Abstract
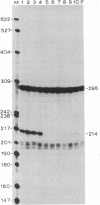
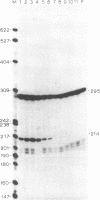
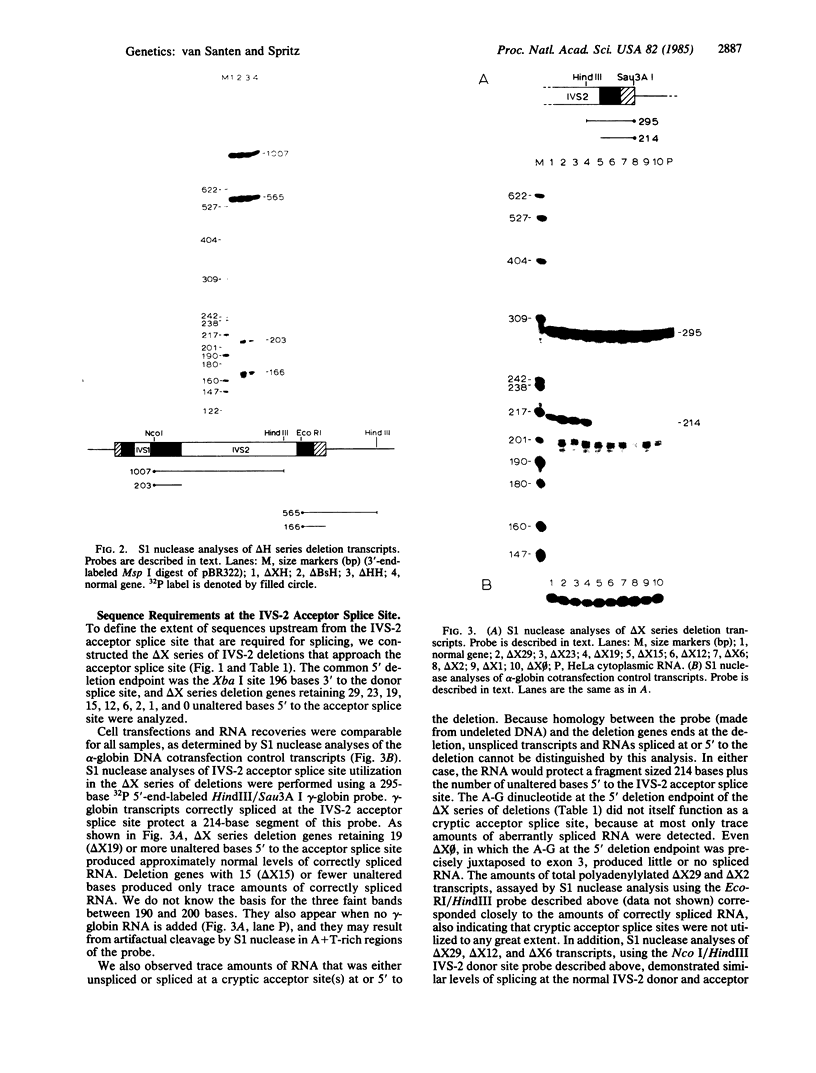
To define the extent of intervening sequence required for splicing higher eukaryotic mRNA precursors in vivo, we constructed deletions within the second intervening sequence of the human G gamma-globin gene that progressively approach the donor or acceptor splice sites. Most of the intervening sequence can be deleted with no effect on splicing. At the donor splice site, 6 bases of intervening sequence are sufficient for accurate and efficient splicing. At the acceptor splice site, 20 bases are sufficient for accurate and efficient splicing, and 16 bases are sufficient for accurate splicing but at a reduced level. However, 15 bases are insufficient for splicing at a significant level. The effect of deletions ending near the acceptor splice site is independent of whether an A-G dinucleotide is introduced into the acceptor splice site region by the deletion.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Antonarakis S. E., Irkin S. H., Cheng T. C., Scott A. F., Sexton J. P., Trusko S. P., Charache S., Kazazian H. H., Jr beta-Thalassemia in American Blacks: novel mutations in the "TATA" box and an acceptor splice site. Proc Natl Acad Sci U S A. 1984 Feb;81(4):1154–1158. doi: 10.1073/pnas.81.4.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benyajati C., Place A. R., Wang N., Pentz E., Sofer W. Deletions at intervening sequence splice sites in the alcohol dehydrogenase gene of Drosophila. Nucleic Acids Res. 1982 Nov 25;10(22):7261–7272. doi: 10.1093/nar/10.22.7261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Busslinger M., Moschonas N., Flavell R. A. Beta + thalassemia: aberrant splicing results from a single point mutation in an intron. Cell. 1981 Dec;27(2 Pt 1):289–298. doi: 10.1016/0092-8674(81)90412-8. [DOI] [PubMed] [Google Scholar]
- Cullen B. R., Kopchick J. J., Stacey D. W. Effect of intron size on splicing efficiency in retroviral transcripts. Nucleic Acids Res. 1982 Oct 11;10(19):6177–6190. doi: 10.1093/nar/10.19.6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felber B. K., Orkin S. H., Hamer D. H. Abnormal RNA splicing causes one form of alpha thalassemia. Cell. 1982 Jul;29(3):895–902. doi: 10.1016/0092-8674(82)90451-2. [DOI] [PubMed] [Google Scholar]
- Fukumaki Y., Ghosh P. K., Benz E. J., Jr, Reddy V. B., Lebowitz P., Forget B. G., Weissman S. M. Abnormally spliced messenger RNA in erythroid cells from patients with beta+ thalassemia and monkey cells expressing a cloned beta+-thalassemic gene. Cell. 1982 Mar;28(3):585–593. doi: 10.1016/0092-8674(82)90213-6. [DOI] [PubMed] [Google Scholar]
- Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981 Jan;23(1):175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- Karn J., Brenner S., Barnett L. Protein structural domains in the Caenorhabditis elegans unc-54 myosin heavy chain gene are not separated by introns. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4253–4257. doi: 10.1073/pnas.80.14.4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang K. M., Spritz R. A. RNA splice site selection: evidence for a 5' leads to 3' scanning model. Science. 1983 Jun 24;220(4604):1351–1355. doi: 10.1126/science.6304877. [DOI] [PubMed] [Google Scholar]
- Langford C. J., Gallwitz D. Evidence for an intron-contained sequence required for the splicing of yeast RNA polymerase II transcripts. Cell. 1983 Jun;33(2):519–527. doi: 10.1016/0092-8674(83)90433-6. [DOI] [PubMed] [Google Scholar]
- Langford C. J., Klinz F. J., Donath C., Gallwitz D. Point mutations identify the conserved, intron-contained TACTAAC box as an essential splicing signal sequence in yeast. Cell. 1984 Mar;36(3):645–653. doi: 10.1016/0092-8674(84)90344-1. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Montell C., Fisher E. F., Caruthers M. H., Berk A. J. Resolving the functions of overlapping viral genes by site-specific mutagenesis at a mRNA splice site. Nature. 1982 Feb 4;295(5848):380–384. doi: 10.1038/295380a0. [DOI] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett R. A., Konarska M. M., Grabowski P. J., Hardy S. F., Sharp P. A. Lariat RNA's as intermediates and products in the splicing of messenger RNA precursors. Science. 1984 Aug 31;225(4665):898–903. doi: 10.1126/science.6206566. [DOI] [PubMed] [Google Scholar]
- Pikielny C. W., Teem J. L., Rosbash M. Evidence for the biochemical role of an internal sequence in yeast nuclear mRNA introns: implications for U1 RNA and metazoan mRNA splicing. Cell. 1983 Sep;34(2):395–403. doi: 10.1016/0092-8674(83)90373-2. [DOI] [PubMed] [Google Scholar]
- Rautmann G., Matthes H. W., Gait M. J., Breathnach R. Synthetic donor and acceptor splice sites function in an RNA polymerase B (II) transcription unit. EMBO J. 1984 Sep;3(9):2021–2028. doi: 10.1002/j.1460-2075.1984.tb02085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J. A precursor of globin messenger RNA. J Mol Biol. 1976 Sep 15;106(2):403–420. doi: 10.1016/0022-2836(76)90093-0. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Krainer A. R., Maniatis T., Green M. R. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell. 1984 Aug;38(1):317–331. doi: 10.1016/0092-8674(84)90553-1. [DOI] [PubMed] [Google Scholar]
- Solnick D. An adenovirus mutant defective in splicing RNA from early region 1A. Nature. 1981 Jun 11;291(5815):508–510. doi: 10.1038/291508a0. [DOI] [PubMed] [Google Scholar]
- Sompayrac L. M., Danna K. J. Efficient infection of monkey cells with DNA of simian virus 40. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7575–7578. doi: 10.1073/pnas.78.12.7575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spritz R. A., Jagadeeswaran P., Choudary P. V., Biro P. A., Elder J. T., deRiel J. K., Manley J. L., Gefter M. L., Forget B. G., Weissman S. M. Base substitution in an intervening sequence of a beta+-thalassemic human globin gene. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2455–2459. doi: 10.1073/pnas.78.4.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimmappaya B., Shenk T. Nucleotide sequence analysis of viable deletion mutants lacking segments of the simian virus 40 genome coding for small t antigen. J Virol. 1979 Jun;30(3):668–673. doi: 10.1128/jvi.30.3.668-673.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman R., Orkin S. H., Maniatis T. Specific transcription and RNA splicing defects in five cloned beta-thalassaemia genes. Nature. 1983 Apr 14;302(5909):591–596. doi: 10.1038/302591a0. [DOI] [PubMed] [Google Scholar]
- Treisman R., Proudfoot N. J., Shander M., Maniatis T. A single-base change at a splice site in a beta 0-thalassemic gene causes abnormal RNA splicing. Cell. 1982 Jul;29(3):903–911. doi: 10.1016/0092-8674(82)90452-4. [DOI] [PubMed] [Google Scholar]
- Volckaert G., Feunteun J., Crawford L. V., Berg P., Fiers W. Nucleotide sequence deletions within the coding region for small-t antigen of simian virus 40. J Virol. 1979 Jun;30(3):674–682. doi: 10.1128/jvi.30.3.674-682.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver R. F., Weissmann C. Mapping of RNA by a modification of the Berk-Sharp procedure: the 5' termini of 15 S beta-globin mRNA precursor and mature 10 s beta-globin mRNA have identical map coordinates. Nucleic Acids Res. 1979 Nov 10;7(5):1175–1193. doi: 10.1093/nar/7.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieringa B., Hofer E., Weissmann C. A minimal intron length but no specific internal sequence is required for splicing the large rabbit beta-globin intron. Cell. 1984 Jul;37(3):915–925. doi: 10.1016/0092-8674(84)90426-4. [DOI] [PubMed] [Google Scholar]
- Wieringa B., Meyer F., Reiser J., Weissmann C. Unusual splice sites revealed by mutagenic inactivation of an authentic splice site of the rabbit beta-globin gene. Nature. 1983 Jan 6;301(5895):38–43. doi: 10.1038/301038a0. [DOI] [PubMed] [Google Scholar]
- Zeitlin S., Efstratiadis A. In vivo splicing products of the rabbit beta-globin pre-mRNA. Cell. 1984 Dec;39(3 Pt 2):589–602. doi: 10.1016/0092-8674(84)90466-5. [DOI] [PubMed] [Google Scholar]