353/ajpath.2010.090821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiviharju-af Hallstrom TM, Jaamaa S, Monkkonen M, Peltonen K, Andersson LC, Medema RH, Peehl DM, Laiho M. Human prostate epithelium lacks Wee1A-mediated DNA damage-induced checkpoint enforcement. Proc Natl Acad Sci U S A. 2007;104:7211–7216. doi: 10.1073/pnas.0609299104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vaira V, Fedele G, Pyne S, Fasoli E, Zadra G, Bailey D, Snyder E, Faversani A, Coggi G, Flavin R, Bosari S, Loda M. Preclinical model of organotypic culture for pharmacodynamic profiling of human tumors. Proc Natl Acad Sci U S A. 2010;107:8352–8356. doi: 10.1073/pnas.0907676107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Golman K, Olsson LE, Axelsson O, Mansson S, Karlsson M, Petersson JS. Molecular imaging using hyperpolarized 13C. Br J Radiol. 2003;76:S118–S127. doi: 10.1259/bjr/26631666. Spec. No. 2. [DOI] [PubMed] [Google Scholar]
- 21.Keshari KR, Kurhanewicz J, Bok R, Larson PEZ, Vigneron DB, Wilson DM. Hyperpolarized 13C dehydroascorbate as an endogenous redox sensor for in vivo metabolic imaging. Proc Natl Acad Sci U S A. 2011;108:18606–18611. doi: 10.1073/pnas.1106920108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gallagher FA, Kettunen MI, Day SE, Hu DE, Ardenkjaer-Larsen JH, in't Zandt R, Jensen PR, Karlsson M, Golman K, Lerche MH, Brindle KM. Magnetic resonance imaging of pH in vivo using hyperpolarized C-13-labelled bicarbonate. Nature. 2008;453:U940–U973. doi: 10.1038/nature07017. [DOI] [PubMed] [Google Scholar]
- 23.Albers MJ, Bok R, Chen AP, Cunningham CH, Zierhut ML, Zhang VY, Kohler SJ, Tropp J, Hurd RE, Yen Y-F, Nelson SJ, Vigneron DB, Kurhanewicz J. Hyperpolarized 13C lactate, pyruvate, and alanine: Noninvasive biomarkers for prostate cancer detection and grading. Cancer Res. 2008;68:8607–8615. doi: 10.1158/0008-5472.CAN-08-0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson DM, Keshari KR, Larson PE, Chen AP, Hu S, Van Criekinge M, Bok R, Nelson SJ, Macdonald JM, Vigneron DB, Kurhanewicz J. Multi-compound polarization by DNP allows simultaneous assessment of multiple enzymatic activities in vivo. J Magn Reson. 2010;205:141–147. doi: 10.1016/j.jmr.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurhanewicz J, Vigneron DB, Brindle K, Chekmenev EY, Comment A, Cunningham CH, Deberardinis RJ, Green GG, Leach MO, Rajan SS, Rizi RR, Ross BD, Warren WS, Malloy CR. Analysis of cancer metabolism by imaging hyperpolarized nuclei: Prospects for translation to clinical research. Neoplasia. 2011;13:81–97. doi: 10.1593/neo.101102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sobel RE, Sadar MD. Cell lines used in prostate cancer research: A compendium of old and new lines—Part 1. J Urol. 2005;173:342–359. doi: 10.1097/01.ju.0000141580.30910.57. [DOI] [PubMed] [Google Scholar]
- 27.Peehl DM. Culture of epithelial cells. Wiley-Liss; New York: 2002. “Human prostatic epithelial cells”. pp. 171–194. [Google Scholar]
- 28.Levin YS, Albers MJ, Butler TN, Spielman D, Peehl DM, Kurhanewicz J. Methods for metabolic evaluation of prostate cancer cells using proton and (13)C HR-MAS spectroscopy and [3-(13)C] pyruvate as a metabolic substrate. Magn Reson Med. 2009;62:1091–1098. doi: 10.1002/mrm.22120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tessem MB, Swanson MG, Keshari KR, Albers MJ, Joun D, Tabatabai ZL, Simko JP, Shinohara K, Nelson SJ, Vigneron DB, Gribbestad IS, Kurhanewicz J. Evaluation of lactate and alanine as metabolic biomarkers of prostate cancer using 1H HR-MAS spectroscopy of biopsy tissues. Magn Reson Med. 2008;60:510–516. doi: 10.1002/mrm.21694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swanson MG, Zektzer AS, Tabatabai ZL, Simko J, Jarso S, Keshari KR, Schmitt L, Carroll PR, Shinohara K, Vigneron DB, Kurhanewicz J. Quantitative analysis of prostate metabolites using 1H HR-MAS spectroscopy. Magn Reson Med. 2006;55:1257–1264. doi: 10.1002/mrm.20909. [DOI] [PubMed] [Google Scholar]
- 31.Keshari KR, Lotz JC, Kurhanewicz J, Majumdar S. Correlation of HR-MAS spectroscopy derived metabolite concentrations with collagen and proteoglycan levels and Thompson grade in the degenerative disc. Spine (PhilaPa 1976) 2005;30:2683–2688. doi: 10.1097/01.brs.0000188256.88859.9c. [DOI] [PubMed] [Google Scholar]
- 32.Keshari KR, Tsachres H, Iman R, Delos Santos L, Tabatabai ZL, Shinohara K, Vigneron DB, Kurhanewicz J. Correlation of phospholipid metabolites with prostate cancer pathologic grade, proliferative status and surgical stage—Impact of tissue environment. NMR Biomed. 2011;24:691–699. doi: 10.1002/nbm.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keshari KR, Kurhanewicz J, Jeffries RE, Wilson DM, Dewar BJ, Van Criekinge M, Zierhut M, Vigneron DB, Macdonald JM. Hyperpolarized (13)C spectroscopy and an NMR-compatible bioreactor system for the investigation of real-time cellular metabolism. Magn Reson Med. 2010;63:322–329. doi: 10.1002/mrm.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson DM, Hurd RE, Keshari K, Van Criekinge M, Chen AP, Nelson SJ, Vigneron DB, Kurhanewicz J. Generation of hyper-polarized substrates by secondary labeling with [1,1-13C] acetic anhydride. Proc Natl Acad Sci U S A. 2009;106:5503–5507. doi: 10.1073/pnas.0810190106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keshari KR, Kurhanewicz J, Macdonald JM, Wilson DM. Generating contrast in hyperpolarized (13)C MRI using ligand–receptor interactions. Analyst. 2012;137:3427–3429. doi: 10.1039/c2an35406c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santos CF, Kurhanewicz J, Tabatabai ZL, Simko JP, Keshari KR, Gbegnon A, Santos RD, Federman S, Shinohara K, Carroll PR, Haqq CM, Swanson MG. Metabolic, pathologic, and genetic analysis of prostate tissues: Quantitative evaluation of histopathologic and mRNA integrity after HR-MAS spectroscopy. NMR Biomed. 2010;23:391–398. doi: 10.1002/nbm.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swanson MG, Vigneron DB, Tabatabai ZL, Males RG, Schmitt L, Carroll PR, James JK, Hurd RE, Kurhanewicz J. Proton HRMAS spectroscopy and quantitative pathologic analysis of MRI/3D-MRSI-targeted postsurgical prostate tissues. Magn Reson Med. 2003;50:944–954. doi: 10.1002/mrm.10614. [DOI] [PubMed] [Google Scholar]
- 38.Warburg O, Wind F, Negelein E. The metabolism of tumors in the body. J Gen Physiol. 1927;8:519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kurhanewicz J, Vigneron DB. Advances in MR spectroscopy of the prostate. Magn Reson Imgaging Clin N Am. 2008;16:697–710. doi: 10.1016/j.mric.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Putluri N, Shojaie A, Vasu VT, Nalluri S, Vareed SK, Putluri V, Vivekanandan-Giri A, Byun J, Pennathur S, Sana TR, Fischer SM, Palapattu GS, Creighton CJ, Michailidis G, Sreekumar A. Metabolomic profiling reveals a role for androgen in activating amino acid metabolism and methylation in prostate cancer cells. PLoS ONE. 2011;6:e21417. doi: 10.1371/journal.pone.0021417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Franklin RB, Feng P, Milon B, Desouki MM, Singh KK, Kajdacsy-Balla A, Bagasra O, Costello LC. hZIP1 zinc uptake transporter down regulation and zinc depletion in prostate cancer. Mol Cancer. 2005;4:32. doi: 10.1186/1476-4598-4-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Desouki MM, Geradts J, Milon B, Franklin RB, Costello LC. hZip2 and hZip3 zinc transporters are down regulated in human prostate adenocarcinomatous glands. Mol Cancer. 2007;6:37. doi: 10.1186/1476-4598-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koochekpour S, Majumdar S, Azabdaftari G, Attwood K, Scioneaux R, Subramani D, Manhardt C, Lorusso GD, Willard SS, Thompson H, Shourideh M, Rezaei K, Sartor O, Mohler JL, Vessella RL. Serum glutamate levels correlate with gleason score and glutamate blockade decreases proliferation, migration, and invasion and induces apoptosis in prostate cancer cells. Clin Cancer Res. 2012;18:5888–5901. doi: 10.1158/1078-0432.CCR-12-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gillies RJ, Morse DL. In vivo magnetic resonance spectroscopy in cancer. Annu Rev Biomed Eng. 2005;7:287–326. doi: 10.1146/annurev.bioeng.7.060804.100411. [DOI] [PubMed] [Google Scholar]
- 45.Chung LW, Huang WC, Sung SY, Wu D, Odero-Marah V, Nomura T, Shigemura K, Miyagi T, Seo S, Shi C, Molitierno J, Elmore J, Anderson C, Isotani S, Edlund M, Hsieh CL, Wang R, Shehata B, Zhau HE. Stromal–epithelial interaction in prostate cancer progression. Clinical genitourinary cancer. 2006;5:162–170. doi: 10.3816/CGC.2006.n.034. [DOI] [PubMed] [Google Scholar]
- 46.Cunha GR, Ricke W, Thomson A, Marker PC, Risbridger G, Hayward SW, Wang YZ, Donjacour AA, Kurita T. Hormonal, cellular, and molecular regulation of normal and neoplastic prostatic development. J Steroid Biochem Mol Biol. 2004;92:221–236. doi: 10.1016/j.jsbmb.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 47.Efstathiou E, Logothetis CJ. A new therapy paradigm for prostate cancer founded on clinical observations. Clin Cancer Res. 2010;16:1100–1107. doi: 10.1158/1078-0432.CCR-09-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ugurbil K, Rottenberg H, Glynn P, Shulman RG. 31P nuclear magnetic resonance studies of bioenergetics and glycolysis in anaerobic Escherichia coli cells. Proc Natl Acad Sci U S A. 1978;75:2244–2248. doi: 10.1073/pnas.75.5.2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kurhanewicz J, Thomas A, Jajodia P, Weiner MW, James TL, Vigneron DB, Narayan P. 31P spectroscopy of the human prostate gland in vivo using a transrectal probe. Magn Reson Med. 1991;22:404–413. doi: 10.1002/mrm.1910220248. [DOI] [PubMed] [Google Scholar]
- 50.Kroemer G, Pouyssegur J. Tumor cell metabolism: Cancer's Achilles’ heel. Cancer Cell. 2008;13:472–482. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 51.DeBerardinis RJ, Thompson CB. Cellular metabolism and disease: What do metabolic outliers teach us? Cell. 2012;148:1132–1144. doi: 10.1016/j.cell.2012.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dimmer KS, Friedrich B, Lang F, Deitmer JW, Broer S. The low-affinity monocarboxylate transporter MCT4 is adapted to the export of lactate in highly glycolytic cells. Biochem J. 2000;350(Pt1):219–227. [PMC free article] [PubMed] [Google Scholar]
- 53.Ros S, Santos CR, Moco S, Baenke F, Kelly G, Howell M, Zamboni N, Schulze A. Functional metabolic screen identifies 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 as an important regulator of prostate cancer cell survival. Cancer Discov. 2012;2:328–343. doi: 10.1158/2159-8290.CD-11-0234. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.