Abstract
Virtually all wild mouse populations carry t haplotypes that cause embryonic lethality or semilethality, distortion of segregation ratios, suppression of crossing-over, and male sterility. The t complex of genes is located on chromosome 17, closely linked to the H-2, the major histocompatibility complex of the mouse. The t haplotypes differ from each other not only in lethal genes they carry but also in their linked H-2 haplotypes. In this study, we compared the class II H-2 genes present on 31 t chromosomes extracted from wild populations in different parts of the world. The comparison was based on the analysis of DNA fragments obtained after digestion with restriction endonucleases. The results reveal the existence of three major groups of class II alleles representing main branches on the evolutionary tree of the t chromosomes. Alleles within each group are similar if not identical, although they are borne by chromosomes that have been separated in time and space. The presence of similar alleles in Mus musculus and Mus domesticus suggests that some of them may have been separated for more than 1 million years. This must also be the minimal age of the t chromosomes but, because at least two of the three main branches appear to be related in their origin, the actual age of t chromosomes could be much greater. The observations support the proposal that H-2 genes evolve slowly.
Full text
PDF

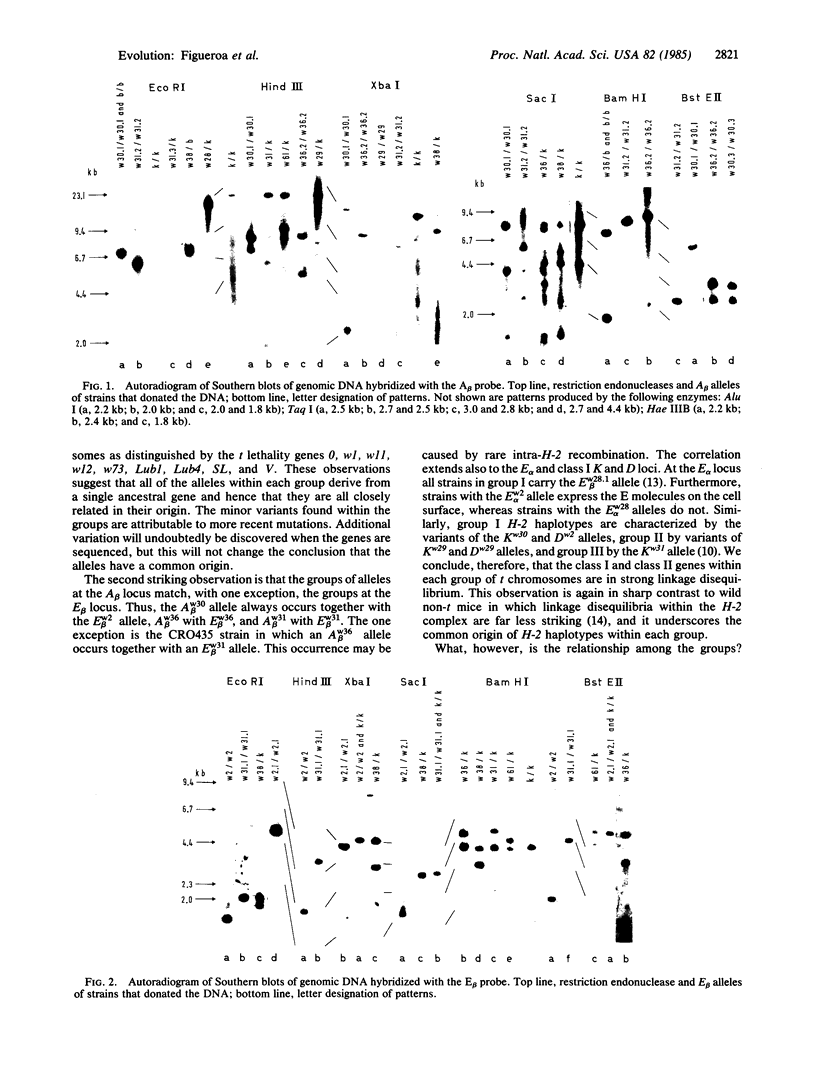
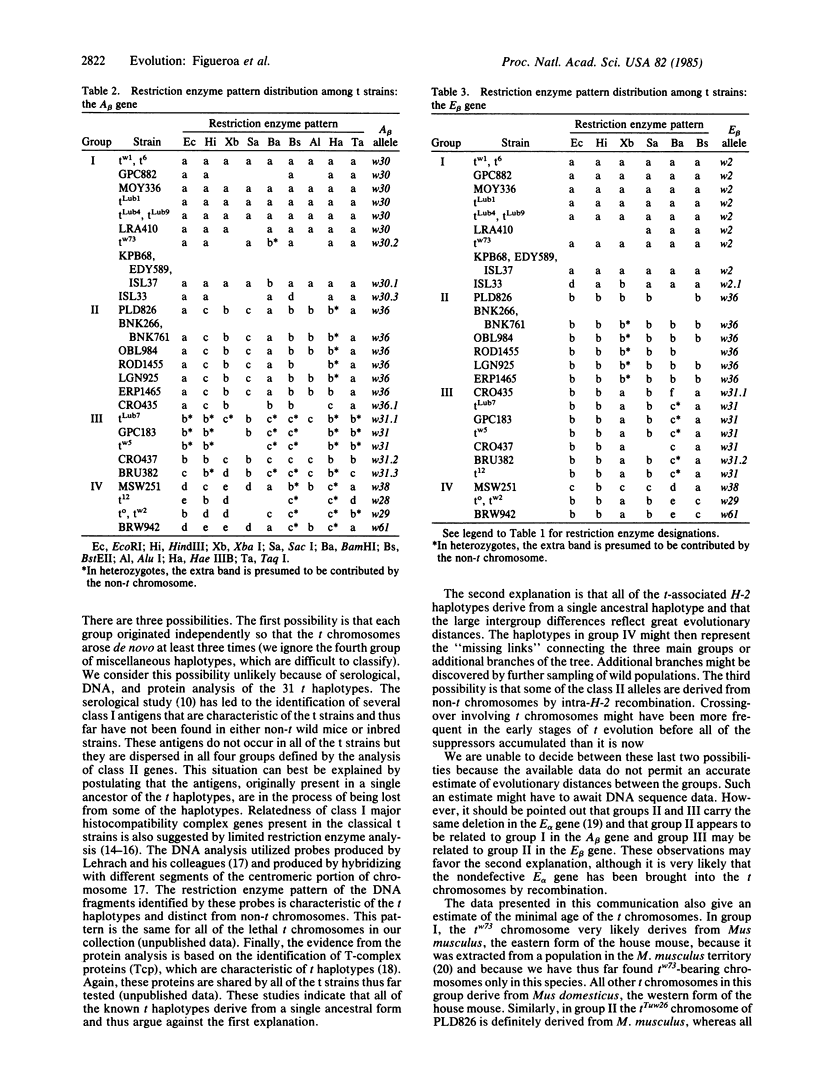

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arden B., Klein J. Biochemical comparison of major histocompatibility complex molecules from different subspecies of Mus musculus: evidence for trans-specific evolution of alleles. Proc Natl Acad Sci U S A. 1982 Apr;79(7):2342–2346. doi: 10.1073/pnas.79.7.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembic Z., Ayane M., Klein J., Steinmetz M., Benoist C. O., Mathis D. J. Inbred and wild mice carry identical deletions in their E alpha MHC genes. EMBO J. 1985 Jan;4(1):127–131. doi: 10.1002/j.1460-2075.1985.tb02326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembić Z., Singer P. A., Klein J. Eo: a history of a mutation. EMBO J. 1984 Jul;3(7):1647–1654. doi: 10.1002/j.1460-2075.1984.tb02025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn L. C., Bennett D., Cookingham J. Polymorphisms for lethal alleles in European populations of Mus musculus. J Mammal. 1973 Nov;54(4):822–830. [PubMed] [Google Scholar]
- Klein J., Figueroa F., Nagy Z. A. Genetics of the major histocompatibility complex: the final act. Annu Rev Immunol. 1983;1:119–142. doi: 10.1146/annurev.iy.01.040183.001003. [DOI] [PubMed] [Google Scholar]
- Klein J., Figueroa F. Polymorphism of the mouse H-2 loci. Immunol Rev. 1981;60:23–57. doi: 10.1111/j.1600-065x.1981.tb00361.x. [DOI] [PubMed] [Google Scholar]
- Klein J., Hammerberg C. The control of differentiation by the T complex. Immunol Rev. 1977 Jan;33:70–104. doi: 10.1111/j.1600-065x.1977.tb00363.x. [DOI] [PubMed] [Google Scholar]
- Nizetić D., Figueroa F., Klein J. Evolutionary relationships between the t and H-2 haplotypes in the house mouse. Immunogenetics. 1984;19(4):311–320. doi: 10.1007/BF00345404. [DOI] [PubMed] [Google Scholar]
- Rogers J. H., Willison K. R. A major rearrangement in the H-2 complex of mouse t haplotypes. Nature. 1983 Aug 11;304(5926):549–552. doi: 10.1038/304549a0. [DOI] [PubMed] [Google Scholar]
- Röhme D., Fox H., Herrmann B., Frischauf A. M., Edström J. E., Mains P., Silver L. M., Lehrach H. Molecular clones of the mouse t complex derived from microdissected metaphase chromosomes. Cell. 1984 Mar;36(3):783–788. doi: 10.1016/0092-8674(84)90358-1. [DOI] [PubMed] [Google Scholar]
- Shin H. S., Stavnezer J., Artzt K., Bennett D. Genetic structure and origin of t haplotypes of mice, analyzed with H-2 cDNA probes. Cell. 1982 Jul;29(3):969–976. doi: 10.1016/0092-8674(82)90460-3. [DOI] [PubMed] [Google Scholar]
- Silver L. M. Genetic organization of the mouse t complex. Cell. 1981 Dec;27(2 Pt 1):239–240. doi: 10.1016/0092-8674(81)90406-2. [DOI] [PubMed] [Google Scholar]
- Silver L. M. Genomic analysis of the H-2 complex region associated with mouse t haplotypes. Cell. 1982 Jul;29(3):961–968. doi: 10.1016/0092-8674(82)90459-7. [DOI] [PubMed] [Google Scholar]
- Silver L. M., Uman J., Danska J., Garrels J. I. A diversified set of testicular cell proteins specified by genes within the mouse t complex. Cell. 1983 Nov;35(1):35–45. doi: 10.1016/0092-8674(83)90205-2. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Steinmetz M., Minard K., Horvath S., McNicholas J., Srelinger J., Wake C., Long E., Mach B., Hood L. A molecular map of the immune response region from the major histocompatibility complex of the mouse. Nature. 1982 Nov 4;300(5887):35–42. doi: 10.1038/300035a0. [DOI] [PubMed] [Google Scholar]
- Sturm S., Figueroa F., Klein J. The relationship between t and H-2 complexes in wild mice. I. The H-2 haplotypes of 20 t-bearing strains. Genet Res. 1982 Aug;40(1):73–88. doi: 10.1017/s0016672300018929. [DOI] [PubMed] [Google Scholar]