Abstract
Background: The HLA-DQB1*06:02 allele across all ethnic groups and the rs5770917 variation between CPT1B and CHKB genes in Japanese and Koreans are common genetic susceptibility factors for narcolepsy. This comprehensive genetic study sought to assess variations in CHKB and CPT1B susceptibility genes and HLA-DQB1*06:02 allele status in Turkish patients with narcolepsy and healthy persons. Methods: CHKB/CPT1B genes were sequenced in patients with narcolepsy (n=37) and healthy persons (n=100) to detect variations. The HLA-DQB1*06:02 allele status was determined by sequence specific polymerase chain reaction. Results: The HLA-DQB1*06:02 allele was significantly more frequent in narcoleptic patients than in healthy persons (p=2×10−7) and in patients with narcolepsy and cataplexy than in those without (p=0.018). The mean of the multiple sleep latency test, sleep-onset rapid eye movement periods, and frequency of sleep paralysis significantly differed in the HLA-DQB1*06:02–positive patients. rs5770917, rs5770911, rs2269381, and rs2269382 were detected together as a haplotype in three patients and 11 healthy persons. In addition to this haplotype, the indel variation (rs144647670) was detected in the 5′ upstream region of the human CHKB gene in the patients and healthy persons carrying four variants together. Conclusion: This study identified a novel haplotype consisting of the indel variation, which had not been detected in previous studies in Japanese and Korean populations, and observed four single-nucleotide polymorphisms in CHKB/CPT1B. The study confirmed the association of the HLA-DQB1*06:02 allele with narcolepsy and cataplexy susceptibility. The findings suggest that the presence of HLA-DQB1*06:02 may be a predictor of cataplexy in narcoleptic patients and could therefore be used as an additional diagnostic marker alongside hypocretin.
Introduction
Narcolepsy is a sleep disorder characterized by excessive daytime sleepiness, cataplexy, and other dissociated manifestations of rapid eye movement (REM) sleep (Aldrich 1992). The frequency of narcolepsy varies between 0.02% and 0.16% in populations, depending on ethnicity (Mignot et al., 1998; Overeem et al., 2001). DQB1*06:02 is the major human leukocyte antigen (HLA) susceptibility allele for narcolepsy across all ethnic groups (Langdon 1984; Mignot et al., 1997; Okun et al., 2002). This haplotype is carried by almost all Japanese individuals with narcolepsy; however, the fact that it is also carried by 10–12% of the general Japanese population indicates that this haplotype alone cannot be responsible for the development of narcolepsy (Mignot et al., 2001). Hypocretin-1 is reduced or undetectable in the cerebrospinal fluid (CSF) of narcoleptic individuals because of reduction in number of hypocretin-producing neurons (Nishino et al., 2001). Consequently, it was suggested that hypocretin-producing cells are destroyed by an autoimmune process in HLA-associated narcolepsy (Lin et al., 2001). Currently, hypocretin is used as a diagnostic marker for narcolepsy. It was also suggested that HLA-typing should precede CSF hypocretin-1 measurements because hypocretin deficiency is rare in HLA-DQB1*06:02–negative patients (Knudsen et al., 2010).
In addition to narcolepsy susceptibility HLA loci, research has found that the variant (rs5770917) located between carnitine palmitoyltransferase 1B (CPT1B) (Mendelian Inheritance in Man [MIM] 601987) and choline kinase B (CHKB) (MIM 612395) is associated with susceptibility to narcolepsy in Japanese and Korean populations, but not in European populations and those of African descent (Miyagawa et al., 2008). However, the susceptibility found in the Japanese and Korean samples was not replicated in Chinese narcolepsy samples (Han et al., 2012). A protein encoded by the CPT1B gene is the rate-controlling enzyme of the long-chain fatty acid (-oxidation pathway in muscle mitochondria (McGarry and Brown, 1997). This enzyme has a role in regulating theta-oscillations during rapid eye movement (REM) sleep in mice (Tafti et al., 2003). The protein encoded by CHKB is the major enzyme in the biosynthesis of phosphatidylcholine/phosphatidylethanolamine in all animal cells and plays a key role in phospholipid biosynthesis. This enzyme has been shown to be a regulator of REM sleep and wakefulness in animal models (Platt and Riedel, 2011). Thus, both genes are candidates for REM sleep regulation (Yoshida et al., 2006; Longo et al., 2006).
Miyagawa et al. suggested that reduced hypocretin activity may be related to decreased β-oxidation, and secondary to lower expression of CPT1B in individuals with the C allele of rs5770917, predisposing them to narcolepsy (Miyagawa et al., 2009). Miyagawa et al. reported that despite the significant association of the rs5770917 genotype and CPT1B mRNA expression levels with narcolepsy, neither of these factors directly affected the serum acylcarnitine level because controls with low CPT1B mRNA expression showed normal acylcarnitine level, suggesting the presence of compensatory factors regulating carnitine fractions (Miyagawa et al., 2011).
The present study examined the variations in the coding regions, exon–intron boundaries, and upstream regions of CHKB and CPT1B susceptibility genes, in addition to the status of the HLA-DQB1*06:02 allele in Turkish patients with narcolepsy with or without cataplexy and in healthy persons. We also investigated the association between HLA-DQB1*06:02 allele status and clinical features of patients with narcolepsy.
Materials and Methods
Patients
This study involved 37 patients with narcolepsy and 100 randomly chosen volunteers with no known narcolepsy symptoms. The Ethical Committee of the Health Institute of the Dokuz Eylul University approved the study, and all participants gave their informed written consent.
Of the patients with narcolepsy, 23 (62.2%) also had cataplexy, and 14 (37.8%) did not. All of the narcoleptic patients were diagnosed according to the International Classification of Sleep Disorders, 2nd Edition (ICSD-2). Only five patients (atypical narcolepsy) were not exactly suitable for ICSD-2 criteria. Patients 3, 4, and 13 with cataplexy did not have two or more sleep-onset REM periods (SOREMPs) despite short latencies during the multiple sleep latency test (MSLT). Patient 5 without cataplexy, who had comorbid conditions and parasomnia, and patient 30 with cataplexy had mean sleep latency (MSL) of more than 8 min despite two SOREMPs during MSLT. The data, which include characteristics of patients, are listed in Tables 1 and 2.
Table 1.
Patients' Clinical-Pathologic Features and DQB1*06:02 Allele Status
| Patient no. | Sex | MSLT (min) | SOREMPs | Cataplexy | Hallucination | Sleep paralysis | Nightmare | Age of onset (yr) | BMI (kg/m2) | HLA-DQB1*06:02 |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | M | 2.4 | 5 | + | − | − | − | 20 | 29.1 | − |
| 2 | F | 0.8 | 4 | + | + | − | + | NA | 21.6 | + |
| 3a | M | 2 | 0 | + | − | + | − | 33 | 30 | + |
| 4a | F | 5.2 | 0 | + | − | − | − | 33 | 21.3 | − |
| 5a | M | 8.2 | 2 | − | − | + | + | NA | 23.3 | − |
| 6 | M | 2.9 | 4 | − | − | − | − | 23 | 26.9 | + |
| 7 | F | 0.4 | 5 | + | − | + | − | NA | 27.7 | + |
| 8 | F | 2.2 | 3 | − | − | − | + | 14 | 27.5 | + |
| 9 | F | 1.7 | 3 | + | − | + | − | 45 | 28.6 | − |
| 10 | M | 1.8 | 5 | + | − | + | + | 21 | 24.4 | + |
| 11 | M | 0.9 | 5 | + | + | + | + | 35 | 24.3 | + |
| 12 | M | 0.8 | 5 | + | + | + | − | 17 | 21.8 | + |
| 13a | F | 6.2 | 0 | + | + | + | + | 27 | 20.3 | − |
| 14 | F | 5.2 | 3 | + | − | + | − | NA | 33.7 | − |
| 15 | F | 1.6 | 4 | + | + | + | + | 37 | 19.5 | + |
| 16 | M | 4 | 4 | + | + | + | + | 13 | 23 | + |
| 17 | M | 0.1 | 2 | + | + | + | + | 36 | 26.1 | + |
| 18 | M | 2.2 | 5 | + | + | + | − | 16 | 29.6 | + |
| 19 | M | 0.7 | 5 | + | − | − | − | 10 | 25.7 | + |
| 20 | F | 5 | 4 | − | − | + | − | 15 | 20 | − |
| 21 | F | 2 | 4 | − | − | + | + | 21 | 22 | − |
| 22 | M | 2 | 3 | − | + | + | + | 25 | 21.2 | − |
| 23 | F | 2 | 4 | − | − | − | − | 27 | 35.4 | − |
| 24 | M | 4.5 | 4 | − | + | − | − | 48 | 37.6 | − |
| 25 | F | 2 | 3 | − | + | − | + | 13 | 19.5 | + |
| 26 | F | 3 | 1 | − | + | + | + | 15 | 24.6 | − |
| 27 | F | 2.2 | 3 | + | + | + | + | 37 | 32 | + |
| 28 | F | 1.7 | 4 | − | + | − | + | 16 | 27.9 | + |
| 29 | M | 6 | 4 | − | − | − | − | 30 | 22.1 | − |
| 30a | F | 9.2 | 2 | + | − | − | − | 25 | 26.5 | − |
| 31 | F | 0.5 | 2 | + | + | − | + | 23 | 22.8 | + |
| 32 | M | 1.1 | 2 | − | − | + | − | 40 | 23.9 | − |
| 33 | M | 4.4 | 3 | − | − | − | + | 22 | 29.9 | − |
| 34 | F | 2.2 | 4 | + | − | + | + | 23 | 34.5 | + |
| 35 | M | 1.3 | 3 | + | + | − | + | 31 | 24.7 | + |
| 36 | F | 1.8 | 5 | + | NA | NA | NA | 39 | 33.3 | + |
| 37 | M | NA | NA | + | + | + | + | 17 | 42.2 | + |
Patients with atypical narcolepsy.
MSLT, multiple sleep latency test; SOREMP, sleep-onset rapid eye movement period; BMI, body mass index; NA, not available; F, female; M, male.
Table 2.
Clinical Characteristics of Patients with Narcolepsy According to HLA-DQB1*06:02 Allele and Cataplexy Status
| Variable | Patients with narcolepsy (n=37) | HLA-DQB1 *06:02+ (n=21) | HLA-DQB1 *06:02− (n=16) | p (OR [95% CI]) | Narcolepsy with cataplexy (n=23) | Narcolepsy without cataplexy (n=14) | p (OR [95% CI]) |
|---|---|---|---|---|---|---|---|
| MSLT (min) | 2.78±2.17 (n=36) | 1.61±0,94 (n=20) | 4.26±2.4 (n=16) | <0.001 | 2.42±2.24 (n=22) | 3.36±2.01 (n=14) | 0.071 |
| SOREMPs (times) | 3.31±1.47 (n=36) | 3.75±1.33 (n=20) | 2.75±1.48 (n=16) | 0.036 | 3.36±1.73 (n=22) | 3.21±0.98 (n=14) | 0.413 |
| Age of onset (yr) | 25.67±10.07 (n=33) | 23.9±9.81 (n=19) | 28.07±10.26 (n=14) | 0.271 | 26.9±9.81 (n=20) | 23.77±10.56 (n=13) | 0.298 |
| BMI (kg/m2) | 26.61±5.45 (n=37) | 26.91±5.45 (n=21) | 26.22±5.61 (n=16) | 0.639 | 27.07±5.5 (n=23) | 25.84±5.48 (n=14) | 0.467 |
| Hallucination, n (%) | 21 (58.3) (n=36) | 12 (60) (n=20) | 9 (56.3) (n=16) | 0.91 (1.17 [0.25–5.45]) | 15 (68.2) (n=22) | 6 (42.9) (n=14) | 0.248 (2.86 [0.58–4.66]) |
| Sleep paralysis, n (%) | 17 (47.2) (n=36) | 13 (65) (n=20) | 4 (25) (n=16) | 0.04 (5.57 [1.07–1.89]) | 12 (54.6) (n=22) | 5 (35.7) (n=14) | 0.447 (2.16 [0.45–0.84]) |
| Nightmare, n (%) | 20 (55.6) (n=36) | 14 (70) (n=20) | 6 (37.5) (n=16) | 0.107 (3.89 [0.79–0.31]) | 12 (54.6) (n=22) | 8 (57.1) (n=14) | 0.848 (0.90 [0.19–4.27]) |
| Age of onset ≤16 yr, n (%) | 8 (24.2) (n=33) | 6 (31.6) (n=19) | 2 (14.3) (n=14) | 0.234a (2.77 [0.38–4.79]) | 3 (15) (n=20) | 5 (38.5) (n=13) | 0.132a (0.28 [0.04–1.89]) |
Because of unavailable data, final sample sizes in each group are listed in parentheses. Mean±standard deviation value significance between groups was calculated by using Mann-Whitney U (one-tailed) test. Other p values were calculated by using chi-square test with Yates correction and Fisher exact test (one-tailed).
Fisher exact test.
OR, odds ratio; CI, confidence interval; MSLT, multiple sleep latency test; SOREMP, sleep-onset REM period; BMI, body mass index.
Identification of sequence variants in the CHKB and CPT1B genes
Genomic DNA was isolated from peripheral blood samples of patients and controls by using the NucleoSpin Blood L kit, in accordance with the manufacturer's instructions (Machenery-Nagel, Germany). Coding regions, exon–intron boundaries, and the upstream regions of the CPT1B and CHKB genes were amplified by using polymerase chain reaction (PCR). All of the primers used for amplification and sequencing were designed by using Oligo Primer Analysis Software (Molecular Biology Insights Inc., Cascade, CO). Primer sequences are given in Supplementary Table S1 (Supplementary Data are available online at www.liebertpub.com/gtmb). Amplified PCR products were sequenced by using an automated sequencer (ABI Genetic Analyzer 3730xi; Applied Biosystems, Life Technologies, Carlsbad, CA).
HLA-DQB1*06:02 allele analysis
The presence or absence of the HLA-DQB1*06:02 allele was determined by PCR using sequence-specific primers 602F (5′-CCCGCAGAGGATTTCGTGTT-3′) and 602R (5′-AACTCCGCCCGGGTCCC-3′) [31]. HGH1F (5′-CAGTGCCTTCCCAACCATTCCCTTA-3′) and HGH1R (5′-ATCCACTCACGGATTTCTGTTGTGTTTC-3′) primers were used as an internal control, which covers the 439–base pair region of the human growth hormone 1 gene. The amplified samples were tested by electrophoresis in a 2% agarose gel to determine the presence or absence of the allele (Fig. 1).
FIG. 1.
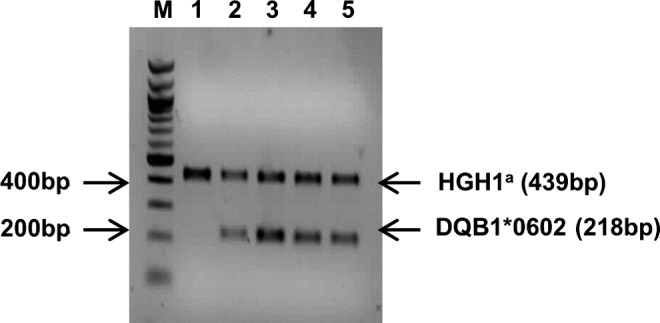
Agarose gel electrophoresis showing the presence or absence of HLA-DQB1*06:02 allele. Lanes 2, 3, 4, and 5 represent HLA-DQB1 *06:02 allele positivity and lane 1 represents HLA-DQB1*06:02 allele negativity. M: 100–base pair DNA ladder. aInternal control of polymerase chain reaction. HGH1, human growth hormone 1.
Statistical analysis
The chi-square with Yates correction test and Fisher exact (one-tailed) test were used to assess the statistical difference of HLA-DQB1*06:02 allele positivity and the single-nucleotide polymorphism (SNP) allele frequencies between groups. A statistically significant difference between the mean values of the groups was calculated by using the Mann-Whitney U (one-tailed) test. Statistical analyses were performed by using SPSS software, version 15.0 (SPSS Inc., Chicago, IL), and Statcalc, version 6 (AcaStat Software, Leesburg, VA). A p value of 0.05 was considered the limit for statistical significance. Odds ratios (ORs) were calculated with 95% confidence intervals (CIs).
Results
HLA-DQB1*06:02 allele status and the clinical features of patients with narcolepsy
Table 1 reports the clinical and pathological features and HLA DQB1*06:02 allele status of patients. Analyses of the HLA-DQB1*06:02 allele status revealed that the rates of those found positive for the HLA-DQB1*06:02 allele were 12% and 56.8% for the control group and narcoleptic patients with and without cataplexy, respectively. The HLA-DQB1*06:02 was significantly more frequent in both types of narcoleptic patients than in controls (OR, 9.63; 95% CI, 3.65–25.93; p=2×10−7) (Table 3). Among the narcoleptic patients, the HLA-DQB1*06:02 allele was significantly more frequent in patients with cataplexy than in those without (OR, 7.08; 95% CI, 1.31–42.57; p=0.018) (Table 3).
Table 3.
HLA-DQB1*06:02 Allele Positivity in Patients with Narcolepsy With and Without Cataplexy and in Controls
| Variable | HLA-DQB1*06:02 + (%) | HLA-DQB1*06:02 − (%) | p | OR (95% CI) |
|---|---|---|---|---|
| Narcolepsy | 21 (56.8) | 16 (43.2) | 2×10−7 | 9.63 (3.65–25.93) |
| Control | 12 (12) | 88 (88) | ||
| Narcolepsy with cataplexy | 17 (73.9) | 6 (26.1) | 0.018 | 7.08 (1.31–42.57) |
| Narcolepsy without cataplexy | 4 (28.6) | 10 (71.4) |
p was calculated by using chi-square test with Yates correction.
Table 2 compares MSLT; SOREMPs; age of onset; body mass index; and presence of hallucinations, sleep paralysis, and nightmares in terms of HLA-DQB1*06:02 allele and cataplexy status.
The sequence variants in CHKB and genes
To determine the variations in coding exons, exon–intron boundaries and the 5′ untranslated regions of CPT1B and CHKB were sequenced in all narcoleptic patients and in the parents of some of these patients. We did not detect any nonconservative mutations. Various common SNPs were found, especially in intronic regions in most patients. However, we focused on previously known associated variations to detect the haplotype of patients. We detected the haplotype that consisted of rs5770911 (C>T), rs2269381 (C>G), rs2269382 (G>A), and rs5770917 (T>C) SNPs in 3 of 37 (8.1%) patients and 11 of 100 (11%) controls. Two patients (patients 3 and 19) and 11 controls had the heterozygous genotype for these SNPs (rs5770917; T/C, rs5770911; C/T, rs2269381; C/G, rs2269382; G/A). Only one patient (patient 13) had the homozygote genotype (rs5770917; C/C, rs5770911; T/T, rs2269381; G/G, rs2269382; A/A) (Table 1). The presence of variant haplotype and allele frequencies of rs5770917 did not significantly differ between patients and controls (p=0.62) (Table 4).
Table 4.
Allele Frequencies in rs5770917 Single-Nucleotide Polymorphism Between Patients with Narcolepsy and Controls
| Allele (%) | ||||
|---|---|---|---|---|
| Group | T | C | p | OR (95% CI) |
| Narcolepsy | 70 (94.6) | 4 (5.4) | 0.62 | 1.02 (0.29–3.94) |
| Control | 189 (94.5) | 11 (5.5) | ||
Fisher exact test (one-tailed test) was used to estimate allele frequencies.
An indel variation in the 5′ upstream of the CHKB gene
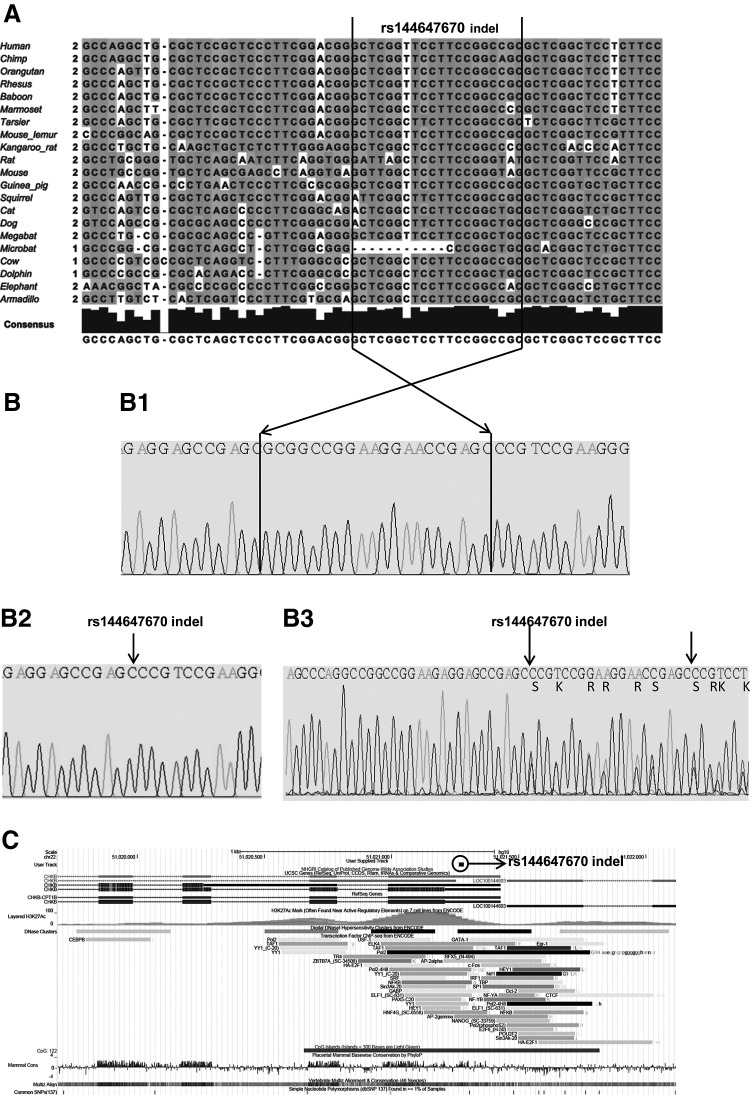
Interestingly, we detected the indel variation (rs144647670) (GRCh37.p5Chr 22 NC_000022.10: g.51021265_51021284del20: c.-76_-57del) in the 5′ upstream regulatory region of the human CHKB1 gene in the narcoleptic patients and controls carrying the rs5770917 (T>C), rs5770911 (C>T), rs2269381 (C>G), and rs2269382 (G>A) variants together. We detected the whole haplotype, including the indel variation, in three narcoleptic patients in our sample (8.1%) and 11 controls (11%). Two patients (patients 3 and 19) and 11 controls were heterozygous, while one patient (patient 13) was homozygous for the four SNPs mentioned above and the indel variation. According to evolutionary conservation analysis, the deleted sequence (-/5′ GCTCGGTTCCTTCCGGCCGC 3′) in the 5′ region upstream of the human CHKB gene has been nearly conserved among 21 mammalian species; however, this has not occurred in the microbat (Myotis lucifugus) (Fig. 2). Figure 2 shows the genomic localization of the regulatory region of the CHKB gene and the track illustrating the 20–base pair deletion (rs144647670) according to the February 2009 human reference sequence (GRCh37/hg19).
FIG. 2.
rs144647670 deletion sequence. (A) Multiple sequence alignment of the region including the deletion (black box) in 5′ upstream of the human CHKB across 21 species. The figure is generated using GRCh37/hg19 [The European Bioinformatics Institute (EMBL-EBI)], a ClustalW method, and Jalview Java alignment editor version 2. (B) The sequence electropherogram of patients carrying the wild type (B1), homozygote (B2), and the heterozygote (B3) deletion region. (C) The genomic localization of the regulatory region of the CHKB gene and track illustration of 20–base pair deletion (rs144647670) according to GRCh37/hg19 (University of California Santa Cruz).
Discussion
To our knowledge, this is the first comprehensive study in which the HLA-DQB1*06:02 allele and two narcolepsy susceptibility genes, CPT1B and CHKB, were analyzed in both patients with narcolepsy and healthy persons of Turkish origin. Although the HLA-DQB1*06:02 allele is not very penetrant, it is an important genetic factor for the susceptibility for narcolepsy (Matsuki et al., 1992; Mignot et al., 2001). Previous studies have provided evidence that this is a disease-modifying gene. It could be used for therapeutic options and as a supplementary diagnostic marker in addition to hypocretin (Mignot et al., 2003; Andlauer et al., 2012). For example, a previous study showed that severity of narcolepsy symptoms varies in a linear manner according to HLA- DQB1*06:02 allele status (Watson et al., 2010). To our knowledge, only two previous studies have investigated HLA-DQB1*06:02 allele status in Turkish patients with narcolepsy (Okun et al., 2002; Pelin et al., 2002). Because the study carried out by Okun et al. covers heterogenous populations, it could not obtain suggestive information regarding the Turkish population. The study carried out by Pelin et al. covered a very limited number of Turkish patients (eight patients).
In our results, HLA-DQB1*06:02 allele status was significantly more frequent in patients with narcolepsy than in controls. Previously published studies have shown that rates for HLA-DQB1*06:02 vary between 12% and 38% of control populations, according to ethnic group (Mignot et al., 1994; Lin et al., 1997). Our data showed that 12% of the controls were positive. In addition, the HLA-DQB1*06:02 allele was significantly more frequent in patients with narcolepsy who had cataplexy (73.9%; n=17 of 23) than in patients without (28.6%; n=4 of 14). The rates for both types of narcolepsy patient were lower than in other populations. In the literature, rates for the HLA-DQB1*06:02 allele are 85–100% for patients with narcolepsy with cataplexy, and 41% of for those without (Mignot et al., 1997; Mignot et al., 2001). When five patients with atypical narcolepsy were excluded from our patient groups, the frequency of the HLA-DQB1*06:02 allele positivity increases in patients with narcolepsy who had and did not have cataplexy, as in other ethnic populations in previously reported studies. Despite the low frequency of HLA-DQB1*06:02 allele positivity, the present study further confirmed the association of the HLA-DQB1*06:02 allele with narcolepsy and cataplexy susceptibility in the Turkish patient group.
Interestingly, three patients who were HLA-DQB1*06:02 allele positive and without cataplexy showed symptoms before 16 years of age. This finding, which reflects previous studies, indicates that HLA-DQB1*06:02 allele status is a potential predictor of cataplexy in narcoleptic patients and could be used as a diagnostic marker in addition to hypocretin. We compared the HLA-DQB1*06:02 allele and cataplexy status with the figures of MSLT; number of SOREMPs ; age of onset; body mass index; and presence of hallucinations, sleep paralysis, and nightmares in patients with narcolepsy (Table 2). The mean MSLT was significantly shorter in HLA-DQB1*06:02–positive patients than in HLA-DQB1*06:02–negative patients (Tables 1 and 2). Similarly, the mean SOREMP was also significantly less in HLA-DQB1*06:02–positive patients than HLA-DQB1*06:02–negative patients (Tables 1 and 2). However, some previous studies have not shown significant differences in MSLT and SOREMPs among the HLA-DQB1*06:02–positive and –negative patients (Woo et al., 2012). In general, however, our results are in line with previous findings (Mignot et al., 1999; Hong et al., 2006; Watson et al., 2010; Andlauer et al., 2012; Mignot et al., 2006).
Although some previous studies have shown no change in the frequency of sleep paralysis according to the HLA-DQB1*06:02 and cataplexy status, our study found significantly higher frequencies in HLA-DQB1*06:02–positive patients (Tables 1 and 2). Statistical analyses displayed no significant correlation when the HLA-DQB1*06:02 allele and cataplexy status were compared with the age of onset, body mass index, or hallucination parameters.
Previously, Miyagava et al., reported that SNP rs5770917, located between the CPT1B/ CHKB genes, was associated with susceptibility to narcolepsy with cataplexy in Japanese and Korean populations but not in European populations or those of African descent. In addition, significantly lower levels of both CPT1B/CHKB messenger RNA expression were observed in heterozygotes with the risk allele, as compared with homozygotes with the major allele (Miyagawa et al., 2008). These researchers suggested that variations in the CPT1B/CHKB susceptibility genes are thought to play a role in sleep regulation and were a common genetic susceptibility factor to narcolepsy in the East Asian population. However, this finding was not replicated in Chinese narcolepsy samples (Han et al., 2012), indicating that the association of SNP harboring between CPT1B and CHKB with narcolepsy susceptibility is still speculative. They also reported that rs5770917 has strong linkage disequilibrium with rs5770911, rs2269381, rs2269382 SNPs according to the HapMap data for individuals of Japanese ancestry (Miyagawa et al., 2008).
We observed the haplotype consisting of rs5770917, rs5770911, rs2269381, and rs2269382 together in three narcoleptic patients in our sample (8.1%) and 11 controls (11%). There were no significant differences between narcoleptic patients and controls or HLA-DQB1*06:02–positive and –negative patients. Two of the patients with narcolepsy with the above-mentioned haplotype were positive for HLA-DQB1*06:02, but, interestingly, none of the 11 controls carrying the same haplotype were positive for HLA-DQB1*06:02. Although the number of patients is small, these findings provide some evidence that while neither the haplotype nor HLA- DQB1*06:02 allele alone is responsible for the development of narcolepsy, the combined presence of these two genetic factors may contribute to narcolepsy development. The interaction between rs5770917 and HLA-DQB1*06:02 was investigated in essential hypersomnias, and no interaction was found. Because of the limited number of patients in this study, it was not possible to evaluate whether there is a significant association between these polymorphisms and HLA-DQB1*06:02 positivity in patients with narcolepsy and control groups.
As another crucial point, we also detected the rs144647670, 20–base pair deletion variations in three narcoleptic patients and in 11 controls in addition to the above-mentioned four SNPs. Although this indel variation was submitted to 1000 Genomes Project, it is not validated and there are no data regarding its frequency. This variation is within the regulatory region in the 5′ upstream region of the CHKB gene (Fig. 2) and thus may lead to reduced gene expression or transcription. Transcriptional regulation is controlled by a cis-acting regulatory sequence in 5′ upstream sequence and posttranscriptional regulation is controlled by short-sequence elements in the 5′ untranslated region of messenger RNA (Morris and Geballe, 2000).
Figure 2 shows the multiple sequence alignment of the region, including the 20–base pair deletion in the 5′ upstream sequence of the human CHKB across 21 species. It shows that the deletion sequence is almost conserved between other species, even in the megabat, but not in the microbat. It has been suggested that megabats and microbats do not share a common flying ancestry, and microbats are paraphyletic (Teeling et al., 2000; Springer et al., 2001; Teeling et al., 2005). The nonconserved region may be responsible for controlling the evolutionarily crucial function or may be relevant with torpor in microbats, which is a type of deep sleep and an evolutionary extension in which an animal lowers its metabolic rate dramatically. Interestingly, Edy et al. showed that the brown bat (microbat) contains several amino acid substitutions absent in other mammalian CPT1B sequences, which may be advantageous for enzyme function at very low temperatures during torpor (Eddy et al., 2006).
Additionally, it is known that CPT1B has a role in regulating theta-oscillations during REM sleep in mice (Tafti et al, 2003); CHKB has a key role in phospholipids biosynthesis and is a regulator of sleep and wakefulness (Platt and Riedel, 2011). Both genes are linked to lipid metabolism and are candidates for REM sleep regulation. These associations have been supported by animal studies in mice with systemic carnitine deficiency (Tafti et al., 2003), those deficient in short-chain acetyl coenzyme A dehydrogenase (Yoshida et al., 2006), and heterozygous CPT1b knockout (CPT1b (+/-) mice (He et al., 2012).
In addition, Eddy et al. showed that selected mitochondrial enzyme activities are elevated and that some genes and proteins are induced during torpor in the brown adipose tissue of the microbat (Eddy et al., 2006). For example, CPT1B was critically important to hibernator energy metabolism. Compared with levels in euthermic brown fat, there was a strong increase in CPT1B protein content in brown fat during hibernation. The primary function of brown adipose tissue is nonshivering thermogenesis. The elevated CPT1B protein content in brown fat in the torpid versus aroused state is consistent with the demand for high rates of lipid oxidation in brown adipose tissue to support thermo genesis, both for low-level heating during torpor and high-level heating during arousal (Eddy et al., 2006).
Interestingly, studies in hypocretin-null mice by Sellayah et al. suggested that obesity associated with hypocretin depletion is linked to brown-fat hypoactivity (Sellayah et al., 2011). They showed that the failure of thermogenesis in hypocretin-null mice was due to the inability of preadipocytes to differentiate. Thus, they traced the integral role of hypocretin in adaptive thermogenesis and body weight regulation via its effects on brown adipose tissue differentiation and function.
Mochizuki et al. revealed the role of hypocretin in thermoregulation. They showed that body temperature is elevated during sleep in hypocretin knockout mice (Mochizuki et al., 2006). Hypocretin deficiency is also associated with obesity despite reduced food intake (Nishino et al., 2000). Loss of hypocretin neurons from the perifornical lateral hypothalamic area not only leads to the disordered sleep patterns of narcolepsy but is also often accompanied by defective energy and metabolic homeostasis, including a high risk of obesity (Kok et al., 2003; Hara et al., 2005), and the potential for altered thermoregulation (Plazzi et al., 2011). Present and previous studies suggest that the factors of hypocretin signaling, brown adipose tissue regulation, and energy homeostasis may be related to the HLA-DQB1*06:02 allele and CPT1B/CHKB haplotype in narcolepsy pathogenesis.
Supplementary Material
Acknowledgments
This work was supported by grants from Dokuz Eylül University Fund (2008.KB.SAG.013 and 2011.KB.SAG.025).
Author Disclosure Statement
No competing financial interests exist.
References
- Aldrich MS. (1992) Narcolepsy. Neurology 42:34–43 [PubMed] [Google Scholar]
- Andlauer O, Moore H, 4th, Hong SC, et al. (2012) Predictors of hypocretin (orexin) deficiency in narcolepsy without cataplexy. Sleep 35:1247–1255F [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SF, Morin P, Jr, Storey KB. (2006) Differential expression of selected mitochondrial genes in hibernating little brown bats, Myotis lucifugus. J Exp Zool A Comp Exp Biol 305:620–630 [DOI] [PubMed] [Google Scholar]
- Han F, Lin L, Li J, et al. (2012) TCRA, P2RY11, and CPT1B/CHKB associations in Chinese narcolepsy. Sleep Med 13:269–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara J, Yanagisawa M, Sakurai T. (2005) Difference in obesity phenotype between orexin-nockout mice and orexin neuron-deficient mice with same genetic background and environmental conditions. Neurosci Lett 380:239–242 [DOI] [PubMed] [Google Scholar]
- He L, Kim T, Long Q, Liu J, et al (2012) Carnitine palmitoyltransferase-1b deficiency aggravates pressure overload-induced cardiac hypertrophy caused by lipotoxicity. Circulation 126:1705–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SC, Lin L, Jeong JH, et al. (2006) A study of the diagnostic utility of HLA typing, CSF hypocretin-1 measurements, and MSLT testing for the diagnosis of narcolepsy in 163 Korean patients with unexplained excessive daytime sleepiness. Sleep 29:1429–1438 [DOI] [PubMed] [Google Scholar]
- Knudsen S, Jennum PJ, Alving J, et al. (2010) Validation of the ICSD-2 criteria for CSF hypocretin-1 measurements in the diagnosis of narcolepsy in the Danish population. Sleep 33:169–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok SW, Overeem S, Visscher TL, et al. (2003) Hypocretin deficiency in narcoleptic humans is associated with abdominal obesity. Obes Res 11:1147–1154 [DOI] [PubMed] [Google Scholar]
- Langdon N, Welsh KI, van Dam M, et al. (1984) Genetic markers in narcolepsy. Lancet 2:1178–1180 [DOI] [PubMed] [Google Scholar]
- Lin L, Hungs M, Mignot E. (2001) Narcolepsy and the HLA region. J Neuroimmunol 117:9–20 [DOI] [PubMed] [Google Scholar]
- Lin L, Jin L, Kimura A, et al. (1997) DQ microsatellite association studies in three ethnic groups. Tissue Antigens 50:507–520 [DOI] [PubMed] [Google Scholar]
- Longo N, Amat di San Filippo C, Pasquali M. (2006) Disorders of carnitine transport and the carnitine cycle. Am J Med Genet C Semin Med Genet 142C:77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuki K, Grumet FC, Lin X, et al. (1992) DQ rather than DR gene marks susceptibility to narcolepsy. Lancet 339:1052. [DOI] [PubMed] [Google Scholar]
- McGarry JD, Brown NF. (1997) The mitochondrial carnitine palmitoyltransferase system. From concept to molecular analysis. Eur J Biochem 244:1–14 [DOI] [PubMed] [Google Scholar]
- Mignot E, Chen W, Black J. (2003) On the value of measuring CSF hypocretin-1 in diagnosing narcolepsy. Sleep 26:646–649 [PubMed] [Google Scholar]
- Mignot E, Hayduk R, Black J, et al. (1997) HLA DQB1*0602 is associated with cataplexy in 509 narcoleptic patients. Sleep 20:1012–1020 [PubMed] [Google Scholar]
- Mignot E, Lin X, Arrigoni J, et al. (1994) DQB1*0602 and DQA1*0102 (DQ1) are better markers than DR2 for narcolepsy in Caucasian and black Americans. Sleep 17: S60–67 [DOI] [PubMed] [Google Scholar]
- Mignot E, Lin L, Finn L, et al. (2006) Correlates of sleep onset REM periods during the Multiple Sleep Latency Test in community adults. Brain 129:1609–1623 [DOI] [PubMed] [Google Scholar]
- Mignot E, Lin L, Rogers W, et al. (2001) Complex HLA-DR and -DQ interactions confer risk of narcolepsy-cataplexy in three ethnic groups. Am J Hum Genet 68:686–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignot E, Young T, Lin L, et al. (1999). Nocturnal sleep and daytime sleepiness in normal subjects with HLA-DQB1*0602. Sleep 22:347–352 [PubMed] [Google Scholar]
- Mignot E, Young T, Lin L, et al. (1998). Reduction of REM sleep latency associated with HLA-DQB1*0602 in normal adults. Lancet 351:727. [DOI] [PubMed] [Google Scholar]
- Miyagawa T, Hohjoh H, Honda Y, et al. (2000) Identification of a telomeric boundary of the HLA region with potential for predisposition to human narcolepsy. Immunogenetics 52:12–18 [DOI] [PubMed] [Google Scholar]
- Miyagawa T, Honda M, Kawashima M, et al. (2009) Polymorphism located between CPT1B and CHKB, and HLA-DRB1*1501-DQB1*0602 haplotype confer susceptibility to CNS hypersomnias (essential hypersomnia). PLoS One 4:e5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagawa T, Kawashima M, Nishida N, et al. (2008) Variant between CPT1B and CHKB associated with susceptibility to narcolepsy. Nat Genet 40:1324–1328 [DOI] [PubMed] [Google Scholar]
- Miyagawa T, Miyadera H, Tanaka S, et al. (2011) Abnormally low serum acylcarnitine levels in narcolepsy patients. Sleep 34: 349–353A [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki T, Klerman EB, Sakurai T, et al. (2006) Elevated body temperature during sleep in orexin knockout mice. Am J Physiol Regul Integr Comp Physiol 291:R533–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris DR, Geballe AP. (2000) Upstream open reading frames as regulators of mRNA translation. Mol Cell Biol 20:8635–8642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino S, Fujiki N, Ripley B, et al. (2001) Decreased brain histamine content in hypocretin/orexin receptor-2 mutated narcoleptic dogs. Neurosci Lett 313:125–128 [DOI] [PubMed] [Google Scholar]
- Nishino S, Ripley B, Overeem S, et al. (2000). Hypocretin (orexin) deficiency in human narcolepsy. Lancet 355:39–40 [DOI] [PubMed] [Google Scholar]
- Okun ML, Lin L, Pelin Z, et al (2002) Clinical aspects of narcolepsy-cataplexy across ethnic groups. Sleep 25:27–35 [DOI] [PubMed] [Google Scholar]
- Overeem S, Mignot E, van Dijk JG, et al. (2001) Narcolepsy: clinical features, new pathophysiologic insights, and future perspectives. J Clin Neurophysiol 18:78–105 [DOI] [PubMed] [Google Scholar]
- Pelin Z, Bozluolcay M, Kaynak D, et al. (2002) Childhood onset of narcolepsy-cataplexy syndrome in Turkey: clinical and genetic study. Turk J Pediatr 44:321–325 [PubMed] [Google Scholar]
- Platt B, Riedel G. (2011) The cholinergic system, EEG and sleep. Behav Brain Res 221:499–504 [DOI] [PubMed] [Google Scholar]
- Plazzi G, Moghadam KK, Maggi LS, et al. (2011) Autonomic disturbances in narcolepsy. Sleep Med Rev 15:187–196 [DOI] [PubMed] [Google Scholar]
- Sellayah D, Bharaj P, Sikder D. (2011) Orexin is required for brown adipose tissue development, differentiation, and function. Cell Metab 14:478–490 [DOI] [PubMed] [Google Scholar]
- Springer MS, Teeling EC, Madsen O, et al. (2001) Integrated fossil and molecular data reconstruct bat echolocation. Proc Natl Acad Sci U S A 98:6241–6246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafti M, Petit B, Chollet D, et al. (2003) Deficiency in short-chain fatty acid beta-oxidation affects theta oscillations during sleep. Nat Genet 34:320–325 [DOI] [PubMed] [Google Scholar]
- Teeling EC, Scall M, Kao DJ, et al. (2000) Molecular evidence regarding the origin of echolocation and flight in bats. Nature 403:188–192 [DOI] [PubMed] [Google Scholar]
- Teeling EC, Springer MS, Madsen O, et al. (2005) A molecular phylogeny for bats illuminates biogeography and the fossil record. Science 307:580–584 [DOI] [PubMed] [Google Scholar]
- The European Bioinformatics Institute (EMBL-EBI), Multiple Sequence Alignment by ClustelW2, Available at http://www.eb.ac.uk/Tools/msa/clustalw2/ (accessed March5, 2013)
- University of California Santa Cruz (UCSC) Human Genome Browser build Feb 2009 GRCh37/hg19. Available at http://genome.ucsc.edu/ (accessed March5, 2013)
- Watson NF, Ton TG, Koepsell TD, et al. (2010) Does narcolepsy symptom severity vary according to HLA-DQB1*0602 allele status? Sleep 33:29–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HI, Joo EY, Hong SB, et al. (2012) Use of PCR with sequence-specific primers for high-resolution human leukocyte antigen typing of patients with narcolepsy. Ann Lab Med 32:57–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida G, Li MX, Horiuchi M, et al. (2006) Fasting-induced reduction in locomotor activity and reduced response of orexin neurons in carnitine-deficient mice. Neurosci Res 55:78–86 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.