Abstract
Sepsis is a complex inflammatory response to infection. Microarray-based gene expression studies of sepsis have illuminated the complex pathogen recognition and inflammatory signaling pathways that characterize sepsis. More recently, gene expression profiling has been used to identify diagnostic and prognostic gene signatures, as well as novel therapeutic targets. Studies in pediatric cohorts suggest that transcriptionally distinct subclasses may account for some of the heterogeneity seen in sepsis. Time series analyses have pointed to rapid and dynamic shifts in transcription patterns associated with various phases of sepsis. These findings highlight current challenges in sepsis knowledge translation, including the need to adapt complex and time-consuming whole-genome methods for use in the intensive care unit environment, where rapid diagnosis and treatment are essential.
Keywords: Sepsis, septic shock, gene expression, microarrays, genomics, bioinformatics
Sepsis and its genomic influences
Since its introduction in the late 1990s, microarray-based gene expression profiling has had a significant impact on the field of medicine. In cancer biology, molecular subtypes of diseases have been identified [1], as well as transcriptional signatures predicting clinical outcome [2], and response to specific therapies [3]. Useful biomarkers have been found that in some cases can obviate the need for genome-wide approaches, enabling the translation of gene expression research into clinical practice. Nonetheless, the impact of genome science remains far from pervasive, especially in the Intensive Care Unit (ICU), where diseases evolve rapidly, resulting in systemic illness, organ failure, and high mortality.
Sepsis, one of the most prevalent diseases in the ICU, is a clinical syndrome characterized by a systemic inflammatory response to infection, typically bacterial in origin. It is defined as documented or suspected infection, in the setting of a subset of four findings that describe the systemic inflammatory response syndrome (SIRS) [4], and can progress rapidly, resulting in organ failure (severe sepsis), or impaired tissue perfusion (septic shock). Sepsis syndromes are both common and dangerous, with the incidence increasing in both adults and children, and mortality rates as high as 10–50% depending on age and disease severity [5,6].
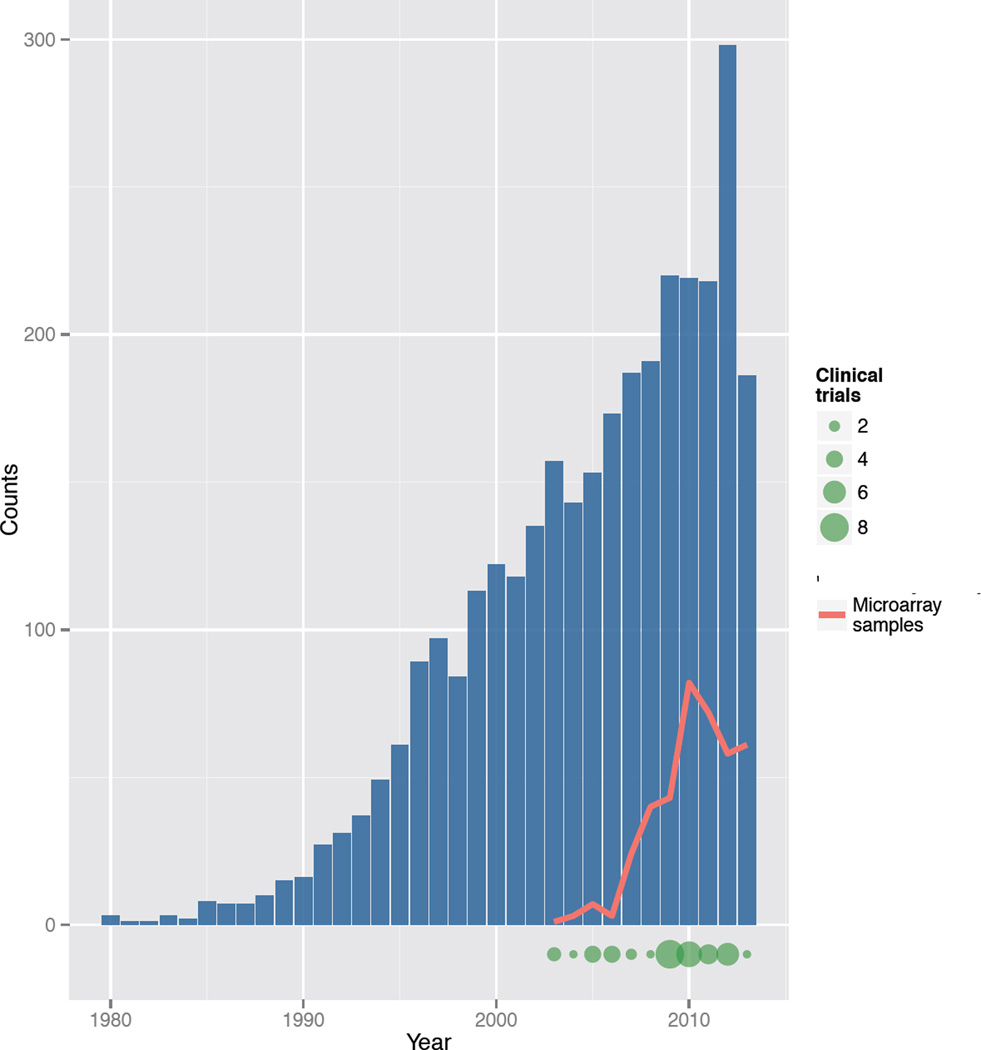
While genetic influences on the pathogenesis of acute conditions like sepsis may be under-appreciated, they are no less striking. In adoptee studies, death from infection has been shown to be 5-fold more heritable than death from cancer [7]. The innate immune response that accounts for the physiologic derangements of bacterial infection is associated with altered expression of more than 3,700 genes [8], making gene expression analysis a potentially useful tool for discovery-oriented studies of the pathogenesis of sepsis and severe infection. Published findings based on this research paradigm are increasing, while expression data are accumulating in publicly available repositories, and a few active clinical trials include a gene expression component (Figure 1).
Figure 1.
Trends in gene expression profiling of sepsis. Bars represent the number of Pubmed citations per year for the search term “gene expression AND sepsis”. Trend line shows the number of individual microarray assays added each year to a publicly available repository of gene expression data (ArrayExpress). The size of the points at the bottom of the plot reflect the number of clinical trials initiated in each year, as identified by a trials registry (clinicaltrials.gov, search terms "gene expression AND sepsis OR septic shock OR severe sepsis").
Goals and challenges of transcriptome research in sepsis
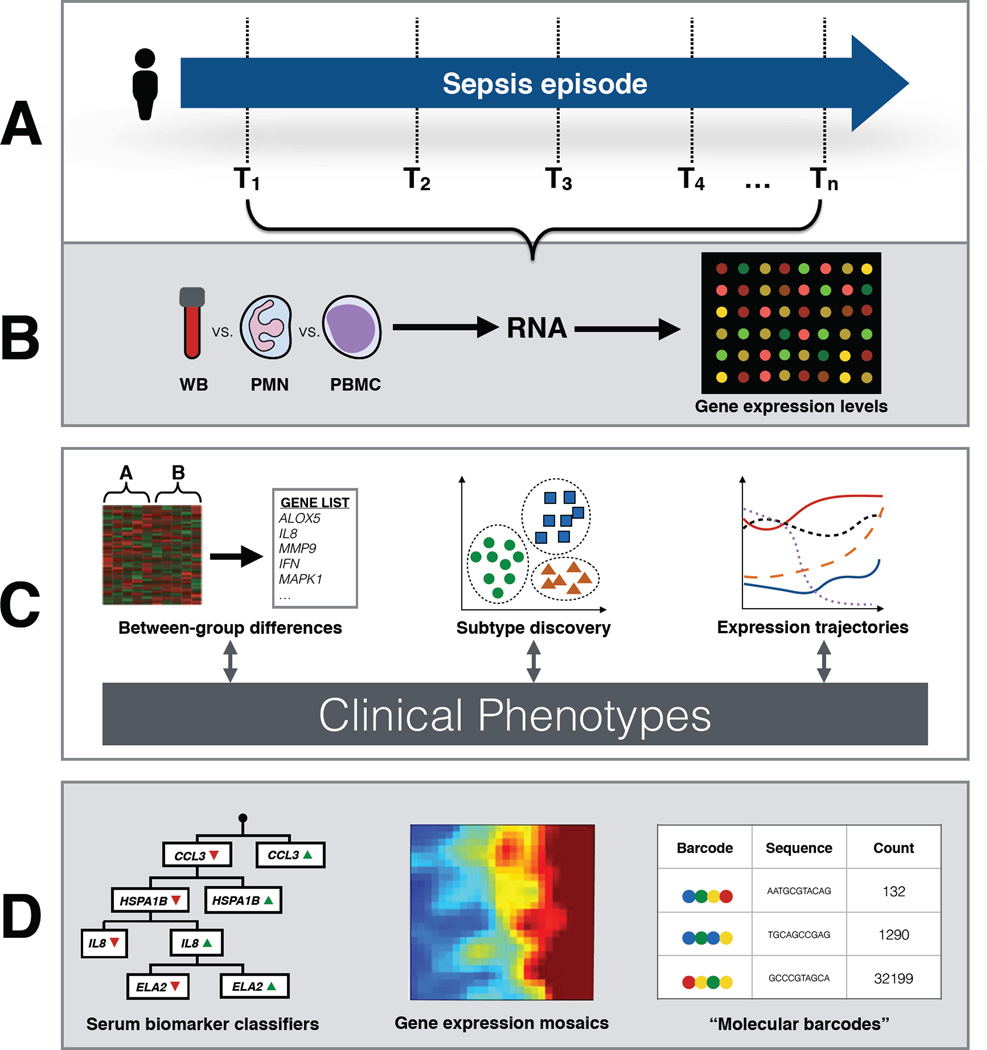
Gene expression analysis of sepsis is distinguished from analyses of cancer and chronic diseases in a number of ways, both conceptual and pragmatic (Figure 2). First, sepsis investigators must make decisions about which tissues to sample and at what time points, which in the absence of additional clinical data or a priori hypotheses, may be arbitrary in nature. Tissue from the source of infection may be difficult to sample directly as biopsies are seldom practical in the critically ill. As an easily accessible compartment of the immune system, whole blood and its various leukocyte fractions have therefore been the source tissue of interest in most gene expression studies of sepsis. The findings from gene expression profiling of blood cells may not accurately reflect expression patterns from immune cells resident in other tissues, such as alveolar macrophages, or splenic lymphocytes, though the significance of this potential discordance remains uncertain [9,10].
Figure 2.
Overview of gene expression profiling in sepsis. (A) As sepsis syndromes are characterized by rapid shifts in gene expression over hours and days, blood samples can be collected for analysis at a variety of time points. Multiple samples taken over the course of resuscitation, stabilization, and convalescence, can be used to generate time series of gene expression. (B) Once samples are collected, RNA can be extracted either directly from whole blood, or from different leukocyte fractions. RNA transcript levels are derived from gene expression microarrays. (C) Bioinformatics pathways can be used to compare gene expression profiles between two or more groups of patients (e.g. sepsis and non-infectious SIRS), resulting in a list of differentially expressed genes, and their associated pathways. Unsupervised machine learning methods including partitional clustering algorithms can be used to identify previously unrecognized sepsis subclasses. Expression data from multiple time points can be analyzed together to generate expression trajectories, which may differ between patients. Interpretation of differences in gene expression is facilitated through comparison with clinical phenotypes derived from patient data collected from electronic medical records or patient registries, or in the context of a clinical trial. (D) Unlike with diseases managed in the outpatient setting, the treatment of sepsis relies on diagnostic testing that can rapidly returns easily interpreted results. High-dimensional gene expression data must therefore be “downsized” to more easily derived and understood signals. Strategies include using serum biomarker assays develop patient classifiers, generating gene expression mosaics that visually represent complex expression signals, and deploying sophisticated multiplex assays that measure a limited number of transcripts using molecular barcoding technology. Abbreviations: whole blood, WB; polymorphonuclear neutrophils, PMN; peripheral blood mononuclear cells, PBMC.
Secondly, sepsis is a dynamic process within a relatively narrow time period. Thus, while genomic changes can occur in tumors over time, large-scale transcriptional shifts in leukocytes have been shown to occur within just a few hours of an inflammatory stimulus [8,11]. In the setting of blunt trauma, a condition with considerable inflammatory features that is often complicated by sepsis, the leukocyte transcriptome is substantially altered to up-regulate inflammatory and pathogen recognition pathways in the days and weeks following injury [12]. These investigations into the functional trajectory of cellular processes constitute a unique method by which to model the dynamic pathophysiology of acute illness. Samples collected repeatedly over the course of an illness episode should therefore ideally be analyzed together rather than in isolation, in an attempt to describe illness trajectory and differentiate responses to treatment. Unlike with trauma, the precise onset of sepsis may be difficult to pinpoint accurately, introducing further complexity in comparing time course gene expression profiles from different patients with sepsis.
Thirdly, while gold standard diagnostic labels can be arrived at for most tumors based on anatomic and molecular pathology findings, the diagnosis of sepsis is predominantly a clinical one. Moreover, the criteria on which the diagnosis is based lack specificity, with more than 40% of cases having negative bacterial cultures [13]. The absence of reliable classification complicates statistical analyses of gene expression data that use supervised methods to detect differences in expression between groups.
Lastly, whole-genome approaches to sepsis research must include strategies for “downsizing” the methods used, from high-dimensional, resource-and time-intensive gene expression assays to rapid, cost-effective diagnostic tests that can be deployed at the point of care. While findings from gene expression studies in cancer may translate to clinical practice via immunohistochemical staining of pathologic specimens, targeted genotyping, or other complex and time-consuming assays, these techniques are impractical in the management of sepsis. Useful assays must reflect the rapidly evolving, dynamic nature of sepsis, the need for quick information in the acute resuscitative phases of this condition, and the necessity that individual samples be analyzed at any time of day or night, with short turnaround time, and without waiting to be batched with other specimens.
Experimental designs
As a primarily immunologic phenomenon, sepsis is often studied by examining leukocytes, including ex vivo immunostimulation experiments. Patients with sepsis syndromes manifest a variety of dynamic shifts in leukocyte populations, often transitioning between states of leukocytosis and leukodepletion, and exhibiting differences in the relative abundance of granulocytes, lymphocytes, and specialized subsets thereof. Each of these cellular subtypes exhibits a distinct gene expression pattern tailored to the specialized function of the respective cell type [14], with expression profiles from whole blood representing a weighted sum of these patterns. As different cellular compartments of the blood perform different immunologic functions in response to infection, leukocyte gene expression in sepsis is cell type specific. The set of genes that distinguish sepsis from non-infectious inflammation in the neutrophils of the innate immune system demonstrates very little overlap with the set of genes similarly identified from the lymphocytes of the adaptive immune system [15,16],
The use of whole blood to derive gene expression data in sepsis has the pragmatic advantage of straightforward sample collection, minimal preprocessing, and limited induction of expression artifacts related to isolation of leukocyte subsets [17]. Most clinical studies in both adults and children have used either whole blood or peripheral blood mononuclear cells (PBMCs). In the case of whole blood studies, statistical methods have been employed to account for the relative abundance of each leukocyte subtype in the sample, and to attribute gene expression findings to specific populations of cells [18].
Insights into molecular mechanisms of sepsis
Initial studies of gene expression in sepsis were largely exploratory in nature, aiming to describe the complex immunologic and inflammatory pathways that characterize this condition. Early insights came from studies of healthy volunteers exposed to bacterial endotoxin [8,11,19], which revealed significant changes in the transcription of more than 3,700 genes as soon as 2 hours after endotoxin exposure. Early after endotoxin exposure, pathogen recognition cell surface receptors, including those from the Toll-like receptor (TLR) family, are upregulated, along with a variety of proinflammatory cytokines and chemokines such as tumor necrosis factor (TNF), interleukin (IL) 1A, IL1B, CXCL1, CXCL2, monocyte chemotactic protein 1 (MCP-1), and IL-8 [8,11]. These changes are accompanied by activation of signal transduction pathways including nuclear factor κB (NFκB), mitogen activated protein kinase (MAPK), janus kinase (JAK), and signaling transducer and activator of transcription (STAT). In parallel, signaling to restrain the immune response is increased, both by the upregulation of suppressor of cytokine signaling genes (e.g. suppressor of cytokine signaling 3; SOCS3) and the downregulation of cytokine expression itself. Expression patterns return to baseline within 24 hours of endotoxin exposure. Unlike with tightly controlled experiments in healthy subjects, clinical studies of gene expression in sepsis must confront substantially more uncertainty, heterogeneity, and complexity in the inflammatory manifestations of infection.
Expression patterns are modulated by a number of factors such as age, gender, and ethnicity, the presence of comorbidities, the timing of inflammatory stimulus, and the patient’s state of immune activation at the time of infection. This is reflected in the results of clinical studies of gene expression in adults with sepsis syndromes. A systematic review of a dozen such studies suggests nearly universal upregulation of the pathogen recognition and signal transduction pathways identified in controlled endotoxin experiments, but a far more mixed picture when it comes to the pro- and anti-inflammatory pathways mediated by genes that govern lymphocyte differentiation, antigen presentation, and cytokine expression [20]. Disagreement between studies in this regard may reflect differences in patient population, in that trauma patients who are otherwise healthy may be predisposed to immunostimulation following injury, while patients with primary sepsis exhibit a greater degree of immunosuppression [18,21].
Gene expression studies have been used to identify novel therapeutic targets in sepsis, based on molecular pathways differentially expressed between cases and controls, or between survivors and non-survivors. Complementing findings from basic science research, animal studies, and clinical trials in humans [22], gene expression studies have highlighted the importance of zinc homeostasis in immune functioning, particularly amongst children with sepsis syndromes [23]. Children admitted to the ICU with septic shock have been shown to exhibit diminished expression of numerous genes that influence or rely upon zinc homeostasis. This pattern that may be more evident in a certain subset of patients, and associated with poor outcomes [24]. Clinical studies of zinc supplementation have demonstrated salutary effects on the incidence and severity of certain infections in both children and the elderly [25–27], but in both adult and pediatric ICU patients, larger randomized trials of mixed micronutrient supplementation that included zinc showed no significant effects on the incidence of infection [28,29].
One particular family of zinc-related proteins has been shown to be consistently overexpressed in sepsis and septic shock. The matrix metalloproteinases (MMPs) are a series of proteases that degrade extracellular matrix, inflammatory cytokines, and other proteins, thereby mediating a variety of immunologic and neoplastic processes [30,31]. Gene expression studies, along with confirmatory serum assays, have shown MMP-8 and MMP-9 to be upregulated in injury and sepsis, correlating in some cases with disease severity [11,32–34]. Consistent with these clinical observations, MMP-8 null mice and wild-type mice treated with a pharmacological inhibitor of MMP-8 have a survival advantage when subjected to a model of sepsis [32].
Influence of demographic factors
While patient age, gender, and ethnicity have been accounted for in traditional clinical and basic science research in sepsis, the importance of these demographic factors in gene expression studies has yet to be fully explored. Nonetheless, there is reason to believe that demographic features exert considerable effects on gene expression patterns in sepsis. Ethnic background is known to be a strong determinant of gene expression in general [35], and there is some evidence to suggest its influence on sepsis in particular. In one small study examining gene expression patterns amongst critically ill patients with ventilator-associated pneumonia (VAP), far more genes varied between ethnic groups (Caucasian or African-American), than between groups sorted by age or gender [36]. Epidemiological studies suggest differences in sepsis incidence and outcomes amongst patients with different ethnic backgrounds, although the extent to which these findings reflect genomic differences is not known [37].
Concurrent with evolving immune function, both early in development and later in life, different genetic responses to sepsis are seen amongst various patient age groups. In one analysis combining results from five separate studies, researchers identified differences in gene expression amongst pediatric patients with septic shock. Full-term neonates (≤28 days) were shown to have gene expression patterns distinct from those of infants (1 month to 1 year), toddlers (2–5 years), and school-aged children (≥6 years) when assessed within 24 hours of ICU admission [38]. Importantly, and in contrast to older children and adults, these results suggest that neonates not only failed to mount a robust inflammatory response, but also in fact demonstrated downregulation of antigen presentation and NF-κB pathway genes, and an overall decrement in immune response to infection. Reduced expression of triggering receptor expressed on myeloid cells-1 (TREM-1) pathway-related genes was also seen, suggesting that neonates may have limited capacity to amplify immune signals related to pathogen recognition, and may be less responsive to novel therapies targeting this pathway in septic shock [39,40]. This study included a relatively small number of neonates (n=17), and the findings would be bolstered by validation in a dedicated prospective study.
At the other end of the age spectrum, there are fewer whole-genome data about the effects of aging on immune functioning in sepsis. In one mouse study examining individual cytokine levels using a cecal ligation and puncture (CLP) model of sepsis, older mice showed more pronounced local and systemic inflammatory responses compared to younger mice with similar survival rates [41]. While gender is known to influence gene expression patterns in other conditions such as ischemic heart diseases, albeit modestly [42], sex-specific differences in gene expression in sepsis remain largely unexplored. Complex interactions between demographic factors are also likely to influence gene expression in sepsis.
Gene expression and sepsis subtypes
One of the most clinically relevant questions following the diagnosis of sepsis is that of which invading pathogen is responsible for the acute infection. Proper knowledge of the inciting cause is useful in selecting appropriate antimicrobial agents, and identifying uncontrolled sources of infection. Sepsis arising from different types of organisms, including gram-positive bacteria, gram-negative bacteria, and fungi, may be clinically indistinguishable, and both the yield and lag time of microbial cultures limit their utility in practice [4,13]. As the molecular pathways underpinning the cellular immune response to these various types of infection have distinctive features, gene expression profiling has been investigated as a means to identify culprit organisms in septic patients.
Early studies in animal models and ex vivo models of human cell types suggest that regardless of the invading pathogen, a core group of co-expressed genes are upregulated in the face of infection, constituting a so-called “common host response” to sepsis [43,44]. This common response, expressed in a variety of cell types, includes activation of inflammatory mediators and signal transduction pathways, as well as negative feedback pathways and apoptotic pathways that put infected cells in a state of ‘high alert’, whereby programmed cell death can be initiated in the event of progressive infection [43]. While targeted experiments suggest that isolated pathways coordinate the immune response to gram-positive and gram-negative infections, microarray experiments suggest considerable overlap between these types of infection. Both TLR2 receptors (associated with the transcriptional response to gram-positive infections), and TLR4 receptors (which bind lipopolysaccharide from gram-negative bacteria) initiate signals that culminate in the common host response [43]. This result is reflected in clinical studies of gene expression in sepsis, which for the most part have shown few differences in expression patterns between patients infected with different types of bacteria [36,45].
In the pediatric population, gene expression data have been used to distinguish bacterial infections from viral infections such as influenza A [46]. Not surprisingly, patients with influenza were identifiable on the basis of increased expression of interferon (IFN) pathways. Up to a third of patients with bacterial sepsis also demonstrated upregulation of IFN genes, suggesting the possibility of a preceding or concurrent viral infection in these cases. Differences were also found between patients with gram-positive infections and those with gram-negative infections. Beyond its focus on pediatric rather than adult patients, this study differed from those that found no differentially expressed genes between gram-positive and gram-negative sepsis. Instead of using neutrophils, investigators generated gene expression profiles from PBMCs which, because of their pleomorphism, might be expected to better reflect pathogen differences. These findings highlight the importance of considering tissue type in designing and interpreting gene expression studies in sepsis.
In addition to determining what type of infection has triggered a sepsis response, it may be equally important to determine the nature of the response mounted by an individual patient. The existence of heretofore unrecognized sepsis subtypes is suggested by the heterogeneity of physiologic and molecular phenotypes encompassed under the non-specific clinical definition of sepsis. This inadvertent case mixing in clinical studies leads to indistinguishable survival curves, overlapping histograms of measured outcomes, and a deficit of actionable evidence. Discovery of sepsis subtypes has thus been identified by some as a key goal in sepsis research [47,48].
This problem is inherently one of unsupervised machine learning, in which patients are grouped not based on clinical labels assigned by investigators, but rather according to similarities across multiple dimensions of gene expression data. Various cluster identification algorithms have been used for this purpose. In one such study, pediatric patients with septic shock were partitioned into distinct gene expression clusters, based on expression levels of genes that differentiated septic patients from a group of non-septic controls [24]. Hierarchical clustering was used with an a priori decision to designate the second-order branch points as distinct clusters. This approach resulted in 3 subclasses of septic shock (subclasses A, B, and C), with nearly 7,000 genes differentially expressed between them. Clinical phenotyping of the subclasses after clustering showed significant differences in important clinical outcomes, with patients in subclass A having more severe manifestations of sepsis, a greater degree of organ injury, and higher mortality. Patients in this subclass also tended to be younger, with a median age of 3.6 months, compared to 4.3 years for subclass B, and 2.0 years for subclass C. From a genomic standpoint, subclass A was characterized by a relative downregulation of adaptive immune pathways and glucocorticoid receptor signaling. In a separate multicenter validation study, 82 patients from an independent cohort were grouped according to their level of expression of the top 100 class-defining genes identified in the first study. Patients from subclass A again showed a greater severity of illness, a trend towards higher mortality, and younger median age [49].
While no prospective studies in adults have been dedicated to sepsis subtype discovery, a post-hoc analysis of gene expression profiles generated in the course of other investigations suggests their existence. Using separate derivation and validation cohorts, patients were clustered using a partitioning around medioids (PAM) algorithm based on expression levels of a subset of genes identified in the literature as being meaningful in sepsis and septic shock [50]. The existence of two distinct clusters was best supported by the data, and while no significant clinical differences were found between subtypes, there were important differences in the expression of genes involved in inflammatory and pathogen recognition pathways, as well as key pharmacogenes involved in the metabolism of drugs used commonly in sepsis.
“Downsizing” genome-wide data for clinical use
Multidimensional gene expression platforms that sample thousands of genes at once are ideally suited for discovery-oriented tasks in sepsis research. Their use in clinical practice, however, is limited by a number of practical considerations. To be useful in the evaluation and treatment of patients with sepsis, assays must be widely available, rapid, repeatable, consistent, and inexpensive. As such, there is a need to “downsize” the high-dimensional data generated by gene expression microarrays, to more targeted signals that can be produced closer to the point of care.
The most reductive approach to this problem is to use gene expression microarray data to identify individual biomarkers that distinguish patient subsets of interest. Typically, this involves examining genes with the highest fold-change difference in expression between patients with sepsis and those with noninfectious SIRS. In one pediatric study, gene expression profiling was used to identify class-predictor genes distinguishing SIRS with negative bacterial cultures, from sepsis with positive bacterial cultures [51]. The most predictive gene was Epstein-Barr virus-induced gene 3 (EBI3), a subunit of IL-27 that, when detected at high levels in the serum, had high specificity and positive predictive value (>90%) for the diagnosis of sepsis. A comparable study in adults showed that serum IL-27 levels had relatively less favorable test characteristics, and was outperformed by procalcitonin, a biomarker already used in clinical practice [52].
Biomarkers identified by gene expression profiling have also been used to stratify patients with sepsis according to mortality risk. The IL8 gene was shown to be upregulated in nonsurvivors of pediatric septic shock, with elevated serum levels of IL-8 significantly increased as compared to survivors and controls [23]. Again this biomarker was shown to have less predictive value in adults with sepsis [53]. Although there were methodological differences between the pediatric and adult studies in the case of both IL-27 and IL-8, these findings again underscore the influence of age in immune functioning, and the importance of considering age in the design and analysis of gene expression studies in sepsis.
In other microarray studies, the chemokine receptor CX3CR1 (fractalkine receptor) was shown to be upregulated in sepsis survivors compared to nonsurvivors [54], and serum levels of its ligand CX3CL1 were found to be elevated in patients with sepsis, as compared to healthy controls [55]. Chemokine ligand 4 (CCL4, also known as MIP-1β) has also been identified as a potential biomarker based on differential gene expression, and shown to have a very high negative predictive value for mortality in pediatric septic shock [56].
Though variously sensitive or specific in certain populations, these single-marker diagnostic strategies do not have well-rounded performance characteristics that would justify their broad use in clinical practice [57]. Moreover, the use of individual biomarkers in isolation in many ways defeats the purpose of using high-throughput technologies like gene expression microarrays to study multifaceted, complex conditions like sepsis. Traditionally identified by knowledge-based approaches predicated on known biologic functions and pathways, the search for biomarkers tends to leave unexamined scores of other proteins that may be useful alone or in combination, but whose biologic function in sepsis has yet to be fully elucidated [58]. One strategy to overcome this bias is to leverage the considerable coverage of gene expression microarrays, to identify candidate biomarkers that appear promising based on statistical, rather than biological features.
In the Pediatric Sepsis Biomarker Risk Model (PERSEVERE) project, investigators used such an approach to derive and validate a panel of serum biomarkers to assign a mortality probability in pediatric sepsis [57–59]. Genes differentially expressed between sepsis survivors, sepsis nonsurvivors, and healthy controls were identified by analysis of variance (ANOVA), and further refined by post hoc pairwise comparisons to identify 137 candidate biomarker genes. These were cross-referenced with a list of 4,397 candidate genes identified using support vector machine (SVM) classifiers based on sepsis mortality, and further reduced by selecting genes whose protein products could be easily measured in the blood [58]. The final panel of 12 biomarkers was subsequently used to derive a classifier based on classification and regression tree (CART) analysis, and validated in a separate patient cohort. The model had adequate sensitivity (91% in the derivation cohort, 89% in the validation cohort), but lacked specificity (86% in the derivation cohort, 64% in the validation cohort), and had an area under the receiver operating characteristic (ROC) curve of 0.759 in the validation cohort [59]. Recently, prospective validation of an updated model yielded a ROC of 0.811 [60] and an analogous model was derived and tested in adult populations [61].
The shortcomings in gene expression derived biomarker performance likely reflect a complex interplay of individual genes and transcripts in sepsis, to say nothing of the post-translational modifications and protein interactions that exert influence on function. Cellular signals in sepsis are dynamic, changing rapidly with inflammatory conditions and an evolving immunologic milieu. The goal of reducing genome-wide signals to individual biomarkers, or even groups of biomarkers used in combination, may therefore be difficult to achieve. Some investigators have instead focused on developing gene signatures that combine signals from dozens or hundreds of genes, to be used as a filter in identifying septic patients from those with noninfectious SIRS, and in predicting outcomes of sepsis syndromes. In one such study, a 138-gene signature distinguished sepsis from SIRS, with sensitivity and specificity in the validation cohort of 81% and 79%, respectively [16].
Translating genome-wide assays for clinical use involves developing tools that can be deployed in the unpredictable and dynamic clinical environment of the emergency department or ICU. New methods of data representation are needed in order to convey key signals contained within high-dimensional data to front-line clinicians both quickly and unambiguously. Along these lines, investigators have tested the use of novel data visualization methods such as “gene expression mosaics” that convey high-dimensional gene expression data in 2-dimensional color patterns [62]. Expression mosaics have been developed in the study of pediatric septic shock subclasses, and have been shown to be useful in both computer-based and clinician-based interpretation of expression patterns [49,63]. Among clinicians without specific training in the interpretation of these patters, gene expression mosaics were sorted according to sepsis subclass (A, B, or C) with overall K value for agreement of 0.81.[63]
In an effort to develop more scalable solutions for gene expression analysis in acute care, investigators have also used multiplexed color-coded probes, so-called “molecular barcodes”, to directly measure mRNA transcript abundance in samples of interest (NanoString nCounter system) [64]. Based on microarray gene expression findings from the leukocyte fractions of 167 trauma patients, researchers derived an expression signature of 63 genes that varied most between patients with uncomplicated, intermediate, and complicated clinical courses following trauma [34]. To create an assay that could reasonably be used in clinical practice, leukocytes were isolated by means of red cell lysis in microfluidics chambers, and samples analyzed using the NanoString platform to evaluate expression of the 63 signature genes. These results were further downsized to a single expression metric, the difference from reference value (DFR), based on a summation of expression differences for each gene, from age-, gender-, and ethnicity-matched controls. Results were generated within 8 to 12 hours, showed good agreement with microarray expression values, and performed better than both microarray-derived DFR, and conventional clinical severity of illness scores.
Temporality of gene expression in sepsis
Most clinical studies of gene expression in sepsis have been based on the analysis of a single time point in the illness trajectory, or on the comparison of an early time point with a later one. However, genomic shifts in response to inflammation are known to occur rapidly, as seen in clinical studies of trauma patients [12], or experimental studies of healthy subjects exposed to endotoxin [8,11]. Unlike in these cases, the time of onset of the inflammatory stimulus in sepsis cannot be accurately known, resulting in considerable uncertainty regarding the timing of sampling with respect to the ebb and flow of the immunologic response. Many studies include protocols to collect samples for gene expression analysis within 24 hours of admission to the ICU, however patients are admitted to the ICU at various stages of sepsis, and may transition from one stage to the next even within the first 24 hours of their stay. The extent to which gene expression is being compared across similar genomic, molecular, and pathophysiologic epochs in these studies is thus uncertain.
The importance of timing in sepsis gene expression analysis is illustrated by one recent study of 5 pediatric patients with severe sepsis and septic shock due to meningococcal meningitis [65]. In this work, expression levels of key genes differed between patients at various time points. Further, the overall contour of expression trajectories of key genes across the entire 48-hour period also differed. These differential trajectories were seen for some genes that have been investigated as biomarkers in sepsis, suggesting that certain biomarkers may be more useful in some patients than others, and may be more useful at certain stages of illness than others.
A number of statistical methods have been developed to analyze time course gene expression data. Approaches include Markov models [66–68], analyses of variance [69], and the use of cubic splines to model changing expression levels over time [70]. Time course gene expression data from trauma and burn patients have been used to develop statistical methods for the analysis of leukocyte gene expression over time, such as the riboleukogram, which uses principal components analysis to graphically represent a patient’s genomic trajectory over time [36,71]. Results from these studies suggest that genomic profiles in sepsis oscillate around a baseline immune attractor state. Early results support an increased between-patient variance in gene expression at the height of the acute inflammatory phase, with differences between individuals diminishing as patients return to a baseline state of health [71]. As these statistical and computational methods evolve, comparison of gene expression trajectories in sepsis may provide even greater insight into the molecular physiology of sepsis than comparison of gene expression at a single time point.
Concluding remarks and future perspectives
The advent of high-throughput genomic technologies such as gene expression microarrays have made possible the study of the complex, dynamic changes in the host transcriptome as it responds to severe infection. Initial studies have added significantly to our understanding of sepsis pathophysiology, identified different molecular phenotypes of sepsis, and suggested novel targets for new sepsis therapies. Nonetheless, conclusions from these initial studies have been far from unanimous. Discrepancies may be attributable to a variety of technical factors, including lack of agreement between microarray platforms in earlier studies, and an abundance of different statistical methods and bioinformatics pathways used to conduct analyses (Table 1). A lack of standardization in terms of timing of sampling and tissue source used further complicates direct comparisons between studies. Importantly, many analyses have been based on small sample sizes, and bear confirming in larger cohorts. As microarray technology and bioinformatics methods evolve, concordance is likely to increase.
Table 1.
Statistical methods and other analytic tools used in gene expression profiling of sepsis
| Analysis | Description | References |
|---|---|---|
| Single time point data | ||
| Univariate Student’s t-test | A t-test is performed for each gene represented in the experiment in order to identify those that are differentially expressed between two groups (eg. “sepsis” and “control”). Correction of significance level is required to reflect testing of multiple hypotheses. | [15,16,45,46] |
| Significance analysis of microarrays (SAM) | Identifies genes that are differentially expressed between two or more groups. SAM uses gene-specific t-tests to assign a score based on differences in expression levels between groups, relative to the standard deviation [73]. A tuning parameter is used to select a tolerable false discovery rate. | [8] |
| K-means clustering | Samples are partitioned into a user-specified number of clusters, according to their proximity to one another in n-dimensional gene space (where is n is the number of genes whose expression levels are used in the analysis). | [9,12,24] |
| Extraction of Differential Gene Expression (EDGE) | Uses an Optimal Discovery Procedure to identify genes that are differentially expressed between user-specified groups [74]. | [12,36] |
| Linear mixed models | Each gene is modelled individually with expression levels as the dependent variables and any number of phenotypic features (group assignment, age, day of sample, etc.) used as independent variables. Differential expression between conditions of interest can be inferred, controlling for potential confounders. | [18] |
| Hierarchical clustering | Similar expression patterns are grouped, forming a dendrogram that can be used to select clusters. Clustering can be done according to similarity between genes, between samples, or both. Results are often depicted as a heatmap. | [23,24,45,46] |
| “Riboleukogram” | This approach uses a mathematical technique similar to principal components analysis, in order to reduce the dimensionality of the data, and compare patients based on average expression vectors over time. | [36,71] |
| Classification and regression trees (CART) | Optimal predictors and cutoff values are determined by means of an algorithm that evaluates all possible combinations. CART has been used to identify a diagnostic panel of serum biomarkers based on findings from whole genome expression profiling. | [59] |
| Multiple time point data | ||
| Timecourse ANOVA (TANOVA) | Accommodates multifactorial data to determine if variations in gene expression over time are related to the condition of interest, or an independent factor (eg. age). | [69] |
| Average time curve | This method involves determining whether the population average time course is best represented by a flat line, suggesting no difference in expression over time, or by a curve (cubic splines), indicating a significant change over time. | [70] |
| Visualization | ||
| Gene expression mosaics | The Gene Expression Dynamics Inspector (GEDI) platform is used to generate a color representation of gene expression patterns based on self-organizing maps [58]. These expression mosaics lend themselves to human pattern recognition, as well as computer-based recognition. | [49,63] |
Additional challenges remain (Box 1), including the need for more comprehensive clinical data by which to annotate gene expression patterns, and for more reliable diagnostic categories with which to label patients with sepsis syndromes. These should not only include basic “case/control” and “survivor/nonsurvivor” categories, but also more nuanced labels related to illness trajectory, and response to therapeutic interventions. New methods continue to be developed including RNA-Seq, sequencing of microRNAs [72], and evaluation of microbial nucleic acid signals.
Box 1: Outstanding Questions.
Which source tissue (whole blood, neutrophils, PBMCs) is best suited to generate the gene expression data needed to address a particular biological hypothesis?
How should time series gene expression data be collected, analyzed, and merged with clinical outcomes data
What are the optimal transcriptome-based definition and classification of sepsis syndromes?
Can gene expression events early in the course of sepsis predict later transcriptional events, response to therapies, and clinical outcome?
What is the role of next-generation sequencing technologies in the transcriptome profiling of sepsis syndromes?
HIGHLIGHTS.
Gene expression analysis has yielded insights into the pathophysiology of sepsis.
Gene expression analysis in sepsis differs conceptually and pragmatically from cancer studies.
Gene expression data must be “downsized” to be useful in the ICU.
Rapid assays are needed to make gene expression useful in the ICU.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Alizadeh AA, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 2.van 't Veer LJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 3.Paez JG. EGFR mutations in lungcvancer: Correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 4.Levy MM, et al. 2001 SCCM/ESICM/ACCP/ATS/SIS international sepsis definitions conference. Crit Care Med. 2003;31:1250–1256. doi: 10.1097/01.CCM.0000050454.01978.3B. [DOI] [PubMed] [Google Scholar]
- 5.Dombrovskiy VY, et al. Rapid increase in hospitalization and mortality rates for severe sepsis in the United States: A trend analysis from 1993 to 2003. Crit Care Med. 2007;35:1244–1250. doi: 10.1097/01.CCM.0000261890.41311.E9. [DOI] [PubMed] [Google Scholar]
- 6.Hartman ME, et al. Trends in the epidemiology of pediatric severe sepsis. Pediatr Crit Care Med. 2013;14:686–693. doi: 10.1097/PCC.0b013e3182917fad. [DOI] [PubMed] [Google Scholar]
- 7.Sørensen TI, et al. Genetic and environmental influences on premature death in adult adoptees. N Engl J Med. 1988;318:727–732. doi: 10.1056/NEJM198803243181202. [DOI] [PubMed] [Google Scholar]
- 8.Calvano SE, et al. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–1037. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- 9.Fitting C, et al. Compartmentalization of tolerance to endotoxin. The J Infect Dis. 2004;189:1295–1303. doi: 10.1086/382657. [DOI] [PubMed] [Google Scholar]
- 10.Ge Y, et al. Relationship of tissue and cellular interleukin-1 and lipopolysaccharide after endotoxemia and bacteremia. J Infect Dis. 1997;176:1313–1321. doi: 10.1086/514127. [DOI] [PubMed] [Google Scholar]
- 11.Talwar S. Gene expression profiles of peripheral blood leukocytes after endotoxin challenge in humans. Physiol Genomics. 2006;25:203–215. doi: 10.1152/physiolgenomics.00192.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao W, et al. A genomic storm in critically injured humans. The J Exp Med. 2011;208:2581–2590. doi: 10.1084/jem.20111354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Phua J, et al. Characteristics and outcomes of culture-negative versus culture-positive severe sepsis. Crit Care. 2013;17:R202. doi: 10.1186/cc12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palmer C, et al. Cell-type specific gene expression profiles of leukocytes in human peripheral blood. BMC Genomics. 2006;7:1–15. doi: 10.1186/1471-2164-7-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang BMP, et al. The use of gene-expression profiling to identify candidate genes in human sepsis. Am J Respir Crit Care Med. 2007;176:676–684. doi: 10.1164/rccm.200612-1819OC. [DOI] [PubMed] [Google Scholar]
- 16.Tang BMP, et al. Gene-expression profiling of peripheral blood mononuclear cells in sepsis. Crit Care Med. 2009;37:882–888. doi: 10.1097/CCM.0b013e31819b52fd. [DOI] [PubMed] [Google Scholar]
- 17.Vartanian K, et al. Gene expression profiling of whole blood: Comparison of target preparation methods for accurate and reproducible microarray analysis. BMC Genomics. 2009;10:2. doi: 10.1186/1471-2164-10-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parnell GP, et al. Identifying key regulatory genes in the whole blood of septic patients to monitor underlying immune dysfunctions. Shock. 2013;40:166–174. doi: 10.1097/SHK.0b013e31829ee604. [DOI] [PubMed] [Google Scholar]
- 19.Prabhakar U, et al. Correlation of protein and gene expression profiles of inflammatory proteins after endotoxin challenge in human subjects. DNA Cell Biol. 2005;24:410–431. doi: 10.1089/dna.2005.24.410. [DOI] [PubMed] [Google Scholar]
- 20.Tang BM, et al. Genome-wide transcription profiling of human sepsis: a systematic review. Crit Care. 2010;14:R237. doi: 10.1186/cc9392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hotchkiss RS, et al. Immunosuppression in sepsis: a novel understanding of the disorder and a new therapeutic approach. Lancet Infect Dis. 2013;13:260–268. doi: 10.1016/S1473-3099(13)70001-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shankar AHA, Prasad ASA. Zinc and immune function: the biological basis of altered resistance to infection. Am J Clin Nutr. 1998;68:447S–463S. doi: 10.1093/ajcn/68.2.447S. [DOI] [PubMed] [Google Scholar]
- 23.Wong HR, et al. Genome-level expression profiles in pediatric septic shock indicate a role for altered zinc homeostasis in poor outcome. Physiol Genomics. 2007;30:146–155. doi: 10.1152/physiolgenomics.00024.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wong HR, et al. Identification of pediatric septic shock subclasses based on genome-wide expression profiling. BMC Medicine. 2009;7:34. doi: 10.1186/1741-7015-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Srinivasan MG, et al. Zinc adjunct therapy reduces case fatality in severe childhood pneumonia: a randomized double blind placebo-controlled trial. BMC Medicine. 2012;10 doi: 10.1186/1741-7015-10-14. 14–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhatnagar S, et al. Zinc as adjunct treatment in infants aged between 7 and 120 days with probable serious bacterial infection: a randomised, double-blind, placebo-controlled trial. Lancet. 2012;379:2072–2078. doi: 10.1016/S0140-6736(12)60477-2. [DOI] [PubMed] [Google Scholar]
- 27.Prasad AS, et al. Zinc supplementation decreases incidence of infections in the elderly: effect of zinc on generation of cytokines and oxidative stress. Am J Clin Nutr. 2007;85:837–844. doi: 10.1093/ajcn/85.3.837. [DOI] [PubMed] [Google Scholar]
- 28.Heyland D, et al. A randomized rrial of glutamine and antioxidants in critically ill patients. N Engl J Med. 2013;368:1489–1497. doi: 10.1056/NEJMoa1212722. [DOI] [PubMed] [Google Scholar]
- 29.Carcillo JA, et al. The randomized comparative pediatric critical illness stress-induced immune suppression (CRISIS) prevention trial. Pediatr Crit Care Med. 2012;13:165–173. doi: 10.1097/PCC.0b013e31823896ae. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vanlaere I, Libert C. Matrix metalloproteinases as drug targets in infections caused by gram-negative bacteria and in septic shock. Clin Microbiol Rev. 2009;22:224–239. doi: 10.1128/CMR.00047-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagase H. Matrix Metalloproteinases. J Biol Chem. 1999;274:21491–21494. doi: 10.1074/jbc.274.31.21491. [DOI] [PubMed] [Google Scholar]
- 32.Solan PD, et al. A novel role for matrix metalloproteinase-8 in sepsis. Crit Care Med. 2012;40:379–387. doi: 10.1097/CCM.0b013e318232e404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yazdan-Ashoori P, et al. Elevated plasma matrix metalloproteinases and their tissue inhibitors in patients with severe sepsis. J Crit Care. 2011;26:556–565. doi: 10.1016/j.jcrc.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 34.Cuenca AG, et al. Development of a genomic metric that can be rapidly used to predict clinical outcome in severely injured trauma patients. Crit Care Med. 2013;41:1175–1185. doi: 10.1097/CCM.0b013e318277131c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spielman RS, et al. Common genetic variants account for differences in gene expression among ethnic groups. Nature Genetics. 2007;39:226–231. doi: 10.1038/ng1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McDunn JE, et al. Plasticity of the systemic inflammatory response to acute infection during critical illness: Development of the riboleukogram. PLoS ONE. 2008;3:e1564. doi: 10.1371/journal.pone.0001564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barnato AE, et al. Racial variation in the incidence, care, and outcomes of severe sepsis. Am J Respir Crit Care Med. 2008;177:279–284. doi: 10.1164/rccm.200703-480OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wynn JL, et al. The influence of developmental age on the early transcriptomic response of children with septic shock. Mol. Med. 2011;17:1146–1156. doi: 10.2119/molmed.2011.00169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bouchon A, et al. TREM-1 amplifies inflammation and is a crucial mediator of septic shock. Nature. 2001;410:1103–1107. doi: 10.1038/35074114. [DOI] [PubMed] [Google Scholar]
- 40.Gibot S. Clinical review: role of triggering receptor expressed on myeloid cells-1 during sepsis. Crit Care. 2005;9:485–489. doi: 10.1186/cc3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turnbull IR, et al. Effects of aging on the immunopathologic response to sepsis. Crit Care Med. 2009;37:1018–1023. doi: 10.1097/CCM.0b013e3181968f3a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fermin DR, et al. Sex and age dimorphism of myocardial gene expression in nonischemic human heart failure. Circulation: Cardiovascular Genetics. 2008;1:117–125. doi: 10.1161/CIRCGENETICS.108.802652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jenner RG, Young RA. Insights into host responses against pathogens from transcriptional profiling. Nature Rev Microbiology. 2005;3:281–294. doi: 10.1038/nrmicro1126. [DOI] [PubMed] [Google Scholar]
- 44.Yu S-L, et al. Differential gene expression in gram-negative and gram-positive sepsis. Am J Respir Crit Care Med. 2004;169:1135–1143. doi: 10.1164/rccm.200211-1278OC. [DOI] [PubMed] [Google Scholar]
- 45.Tang BMP, et al. Gene-expression profiling of gram-positive and gram-negative sepsis in critically ill patients. Crit Care Med. 2008;36:1125–1128. doi: 10.1097/CCM.0b013e3181692c0b. [DOI] [PubMed] [Google Scholar]
- 46.Ramilo O, et al. Gene expression patterns in blood leukocytes discriminate patients with acute infections. Blood. 2007;109:2066–2077. doi: 10.1182/blood-2006-02-002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marshall JC. Sepsis: rethinking the approach to clinical research. J Leukocyte Biol. 2007;83:471–482. doi: 10.1189/jlb.0607380. [DOI] [PubMed] [Google Scholar]
- 48.Vincent J-L, et al. Sepsis definitions: time for change. Lancet. 2013;381:774–775. doi: 10.1016/S0140-6736(12)61815-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wong HR, et al. Validation of a gene expression-based subclassification strategy for pediatric septic shock. Crit Care Med. 2011;39:2511–2517. doi: 10.1097/CCM.0b013e3182257675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maslove DM, et al. Identification of sepsis subtypes in critically ill adults using gene expression profiling. Crit Care. 2012;16:R183. doi: 10.1186/cc11667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wong HR, et al. Interleukin-27 is a novel candidate diagnostic biomarker for bacterial infection in critically ill children. Crit Care. 2012;16:R213–R213. doi: 10.1186/cc11847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wong HR, et al. Interleukin 27 as a sepsis diagnostic biomarker in critically ill adults. Shock. 2013;40:382–386. doi: 10.1097/SHK.0b013e3182a67632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Calfee CS, et al. Plasma interleukin-8 is not an effective risk stratification tool for adults with vasopressor-dependent septic shock. Crit Care Med. 2010;38:1436–1441. doi: 10.1097/CCM.0b013e3181de42ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pachot A, et al. Systemic transcriptional analysis in survivor and non-survivor septic shock patients: a preliminary study. Immunol Lett. 2006;106:63–71. doi: 10.1016/j.imlet.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 55.Pachot A, et al. Decreased expression of the fractalkine receptor CX3CR1 on circulating monocytes as new feature of sepsis-induced immunosuppression. J Immunol. 2008;180:6421–6429. doi: 10.4049/jimmunol.180.9.6421. [DOI] [PubMed] [Google Scholar]
- 56.Nowak JE, et al. Admission chemokine (C-C motif) ligand 4 levels predict survival in pediatric septic shock. Pediatr Crit Care Med. 2010;11:213–216. doi: 10.1097/PCC.0b013e3181b8076c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Standage SW, Wong HR. Biomarkers for pediatric sepsis and septic shock. Expert Rev Anti Infect Ther. 2011;9:71–79. doi: 10.1586/eri.10.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kaplan JM, Wong HR. Biomarker discovery and development in pediatric critical care medicine. Pediatr Crit Care Med. 2011;12:165–173. doi: 10.1097/PCC.0b013e3181e28876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wong HR, et al. The pediatric sepsis biomarker risk model. Crit Care. 2012;16:R174. doi: 10.1186/cc11652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wong HR, et al. Testing the prognostic accuracy of the updated pediatric sepsis biomarker risk model. PLoS ONE. 2104 doi: 10.1371/journal.pone.0086242. (In press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wong HR, et al. A multi-biomarker-based outcome risk stratification model for adult septic shock. Crit Care Med. 2014 Dec 11; doi: 10.1097/CCM.0000000000000106. [Epub ahead of print] PMID: 24335447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eichler GS, et al. Gene Expression Dynamics Inspector (GEDI): for integrative analysis of expression profiles. Bioinformatics. 2003;19:2321–2322. doi: 10.1093/bioinformatics/btg307. [DOI] [PubMed] [Google Scholar]
- 63.Wong HR, et al. Toward a clinically feasible gene expression-based subclassification strategy for septic shock: Proof of concept. Crit Care Med. 2010;38:1955–1961. doi: 10.1097/CCM.0b013e3181eb924f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Geiss GKG, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 65.Kwan A, et al. Transcriptional instability during evolving sepsis may limit biomarker nased risk stratification. PLoS ONE. 2013;8:e60501. doi: 10.1371/journal.pone.0060501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zeng Y, Garcia-Frias J. A novel HMM-based clustering algorithm for the analysis of gene expression time-course data. Comput Stat Data Anal. 2006;50:2472–2494. [Google Scholar]
- 67.Geng H, et al. Applications of hidden markov models in microarray gene expression data. In: Dymarski P, editor. Hidden Markov Models, Theory and Applications. 2011. pp. 249–268. In Tech. [Google Scholar]
- 68.Ming Y, Kendziorski C. Hidden Markov Models for Microarray Time Course Data in Multiple Biological Conditions. J Am Stat Assoc. 2006;101:1323–1332. [Google Scholar]
- 69.Zhou B, et al. Analysis of factorial time-course microarrays with application to a clinical study of burn injury. Proc Natl Acad Sci U S A. 2010;107:9923–9928. doi: 10.1073/pnas.1002757107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Storey JD, et al. Significance analysis of time course microarray experiments. Proc Natl Acad Sci U S A. 2005;102:12837–12842. doi: 10.1073/pnas.0504609102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Polpitiya AD, et al. Using systems biology to simplify complex disease: Immune cartography. Crit Care Med. 2009;37:S16–S21. doi: 10.1097/CCM.0b013e3181920cb0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ma Y, et al. Genome-Wide Sequencing of Cellular microRNAs Identifies a Combinatorial Expression Signature Diagnostic of Sepsis. PLoS ONE. 2013;8:e75918. doi: 10.1371/journal.pone.0075918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tusher VG, et al. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Storey JD, et al. The optimal discovery procedure for large-scale significance testing, with applications to comparative microarray experiments. Biostatistics. 2007;8:414–432. doi: 10.1093/biostatistics/kxl019. [DOI] [PubMed] [Google Scholar]