Abstract
Circadian clocks integrate environmental signals with internal cues to coordinate diverse physiological outputs so that they occur at the most appropriate season or time of day. Recent studies using systems approaches, primarily in Arabidopsis, have expanded our understanding of the molecular regulation of the central circadian oscillator and its connections to input and output pathways. Similar approaches have also begun to reveal the importance of the clock for key agricultural traits in crop species. In this review, we discuss recent developments in the field, including: a new understanding of the molecular architecture underlying the plant clock; mechanistic links between clock components and input and output pathways; and our growing understanding of the importance of clock genes for agronomically important traits.
Keywords: Circadian clock, Arabidopsis, systems biology, input, output, agricultural traits
Complexity of the circadian clock
Circadian clocks enhance growth and fitness by coordinating numerous biological events with the environment [1]. Conceptually, the circadian system is composed of three major parts: a self-sustaining central oscillator (or clock), input pathways that integrate oscillator function with environmental timing cues, and output pathways that control diverse processes. The influence of the clock on plant life is inescapable because the abundance of thousands of transcripts and the timing of multiple processes that occur throughout the plant life cycle are clock-regulated [2]. Both the molecular mechanisms underlying the circadian clock and its interactions with environmental sensing and physiological output pathways involve sophisticated and complex networks and, thus, represent appealing problems for systems biology approaches.
Recent large-scale approaches have considerably advanced our understanding of the plant clock. Genomic, transcriptomic, and proteomic approaches have led to the identification of novel clock components and downstream targets [3–11], helping to reveal the network structure of the clock system. In addition to experimental approaches, mathematical modeling and computational techniques have helped elucidate how known components function both within the oscillator network and as links to other physiological pathways and have in some cases led to the prediction of unknown players [12–16]. These developments have not only improved our insight into how the plant clock mechanism works but also how it is connected to input and output pathways. In this review, we focus on new findings that provide a revised view of the architecture of the clock, increase our understanding of the connections between clock components and input and output pathways, and demonstrate the importance of clock genes for yield-related traits in diverse crop species.
Clock mechanism
To date, more than 20 clock or clock-associated components have been identified in Arabidopsis (Arabidopsis thaliana) (Table 1), and homologs of these genes in other plant species have begun to be characterized. In plants, as in other eukaryotes, the clock is composed of interlocking transcription–translation feedback loops. However, the architecture of the Arabidopsis circadian oscillator has been found to be significantly more complex than that of other model eukaryotes [3, 17]. Different clock proteins act at distinct times throughout the day and night to reciprocally regulate expression of other clock genes at both the transcriptional and post-transcriptional levels.
Table 1.
Molecular functions of clock genes in Arabidopsis
| Gene | AGI | Time of protein activity | Clock phenotypes of loss-of-function mutant | Molecular function | Refs. |
|---|---|---|---|---|---|
| Genes believed to function primarily within the clock system | |||||
| CCA1 | AT2G46830 | Morning | Short period (partially redundant with LHY) | Transcription factor; primarily represses genes with EE | [1] |
| LHY | AT1G01060 | Morning | Short period (partially redundant with CCA1) | Transcription factor; primarily represses genes with EE | [1] |
| PRR9 | AT2G46790 | After dawn | Long period (partially redundant with PRR7 and PRR5) | Transcription factor; represses expression of CCA1 and LHY | [27, 103] |
| PRR7 | AT5G02810 | After dawn | Long period (partially redundant with PRR9 and PRR5) | Transcription factor; represses expression of CCA1 and LHY | [27, 103] |
| RVE8 (LCL5) | AT3G09600 | Afternoon | Long period (partially redundant with RVE4 and RVE6) | Transcription factor; promotes expression of genes with EE | [3, 9, 10] |
| RVE4 | AT5G02840 | Presumably afternoon | No obvious period or phase change (partially redundant with RVE6 and RVE8) | Transcription factor; promotes expression of genes with EE | [3] |
| RVE6 | AT5G52660 | Presumably afternoon | No obvious period or phase change (partially redundant with RVE4 and RVE8) | Transcription factor; promotes expression of genes with EE | [3] |
| PRR5 | AT5G24470 | Afternoon | Short period (partially redundant with PRR9 and PRR7) | Transcription factor; represses expression of CCA1 and LHY; regulates TOC1 nuclear localization | [6, 27, 52] |
| PRR3 | AT5G60100 | Evening | Short period; tissue specific function | Putative transcription factor; modulates TOC1 stability | [33] |
| TOC1 (PRR1) | AT5G61380 | Evening | Short period | Transcription factor; represses expression of CCA1 and LHY | [4, 11, 14] |
| CHE | AT5G08330 | Evening | No obvious period or phase change | Transcription factor; modulates CCA1 expression | [8] |
| LUX | AT3G46640 | Evening | Arrhythmic under constant light | Transcription factor; represses expression of PRR9 and output genes | [7, 30] |
| BOA (NOX) | AT5G59570 | Evening | Both no phenotype and short period phenotypes have been reported | Transcription factor; activates expression of CCA1 and regulates output genes | [7, 22] |
| ELF3 | AT2G25930 | Evening | Arrhythmic under constant light | Transcriptional regulator; represses expression of PRR9 and output genes | [7, 29] |
| ELF4 | AT2G40080 | Evening | Arrhythmic under constant light | Transcriptional regulator; represses expression of PRR9 and output genes | [7, 31] |
| ZTL | AT5G57360 | Evening | Long period (partially redundant with FKF1 and LKP2) | F-box protein, blue-light receptor; regulates TOC1 protein stability | [47, 48] |
| Genes with multiple functions in addition to circadian regulation | |||||
| CKB4 | AT2G44680 | – | Not reported | Regulatory β subunit of CK2 kinase | [104] |
| FKF1 | AT1G68050 | – | Not obvious (partially redundant with ZTL and LKP2) | F-box protein, blue-light receptor; regulates CO expression | [47] |
| GI | AT1G22770 | – | Short period under most conditions | Nuclear protein, physically interacts with diverse proteins | [12, 44, 46, 50, 105, 106] |
| LKP2 | AT2G18915 | – | Not obvious (partially redundant with ZTL and FKF1) | F-box protein, blue-light receptor; regulates CO expression | [47] |
| PRMT5 | AT4G31120 | – | Long period | Methyl transferase that demethylates a splicing factor | [38, 39] |
| SKIP | AT1G77180 | – | Long period | SNW/Ski-interacting protein, a component of the spliceosome | [40] |
| STIPL1 | AT1G17070 | – | Long period | Splicing factor | [41] |
| Genes with unknown roles within the circadian system | |||||
| LNK1 | AT5G64170 | – | Not obvious (redundant with LNK2) | Nuclear protein with unknown function | [28] |
| LNK2 | AT3G54500 | – | Long period (redundant with LNK1) | Presumably nuclear protein | [28] |
| FIO1 | AT2G21070 | – | Long period | Nuclear protein with unknown function | [107] |
| JMJD5 | AT3G20810 | – | Short period | Putative histone demethylase | [108, 109] |
| LIP1 | AT5T64813 | – | Short period | Small GTPase | [110] |
| LWD1 | AT1G12910 | – | Not obvious (redundant with LWD2) | Nuclear protein; transcription factor? | [111, 112] |
| LWD2 | AT3G26640 | – | Not obvious (redundant with LWD1) | Nuclear protein; transcription factor? | [111, 112] |
| TEJ | AT2G31870 | – | Long period | Poly (ADP-ribose) glycohydrolase | [113] |
| TIC | AT3G22380 | – | Short period | Nuclear protein with unknown function | [114, 115] |
| XCT | AT2G21150 | – | Short period | Nuclear protein with unknown function | [116] |
Transcriptional regulation
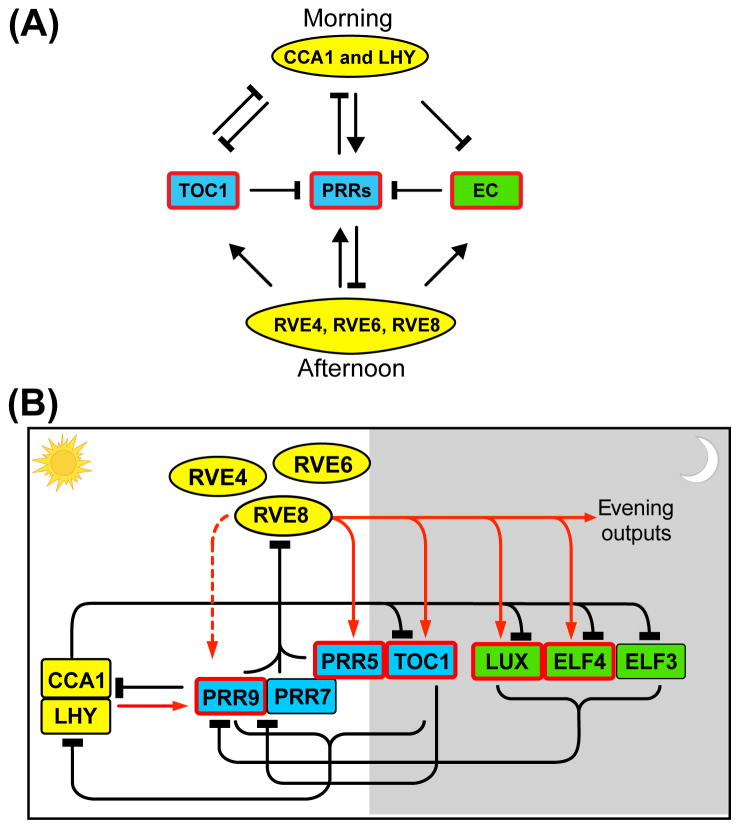
Many clock components in Arabidopsis regulate the transcription of other, differentially phased, clock genes. As detailed below, recent work has led to the identification of new clock components and to the reassignment of function of previously known proteins. A simplified schematic of the network structure is shown in Figure 1A and a more complete diagram is shown in Figure 1B. The clock components described below are also listed in Table 1.
Figure 1.
Transcriptional regulation of the clock in Arabidopsis.(A) A simplified model for transcriptional regulation among classes of clock components. CCA1, LHY and RVE components are shown in yellow; pseudo response regulator (PRR) components are shown in blue; evening complex (EC) components are shown in green. Members of each group that contain the evening element(s) in their promoter regions are marked with red boxes. Arrows indicate activation; perpendicular bars indicate repression. In the morning, CCA1 and LHY repress most EE-regulated clock genes; whereas in the afternoon, RVE4, RVE6, and RVE8 activate EE-regulated clock genes. These EE-controlled components also feed back to regulate expression of CCA1, LHY, RVE4, RVE6, RVE8 and each other. (B) A more detailed model for transcriptional regulation among the clock components. The relative timing of action for each component during a day–night cycle is shown from left to right. White area indicates subjective day; gray area indicates subjective night. Yellow, blue and green colors indicate members of each family described above. Genes with EE(s) in their promoter regions are marked with red boxes. Solid red arrows indicate activation; broken red arrow indicates conditional activation; perpendicular bars indicate repression.
Morning-phased components (CCA1 and LHY)
CIRCADIAN CLOCK-ASSOCIATED 1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) are two MYB-like transcription factors. Their transcript and protein levels are highly abundant in the morning and these proteins physically interact with each other [18, 19]. They have long been known to repress expression of an evening-phased component, TIMING OF CAB EXPRESSION 1 (TOC1), also known as PSEUDO-RESPONSE REGULATOR 1 (PRR1), via binding to a motif in the TOC1 promoter called the evening element (EE) [20, 21]. TOC1 in turn directly regulates expression of CCA1 and LHY, forming the first described plant clock transcriptional feedback loop (see below).
Recently, CCA1 and LHY have also been shown to repress several other evening genes, including GIGANTEA (GI), LUX ARRHYTHMO (LUX), BROTHER OF LUX ARRHYTHMO (BOA, also known as NOX, meaning ‘night’ in Latin), EARLY FLOWERING 3 (ELF3) and ELF4 [22–24]. The repression of evening genes by CCA1 and LHY is dependent on DEETIOLATED1 (DET1), a key repressor in photomorphogenesis [25]. Besides functioning as transcriptional repressors, genetic data suggest that CCA1 and LHY may also function as activators for the day-phased clock genes PRR9 and PRR7 [24].
Day-phased components (PRR9, PRR7, LNK1 and LNK2)
PRR9 and PRR7, together with their homologs PRR5 and PRR1 (TOC1), play key roles in the clock. PRR9 is expressed earliest, just after dawn, followed by PRR7, PRR5, PRR3 and finally TOC1 in the evening [26]. PRR9 and PRR7, together with PRR5, play partially redundant roles in repressing expression of the morning genes CCA1 and LHY [27]. Recently, the PRRs have been shown to also repress expression of REVEILLE8 (RVE8, also known as LHY and CCA1-like 5, LCL5) (see below) [6, 9]. The importance of the PRRs to clock function is underscored by the arrhythmic phenotype of prr9 prr7 prr5 triple mutants [26].
A small family of genes with clock-regulated and light-induced expression has recently been implicated in clock function. NIGHT LIGHT-INDUCIBLE AND CLOCK-REGULATED GENE 1 (LNK1) and LNK2 transcripts peak near the middle of the subjective day and loss of these genes causes a long period phenotype and downregulation of many clock-regulated genes with afternoon and evening phases [28]. Transcript levels of PRR5 and ELF4 are significantly reduced in lnk1 lnk2 double mutants, suggesting that LNK1 and LNK2 may act as transcriptional activators for these genes in the afternoon [28].
Afternoon-phased components (RVE8, and probably RVE4 and RVE6)
Several genes homologous to CCA1 and LHY have recently been identified as important clock components [3, 10]. Several of these RVE transcription factors have dawn-phased clock-regulated gene expression, which suggests that they might provide an important signal to plants at dawn (hence their name ‘REVEILLE’, which evokes the bugle call starting the military day). Like CCA1 and LHY, RVE8 binds to the EE [9], a motif overrepresented among promoters of clock-regulated genes that have an evening phase [20]. However, unlike CCA1 and LHY, which act as repressors of the EE, experiments with plants expressing an inducible form of RVE8 demonstrated that RVE8 induces hundreds of evening genes that contain EEs in their promoters. The direct nature of this regulation was shown for many evening-phased EE-containing clock genes, including PRR5, TOC1, GI, LUX and ELF4, since their expression increased upon RVE8 induction even in the presence of an inhibitor of translation [3, 10]. In addition, RVE8 has been shown to associate with the PRR5 and TOC1 promoters [3, 10]. Two close homologs of RVE8, RVE4 and RVE6, also play partially redundant roles with RVE8 within the circadian system. Plants mutant for all three of these genes exhibit a more extreme long-period phenotypes than the rve8 single mutant and the rve4 rve6 rve8 triple mutant has lost the predominant afternoon-phased EE-binding activity [3]. This finding, along with the observation that RVE8 protein levels peak in the subjective afternoon [9], indicates that at least some RVEs are afternoon-phased clock components and suggests that a better name for them might have been ‘SUNSET’ (a bugle call signaling the end of the official day).
Evening-phased components (PRR5, TOC1, CHE, LUX, BOA (NOX), ELF3 and ELF4)
As mentioned above, PRR5 forms a negative feedback loop with RVE8 [9] and plays a partially redundant role with PRR9 and PRR7 to repress the morning genes CCA1 and LHY [27]. The role of TOC1 in the regulation of CCA1 and LHY was ambiguous until recently. Experimental and computational modeling work now indicate that TOC1 represses CCA1 and LHY expression, similar to the roles of the TOC1 homologs PRR9, PRR7 and PRR5 [4, 11, 14]. Together, these four PRR proteins ensure that CCA1 and LHY are only expressed during a small fraction of each day. In addition, CCA1 HIKING EXPEDITION (CHE) interacts with TOC1 to help repress CCA1 expression in an as-yet undefined manner [8]. Finally, LUX (a MYB-like transcription factor), ELF3 and ELF4 (two unrelated novel nuclear proteins) interact to form the ‘evening complex’ that represses expression of the day-phased clock gene PRR9 [7, 29–31]. Mutation of any member of the evening complex causes plants to become arrhythmic [24]. A homolog of LUX, alternatively called BOA or NOX, has been reported to form a complex with ELF3 and ELF4 (like LUX) and also directly promotes CCA1 expression [7, 22].
The EE has emerged as an essential regulatory nexus for central clock oscillation. Most clock components either regulate the EE (CCA1 and LHY as repressors and RVE4, RVE6, and RVE8 as activators) [3, 4, 9, 21] or are regulated by other clock components through EE in their promoters (PRR5, TOC1, LUX, ELF4, GI and PRR9) (Figure 1) [3, 9, 24]. This reciprocal regulation between clock components ensures the ordered expression of clock genes and forms a complex interconnected network comprising the plant circadian oscillator. Emerging data indicate that regulation of histone modifications also plays an important role in this transcriptional network [32]. As if this picture were not complicated enough already, there are also cell- and tissue-specific variations of the oscillator network described above [33–35]. For example, PRR3 is a modulator of TOC1 stability that acts primarily in the vasculature [33] and different cell types in leaves have slightly different free-running periods and expression levels of some clock genes [34].
Post-transcriptional regulation
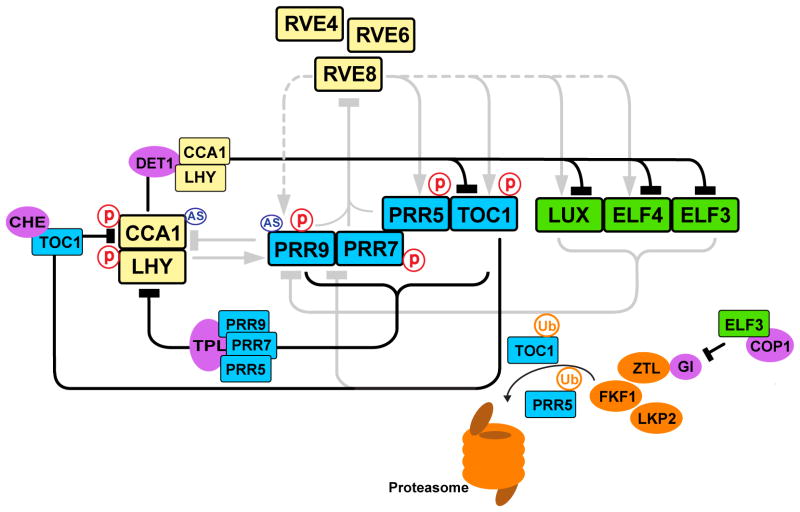
In addition to transcriptional regulation, many post-transcriptional regulatory mechanisms are key to the plant circadian oscillator (Figure 2).
Figure 2.
Post-transcriptional regulation of the clock in Arabidopsis. CCA1, LHY and RVE components are shown in yellow; pseudo response regulator components are shown in blue; evening complex components are shown in green. F-box proteins that are a part of E3 ubiquitin ligase complex are shown in orange. Other interacting proteins are shown in magenta. Abbreviations: AS, alternative splicing; P, protein phosphorylation; Ub, ubiquitination.
Alternative splicing
Alternative splicing has been reported for many clock genes (CCA1, LHY, PRR9, PRR7, PRR5, PRR3, TOC1, RVE4 and RVE8) [36, 37]. PROTEIN ARGININE METHYLTRANSFEREASE 5 (PRMT5) regulates the alternative splicing of PRR9, presumably via its methylation of splicing factors, and prmt5 mutants have a long circadian period [38, 39]. SNW/Ski-interacting protein (SKIP), a component of the spliceosome, also regulates alternative splicing of PRR9 and other clock genes including PRR7, CCA1, LHY, and TOC1 [40]. Similar to PRMT5, loss of SKIP causes a long period phenotype[ 40]. A third protein involved in mRNA splicing has also been recently implicated in clock function. Mutation of SPLICEOSOMAL TIMEKEEPER LOCUS1 (STIPL1), a homolog of a human spliceosomal proteins involved in spliceosome disassembly, causes a long period phenotype [41]. As reported for skip mutants, the accumulation of splicing variants of several clock gene transcripts is altered in stipl1 mutants, making it difficult to conclude whether altered splicing of one or more genes causes the clock phenotype [41].
Further evidence of a role for alternative splicing in the circadian oscillator comes from work on CCA1. A splice variant CCA1 transcript with a retained fourth intron produces a protein called CCA1β that lacks the MYB-like DNA-binding domain. CCA1β can interfere with CCA1 function by interacting with full-length CCA1 (CCA1α) or LHY to form nonfunctional heterodimers [42]. CCA1pro::CCA1α cca1-2 plants that do not express CCA1β have a long period, whereas 35S::CCA1β has a short period similar to cca1 lhy double mutants [42]. Hence this naturally produced CCA1β modulates activity of full-length CCA1α protein and fine-tunes the pace of the clock.
Protein–protein interactions and regulation of protein stability
Several clock components are known to interact with other proteins that are essential for their regulatory activity. As mentioned earlier, the repressive activity of CCA1 and LHY on evening genes relies on DET1 [25]. Similarly, PRR9, PRR7 and PRR5 physically interact with a transcriptional co-repressor, TOPLESS, to inhibit expression of CCA1 and LHY [43]. GI has been shown to have separable functions in the cytoplasm and then nucleus [44, 45] and its partitioning between these cellular compartments is regulated by physical interactions with ELF4 [46].
A common theme in all eukaryotic circadian systems is the regulated degradation of clock proteins. A key enzyme involved in regulated protein degradation in the plant clock is ZEITLUPE (ZTL), a F-box protein with a blue light photosensing LOV (light, oxygen, and voltage) domain and a KELCH protein-protein interaction domain. Along with its two homologs, FLAVIN BINDING, KELCH REPEAT, F-BOX (FKF1) and LOV KELCH PROTEIN 2 (LKP2), ZTL targets TOC1 and PRR5 for ubiquitination and subsequent proteasome degradation [47, 48]. GI physically interacts with ZTL and stabilizes it, while ZTL also controls GI stability and nucleocytoplasmic partitioning [49]. Finally, the physical interaction between ELF3, the E3-ubiquitin ligase CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1) and GI promotes GI degradation [50].
Some post-translational modifications also affect clock protein stability and interactions with other proteins. Elevated phosphorylation of all of the PRR proteins is observed before their degradation by the proteasome [51], suggesting this may be a means to control their stability. Indeed, phosphorylation of PRR5 and TOC1 promotes their interactions with ZTL and enhances their subsequent degradation [52]. PRR5 also interacts with TOC1, enhancing its accumulation in the nucleus and preventing it from being targeted for degradation by ZTL, which is exclusively found in the cytoplasm [52].
Interactions between inputs and the clock
The central clock can be synchronized with the environment by input pathways that sense external timing cues. Light and temperature are the two most studied entraining signals. Light influences the clock in many ways, affecting transcription, messenger RNA stability and translation rate of several clock genes [53] and the protein stability of several others [54]. Reciprocally, several clock components are involved in the regulation of light input pathways [55].
Recent studies have provided clues as to how light mediates these effects on the clock. In addition to ZTL, the blue light circadian photoreceptor that regulates TOC1 protein stability (see above), plants also possess phytochromes (reversible red and far-red light receptors) and cryptochromes (blue light receptors). Both phytochromes and cryptochromes are involved in the entrainment of the clock and contribute to fluence rate-dependent shortening of the period [55]. Recent studies on phyA phyB phyC phyD phyE quintuple mutant have revealed that phytochromes do not simply increase the pace of the clock in a light-dependent manner [56, 57]. As expected based on the period-shortening effects of light, the phyA phyB phyC phyD phyE mutant has a longer period than wild type at relatively high fluence rates of continuous red light. However, at lower fluence rates (<10 μmol m−2 s−1) the phyA phyB phyC phyD phyE mutant has a shorter period than that of wild type [56]. To explain this fluence-rate dependent effect, it has been proposed that the Pfr (far-red light absorbing) form of phytochromes decreases the pace of the clock whereas the Pr (red-light absorbing) form accelerates clock pace [56].
To more closely examine how light inputs shape the clock system, plants were entrained with far-red light so that PHYA is expected to be the only active photoreceptor [58]. Surprisingly, clock gene expression is significantly altered under far-red light with elevated expression of evening genes and decreased expression of morning genes [58]. Through mutant analysis, ELF4 was identified as a potential target involved in these far-red effects [58]. In addition, a study has shown that three positive regulators of PHYA signaling pathways, Far-red Elongated Hypocotyl 3 (FHY3), Far-red Impaired Response 1 (FAR1) and LONG HYPOCOTYL 5 (HY5) activate ELF4 directly during the day [59], thus providing a clue as to how far-red light signaling affects the central oscillator.
Although daily changes in temperature can entrain the circadian clock [1], clock pace is largely invariant when plants are grown across a range of different physiologically relevant temperatures, a phenomenon called temperature compensation. PRR7 and PRR9 have been shown to be essential for this process, preferentially repressing expression of CCA1 and LHY at elevated temperatures [60]. In addition, several other clock genes show altered expression after short or long exposure to cold temperatures [36, 61], perhaps playing a role in temperature compensation. Surprisingly, a recent study has implicated the cryptochrome blue light photoreceptors in temperature compensation [13]. Using experimental and computational modeling approaches, the authors of this study suggest that light and temperature pathways may converge on common target genes within the circadian oscillator [13].
Temperature also regulates the alternative splicing of clock genes, including CCA1, LHY, PRR9, PRR7, PRR5, PRR3, and TOC1 [36, 37]. As mentioned above, CCA1 has at least two physiologically relevant splicing variants: CCA1α and CCA1β, which encode full-length and truncated CCA1 proteins, respectively. Under low temperatures, CCA1β production is suppressed [42]. Intriguingly, elevated CCA1α levels seem to be essential for cold tolerance because 35S::CCA1α plants have better freezing tolerance whereas 35S::CCA1β plants are hypersensitive to freezing [42]. Thus temperature-regulated splicing of clock genes is implicated not only in oscillator function but also in plant adaptation to temperature extremes. The circadian system is also involved in many other environmental response pathways, with a large fraction of clock-regulated output genes regulated by various stresses (see below) [2].
Interactions between the clock and outputs
Recently, several genome-wide studies have shown that central clock components, including TOC1, PRR5, PRR7, and RVE8, directly control groups of genes enriched for specific circadian phases and biological processes [3–6]. Current systems approaches have expanded our understanding of how these processes, including growth, metabolism, and abiotic and biotic stresses, are influenced by the circadian system.
Growth
The clock generates a large number of circadian rhythms that optimize growth and development [62]. One particularly well-studied growth process is the diurnal control of hypocotyl elongation, with daily rhythms in growth that are influenced by external cues and by the internal clock. There are at least four pathways coordinated by the clock that are involved in rhythmic hypocotyl growth, including light, sucrose, hormone signaling pathways, and the direct action of clock components themselves. Together these four pathways converge upon PHYTOCHROME INTERACTING FACTOR4 (PIF4) and/or PIF5, basic helix–loop–helix (bHLH) transcription factors that promote hypocotyl growth. First, the evening complex clock proteins (ELF3, ELF4 and LUX) directly repress expression of PIF4 and PIF5 in the evening, thus generating circadian rhythms in PIF4 and PIF5 transcript levels with a peak phase in the late night and early morning [7, 23]. In addition, light and sucrose control PIF protein stability: PHYB promotes PIF4 and PIF5 degradation under red light [63], whereas high sucrose levels stabilize PIF5 [64]. Given that the clock influences phytochrome signaling [55, 65] and sugar metabolism (see below), the circadian system influences PIF4 and PIF5 abundance at multiple levels.
Another important regulator of daily rhythms in hypocotyl elongation is the hormone gibberellic acid (GA) [66]. The clock-controlled expression of GA 20ox1 (a GA biosynthetic enzyme) and GA-INSENSITIVE DWARF1 (GID1, which encodes a GA receptor) contribute to higher abundance of a GA–GID1 complex around dawn, which promotes degradation of DELLA transcription factors at this time [66, 67]. DELLA proteins are crucial repressors of the GA signaling pathway and block PIF4 activity by binding to the PIF4 DNA binding domain [68]. Degradation of DELLA proteins in the morning therefore helps fine-tune the daily timing of PIF4 activity and the expression of PIF4 targets that promote hypocotyl growth [68].
The hormone auxin is also involved in hypocotyl growth regulation. The clock regulates plant sensitivity to many stimuli in a time-of-day-specific manner, a phenomenon called ‘gating’. Auxin-induced gene expression is gated by the clock and the time of maximal auxin responsiveness correlates with the time of maximal clock-regulated hypocotyl growth [69]. The reduced auxin-induced growth of pif4 pif5 double mutants suggests that auxin regulates growth at least partially through PIF4 and PIF5 [70]. There are multiple connections between these bHLH factors and auxin signaling: PIF4 has been shown to influence auxin production [71, 72] and PIF4 and PIF5 both affect auxin signaling downstream of biosynthesis [70, 73, 74]. In addition, a clock-controlled transcription factor, RVE1, also regulates hypocotyl growth by controlling auxin biosynthesis genes in a PIF4 and PIF5-independent manner [75]. It seems likely that the circadian network influences growth in additional ways that have yet to be discovered. Further systems biology studies are needed to fully elucidate how diverse growth-regulatory pathways are coordinated with each other and with the circadian system.
Metabolism
Photosynthesis and carbohydrate metabolism have long been known to be clock-regulated. Many transcripts involved in photosynthesis, including both those encoded by the nuclear and chloroplast genomes, are controlled by the clock [76]. Circadian regulation of these two genomes is coordinated by SIGMA FACTOR 5 (SIG5), a subunit of the plastid RNA polymerase. SIG5 is a nuclear gene expressed with a circadian rhythm; upon import into the chloroplast, it generates circadian expression of several chloroplast genes involved in photosynthesis [77]. Daily rhythms in photosynthetic capacity, light availability, and sugar metabolism together lead to the daily accumulation and mobilization of starch, the major form in which fixed carbon is stored by plants. Plants accumulate transitory starch reserves during the day and break down these reserves during the night to support growth and metabolism. Intriguingly, the rate of starch breakdown is tightly controlled so that 95% of starch is consumed by the anticipated time of dawn, even when the time of lights off is altered in unexpected ways [78]. The authors of a recent study have suggested that this is achieved by biological molecules that carry out a sort of arithmetic division, calculating both the amount of starch that has accumulated by the end of the day and the time until dawn to determine the rate of starch degradation [15].
Many clock-controlled outputs also serve as clock input signals, feeding back on the oscillator mechanism. For example, sucrose can serve as an entrainment signal to reset the clock in constant darkness [12]. Sucrose also affects the amplitude of clock gene expression and modulates period length [79]. More recently, it has been demonstrated that the production of sugar through photosynthesis can reset the clock, via a pathway that requires PRR7 [80]. Similarly, not only is iron homeostasis controlled by the clock but application of iron also feeds back to regulate clock pace [81–83]. Glucosinolates, clock-controlled metabolites involved in plant defense, also fine-tune the pace of the clock [16]. It will be interesting to investigate the mechanisms by which these nutrients and metabolites interact with the central clock.
Abiotic and biotic stresses
Given the predictable nature of daily changes in the physical environment, it is not surprising that the circadian clock helps plants adapt to abiotic stresses. Perhaps more unexpectedly, several recent studies have also reported that the clock has a role in the response to biotic stresses.
It has long been known that the clock influences plant response to cold [84]. The major players in cold acclimation are the C-REPEAT BINDING FACTOR 1 (CBF1), CBF2 and CBF3 transcription factors that activate downstream cold-regulated (COR) genes. Transcript levels of CBFs are clock-regulated, peaking at midday, and cold-induction of these genes is gated by the clock [84]. Several clock components, including CCA1, PRR5, PRR7, and TOC1, have been reported to bind to promoters of some or all these CBFs [4, 6, 85]. CCA1 activates CBFs and promotes freezing tolerance [85] whereas PRR5 and PRR7 repress the expression of CBFs and inhibit freezing tolerance [86]. In addition to regulation of cold acclimation, CCA1 is also involved in other stress responses. Recently, reactive oxygen species (ROS) homeostasis was reported to be clock-controlled; this is likely to be through regulated expression of ROS genes by CCA1 [87]. Like several metabolites mentioned above, exogenous ROS also act as an input signal that affects clock pace [87].
Many genes implicated in plant defense against pathogens contain an EE in their promoters and are controlled by the clock [88]. Disruption of CCA1 in Arabidopsis results in decreased resistance to downy mildew at dawn (when infection is most likely to occur), whereas overexpression of CCA1 results in enhanced resistance [88]. Similarly, Arabidopsis is most resistant to Pseudomonas syringae in the subjective morning, a time when pathogen-associated molecular pattern (PAMP)-triggered immunity genes are highly expressed [89]. The circadian clock also provides enhanced defense against herbivores by promoting accumulation of the defense hormone jasmonate at times of likely herbivore attack [90]. Interestingly, metabolites involved in defense against herbivores continue to show daily rhythms in a variety of fruits and vegetables even after harvest [91], highlighting the importance of the clock throughout the process of food production. Finally, recent work has shown that not only does the clock affect immune responses but that immune responses can also affect clock pace, demonstrating crosstalk between these pathways [92].
Clock genes and agricultural traits
Although the plant clock has been best characterized in Arabidopsis, its importance in plants of agronomic importance is becoming clear [93]. Many species of crops have undergone whole genome duplication followed by considerable gene loss and diploidization. Studies in the crop Brassica rapa have shown that clock genes, including PRR and RVE family genes, were preferentially retained during this process [94], perhaps because of gene dosage constraints.
Numerous studies have shown that the circadian clock plays an essential role in agricultural traits. For example, photoperiodic control of flowering time is a key factor that determines crop performance. The Photoperiod-1 gene, a homolog of PRR7, is involved in flowering regulation in barley (Hordeum vulgare) and wheat (Triticum aestivum) [95, 96]. Recently rice (Oryza sativa) ELF3 (OsELF3) has been reported to control heading date under long days [97]. Two studies have also reported that a commercial barley variety bred for short-growing seasons carries a mutation in a homolog of ELF3 [98, 99]. The accelerated transition from vegetative to reproductive growth enables this barley variety to be cultivated in regions with short growing seasons. Early maturity in another barley variety is likely to be due to variation in a homolog of the Arabidopsis clock gene LUX [100]. Further, studies in wheat and sorghum (Sorghum bicolor) have also identified key photoperiodic flowering genes that are controlled by the clock [95, 101]. In addition to breeding, transgenic approaches toward yield improvement via manipulation of clock genes have also been successful. Constitutive expression of an Arabidopsis clock-regulated B-box domain gene (AtBBX32) in soybean (Glycine max) altered expression of clock gene homologs and significantly enhanced yield, perhaps by altering the timing of crucial phases of reproductive development [102].
Concluding remarks
The circadian clock is a complicated system that coordinates external stimuli and an internal timing mechanism to optimize growth and development. Further investigation into the functions of known clock components and the elucidation of interactions between the clock and input and output pathways will help us to understand how plants thrive in a predictably changing environment. As illustrated by recent studies in crop plants, these insights are likely to have important implications for agricultural improvement.
Highlights.
The circadian system is a complex regulatory network
Multiple links exist between the clock and other cellular pathways and signaling networks
Recent systems approaches have advanced our understanding of the clock system
Insights into the Arabidopsis clock have been applied to several crop species
Acknowledgments
We apologize to colleagues whose work could not be cited due to space limitations. We thank S. Brady and V. Sundaresan for careful reading of the manuscript. Work in the Harmer laboratory is supported by the National Institute of General Medical Science of the National Institutes of Health under award number R01GM069418 and the National Science Foundation under award number IOS 1238040.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Harmer SL. The circadian system in higher plants. Annu Rev Plant Biol. 2009;60:357–377. doi: 10.1146/annurev.arplant.043008.092054. [DOI] [PubMed] [Google Scholar]
- 2.Covington MF, et al. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 2008;9:R130. doi: 10.1186/gb-2008-9-8-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hsu PY, et al. Accurate timekeeping is controlled by a cycling activator in Arabidopsis. eLife. 2013;2:e00473. doi: 10.7554/eLife.00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang W, et al. Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science. 2012;336:75–79. doi: 10.1126/science.1219075. [DOI] [PubMed] [Google Scholar]
- 5.Liu T, et al. Direct regulation of abiotic responses by the Arabidopsis circadian clock component PRR7. Plant J. 2013;76:101–114. doi: 10.1111/tpj.12276. [DOI] [PubMed] [Google Scholar]
- 6.Nakamichi N, et al. Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc Natl Acad Sci U S A. 2012;109:17123–17128. doi: 10.1073/pnas.1205156109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nusinow DA, et al. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature. 2011;475:398–402. doi: 10.1038/nature10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pruneda-Paz JL, et al. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science. 2009;323:1481–1485. doi: 10.1126/science.1167206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rawat R, et al. REVEILLE8 and PSEUDO-REPONSE REGULATOR5 form a negative feedback loop within the Arabidopsis circadian clock. PLoS genetics. 2011;7:e1001350. doi: 10.1371/journal.pgen.1001350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farinas B, Mas P. Functional implication of the MYB transcription factor RVE8/LCL5 in the circadian control of histone acetylation. Plant J. 2011;66:318–329. doi: 10.1111/j.1365-313X.2011.04484.x. [DOI] [PubMed] [Google Scholar]
- 11.Gendron JM, et al. Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc Natl Acad Sci U S A. 2012;109:3167–3172. doi: 10.1073/pnas.1200355109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dalchau N, et al. The circadian oscillator gene GIGANTEA mediates a long-term response of the Arabidopsis thaliana circadian clock to sucrose. Proc Natl Acad Sci U S A. 2011;108:5104–5109. doi: 10.1073/pnas.1015452108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gould PD, et al. Network balance via CRY signalling controls the Arabidopsis circadian clock over ambient temperatures. Mol Syst Biol. 2013;9:650. doi: 10.1038/msb.2013.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pokhilko A, et al. The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Mol Syst Biol. 2012;8:574. doi: 10.1038/msb.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scialdone A, et al. Arabidopsis plants perform arithmetic division to prevent starvation at night. eLife. 2013;2:e00669. doi: 10.7554/eLife.00669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kerwin RE, et al. Network quantitative trait loci mapping of circadian clock outputs identifies metabolic pathway-to-clock linkages in Arabidopsis. Plant Cell. 2011;23:471–485. doi: 10.1105/tpc.110.082065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang EE, Kay SA. Clocks not winding down: unravelling circadian networks. Nature reviews Molecular cell biology. 2010;11:764–776. doi: 10.1038/nrm2995. [DOI] [PubMed] [Google Scholar]
- 18.Lu SX, et al. CIRCADIAN CLOCK ASSOCIATED1 and LATE ELONGATED HYPOCOTYL function synergistically in the circadian clock of Arabidopsis. Plant Physiol. 2009;150:834–843. doi: 10.1104/pp.108.133272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yakir E, et al. Posttranslational regulation of CIRCADIAN CLOCK ASSOCIATED1 in the circadian oscillator of Arabidopsis. Plant Physiol. 2009;150:844–857. doi: 10.1104/pp.109.137414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harmer SL, et al. Orchestrated Transcription of Key Pathways in Arabidopsis by the Circadian Clock. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- 21.Alabadi D, et al. Reciprocal Regulation Between TOC1 and LHY/CCA1 Within the Arabidopsis Circadian Clock. Science. 2001;293:880–883. doi: 10.1126/science.1061320. [DOI] [PubMed] [Google Scholar]
- 22.Dai S, et al. BROTHER OF LUX ARRHYTHMO is a component of the Arabidopsis circadian clock. Plant Cell. 2011;23:961–972. doi: 10.1105/tpc.111.084293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu SX, et al. CCA1 and ELF3 Interact in the control of hypocotyl length and flowering time in Arabidopsis. Plant Physiol. 2012;158:1079–1088. doi: 10.1104/pp.111.189670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagel DH, Kay SA. Complexity in the wiring and regulation of plant circadian networks. Current biology : CB. 2012;22:R648–657. doi: 10.1016/j.cub.2012.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lau OS, et al. Interaction of Arabidopsis DET1 with CCA1 and LHY in mediating transcriptional repression in the plant circadian clock. Molecular cell. 2011;43:703–712. doi: 10.1016/j.molcel.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farre EM, Liu T. The PRR family of transcriptional regulators reflects the complexity and evolution of plant circadian clocks. Current opinion in plant biology. 2013;16:621–629. doi: 10.1016/j.pbi.2013.06.015. [DOI] [PubMed] [Google Scholar]
- 27.Nakamichi N, et al. PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell. 2010;22:594–605. doi: 10.1105/tpc.109.072892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rugnone ML, et al. LNK genes integrate light and clock signaling networks at the core of the Arabidopsis oscillator. Proc Natl Acad Sci U S A. 2013;110:12120–12125. doi: 10.1073/pnas.1302170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dixon LE, et al. Temporal repression of core circadian genes is mediated through EARLY FLOWERING 3 in Arabidopsis. Current biology : CB. 2011;21:120–125. doi: 10.1016/j.cub.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Helfer A, et al. LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Current biology : CB. 2011;21:126–133. doi: 10.1016/j.cub.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herrero E, et al. EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell. 2012;24:428–443. doi: 10.1105/tpc.111.093807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malapeira J, et al. Ordered changes in histone modifications at the core of the Arabidopsis circadian clock. Proc Natl Acad Sci U S A. 2012;109:21540–21545. doi: 10.1073/pnas.1217022110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Para A, et al. PRR3 Is a vascular regulator of TOC1 stability in the Arabidopsis circadian clock. Plant Cell. 2007;19:3462–3473. doi: 10.1105/tpc.107.054775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yakir E, et al. Cell autonomous and cell-type specific circadian rhythms in Arabidopsis. Plant J. 2011;68:520–531. doi: 10.1111/j.1365-313X.2011.04707.x. [DOI] [PubMed] [Google Scholar]
- 35.James AB, et al. The circadian clock in Arabidopsis roots is a simplified slave version of the clock in shoots. Science. 2008;322:1832–1835. doi: 10.1126/science.1161403. [DOI] [PubMed] [Google Scholar]
- 36.James AB, et al. Alternative splicing mediates responses of the Arabidopsis circadian clock to temperature changes. Plant Cell. 2012;24:961–981. doi: 10.1105/tpc.111.093948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Filichkin SA, Mockler TC. Unproductive alternative splicing and nonsense mRNAs: a widespread phenomenon among plant circadian clock genes. Biology direct. 2012;7:20. doi: 10.1186/1745-6150-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hong S, et al. Type II protein arginine methyltransferase 5 (PRMT5) is required for circadian period determination in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2010;107:21211–21216. doi: 10.1073/pnas.1011987107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanchez SE, et al. A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature. 2010;468:112–116. doi: 10.1038/nature09470. [DOI] [PubMed] [Google Scholar]
- 40.Wang X, et al. SKIP is a component of the spliceosome linking alternative splicing and the circadian clock in Arabidopsis. Plant Cell. 2012;24:3278–3295. doi: 10.1105/tpc.112.100081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jones MA, et al. Mutation of Arabidopsis spliceosomal timekeeper locus1 causes circadian clock defects. Plant Cell. 2012;24:4066–4082. doi: 10.1105/tpc.112.104828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seo PJ, et al. A self-regulatory circuit of CIRCADIAN CLOCK-ASSOCIATED1 underlies the circadian clock regulation of temperature responses in Arabidopsis. Plant Cell. 2012;24:2427–2442. doi: 10.1105/tpc.112.098723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang L, et al. Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc Natl Acad Sci U S A. 2013;110:761–766. doi: 10.1073/pnas.1215010110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim WY, et al. ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature. 2007;449:356–360. doi: 10.1038/nature06132. [DOI] [PubMed] [Google Scholar]
- 45.Sawa M, Kay SA. GIGANTEA directly activates Flowering Locus T in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2011;108:11698–11703. doi: 10.1073/pnas.1106771108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim Y, et al. ELF4 regulates GIGANTEA chromatin access through subnuclear sequestration. Cell reports. 2013;3:671–677. doi: 10.1016/j.celrep.2013.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baudry A, et al. F-box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression. Plant Cell. 2010;22:606–622. doi: 10.1105/tpc.109.072843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mas P, et al. Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature. 2003;426:567–570. doi: 10.1038/nature02163. [DOI] [PubMed] [Google Scholar]
- 49.Kim J, et al. The F-box protein ZEITLUPE controls stability and nucleocytoplasmic partitioning of GIGANTEA. Development. 2013;140:4060–4069. doi: 10.1242/dev.096651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu JW, et al. COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Molecular cell. 2008;32:617–630. doi: 10.1016/j.molcel.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fujiwara S, et al. Post-translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo-response regulator proteins. The Journal of biological chemistry. 2008;283:23073–23083. doi: 10.1074/jbc.M803471200. [DOI] [PubMed] [Google Scholar]
- 52.Wang L, et al. PRR5 regulates phosphorylation, nuclear import and subnuclear localization of TOC1 in the Arabidopsis circadian clock. The EMBO journal. 2010;29:1903–1915. doi: 10.1038/emboj.2010.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Staiger D, Green R. RNA-based regulation in the plant circadian clock. Trends in plant science. 2011;16:517–523. doi: 10.1016/j.tplants.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 54.Ito S, et al. LOV domain-containing F-box proteins: light-dependent protein degradation modules in Arabidopsis. Mol Plant. 2012;5:573–582. doi: 10.1093/mp/sss013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fankhauser C, Staiger D. Photoreceptors in Arabidopsis thaliana: light perception, signal transduction and entrainment of the endogenous clock. Planta. 2002;216:1–16. doi: 10.1007/s00425-002-0831-4. [DOI] [PubMed] [Google Scholar]
- 56.Hu W, et al. Unanticipated regulatory roles for Arabidopsis phytochromes revealed by null mutant analysis. Proc Natl Acad Sci U S A. 2013;110:1542–1547. doi: 10.1073/pnas.1221738110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Strasser B, et al. Arabidopsis thaliana life without phytochromes. Proc Natl Acad Sci US A. 2010;107:4776–4781. doi: 10.1073/pnas.0910446107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wenden B, et al. Light inputs shape the Arabidopsis circadian system. Plant J. 2011;66:480–491. doi: 10.1111/j.1365-313X.2011.04505.x. [DOI] [PubMed] [Google Scholar]
- 59.Li G, et al. Coordinated transcriptional regulation underlying the circadian clock in Arabidopsis. Nature cell biology. 2011;13:616–622. doi: 10.1038/ncb2219. [DOI] [PubMed] [Google Scholar]
- 60.Salome PA, et al. The role of the Arabidopsis morning loop components CCA1, LHY, PRR7, and PRR9 in temperature compensation. Plant Cell. 2010;22:3650–3661. doi: 10.1105/tpc.110.079087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bieniawska Z, et al. Disruption of the Arabidopsis circadian clock is responsible for extensive variation in the cold-responsive transcriptome. Plant Physiol. 2008;147:263–279. doi: 10.1104/pp.108.118059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Farre EM. The regulation of plant growth by the circadian clock. Plant Biol (Stuttg) 2012;14:401–410. doi: 10.1111/j.1438-8677.2011.00548.x. [DOI] [PubMed] [Google Scholar]
- 63.Lorrain S, et al. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 2008;53:312–323. doi: 10.1111/j.1365-313X.2007.03341.x. [DOI] [PubMed] [Google Scholar]
- 64.Stewart JL, et al. PIF genes mediate the effect of sucrose on seedling growth dynamics. PloS one. 2011;6:e19894. doi: 10.1371/journal.pone.0019894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yamashino T. From a repressilator-based circadian clock mechanism to an external coincidence model responsible for photoperiod and temperature control of plant architecture in Arabodopsis thaliana. Bioscience, biotechnology, and biochemistry. 2013;77:10–16. doi: 10.1271/bbb.120765. [DOI] [PubMed] [Google Scholar]
- 66.Arana MV, et al. Circadian oscillation of gibberellin signaling in Arabidopsis. Proc Natl Acad Sci U S A. 2011;108:9292–9297. doi: 10.1073/pnas.1101050108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gao XH, et al. An updated GA signaling ‘relief of repression’ regulatory model. Mol Plant. 2011;4:601–606. doi: 10.1093/mp/ssr046. [DOI] [PubMed] [Google Scholar]
- 68.de Lucas M, et al. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451:480–484. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- 69.Covington MF, Harmer SL. The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS Biol. 2007;5:e222. doi: 10.1371/journal.pbio.0050222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nozue K, et al. Genomic analysis of circadian clock-, light-, and growth-correlated genes reveals PHYTOCHROME-INTERACTING FACTOR5 as a modulator of auxin signaling in Arabidopsis. Plant Physiol. 2011;156:357–372. doi: 10.1104/pp.111.172684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Franklin KA, et al. Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc Natl Acad Sci U S A. 2011;108:20231–20235. doi: 10.1073/pnas.1110682108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sun J, et al. PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating arabidopsis hypocotyl growth. PLoS genetics. 2012;8:e1002594. doi: 10.1371/journal.pgen.1002594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hornitschek P, et al. Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 2012;71:699–711. doi: 10.1111/j.1365-313X.2012.05033.x. [DOI] [PubMed] [Google Scholar]
- 74.Kunihiro A, et al. Phytochrome-interacting factor 4 and 5 (PIF4 and PIF5) activate the homeobox ATHB2 and auxin-inducible IAA29 genes in the coincidence mechanism underlying photoperiodic control of plant growth of Arabidopsis thaliana. Plant & cell physiology. 2011;52:1315–1329. doi: 10.1093/pcp/pcr076. [DOI] [PubMed] [Google Scholar]
- 75.Rawat R, et al. REVEILLE1, a Myb-like transcription factor, integrates the circadian clock and auxin pathways. Proc Natl Acad Sci U S A. 2009;106:16883–16888. doi: 10.1073/pnas.0813035106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Michael TP, et al. Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS genetics. 2008;4:e14. doi: 10.1371/journal.pgen.0040014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Noordally ZB, et al. Circadian control of chloroplast transcription by a nuclear-encoded timing signal. Science. 2013;339:1316–1319. doi: 10.1126/science.1230397. [DOI] [PubMed] [Google Scholar]
- 78.Graf A, et al. Circadian control of carbohydrate availability for growth in Arabidopsis plants at night. Proc Natl Acad Sci U S A. 2010;107:9458–9463. doi: 10.1073/pnas.0914299107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Knight H, et al. Sensitive to freezing6 integrates cellular and environmental inputs to the plant circadian clock. Plant Physiol. 2008;148:293–303. doi: 10.1104/pp.108.123901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Haydon MJ, et al. Photosynthetic entrainment of the Arabidopsis thaliana circadian clock. Nature. 2013;502:689–692. doi: 10.1038/nature12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Salome PA, et al. Circadian clock adjustment to plant iron status depends on chloroplast and phytochrome function. The EMBO journal. 2013;32:511–523. doi: 10.1038/emboj.2012.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen YY, et al. Iron is involved in the maintenance of circadian period length in Arabidopsis. Plant Physiol. 2013;161:1409–1420. doi: 10.1104/pp.112.212068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hong S, et al. Reciprocal interaction of the circadian clock with the iron homeostasis network in Arabidopsis. Plant Physiol. 2013;161:893–903. doi: 10.1104/pp.112.208603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Eriksson ME, Webb AA. Plant cell responses to cold are all about timing. Current opinion in plant biology. 2011;14:731–737. doi: 10.1016/j.pbi.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 85.Dong MA, et al. Circadian clock-associated 1 and late elongated hypocotyl regulate expression of the C-repeat binding factor (CBF) pathway in Arabidopsis. Proc Natl Acad Sci U S A. 2011;108:7241–7246. doi: 10.1073/pnas.1103741108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nakamichi N, et al. Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant & cell physiology. 2009;50:447–462. doi: 10.1093/pcp/pcp004. [DOI] [PubMed] [Google Scholar]
- 87.Lai AG, et al. CIRCADIAN CLOCK-ASSOCIATED 1 regulates ROS homeostasis and oxidative stress responses. Proc Natl Acad Sci U S A. 2012;109:17129–17134. doi: 10.1073/pnas.1209148109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang W, et al. Timing of plant immune responses by a central circadian regulator. Nature. 2011;470:110–114. doi: 10.1038/nature09766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bhardwaj V, et al. Defence responses of Arabidopsis thaliana to infection by Pseudomonas syringae are regulated by the circadian clock. PloS one. 2011;6:e26968. doi: 10.1371/journal.pone.0026968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Goodspeed D, et al. Arabidopsis synchronizes jasmonate-mediated defense with insect circadian behavior. Proc Natl Acad Sci U S A. 2012;109:4674–4677. doi: 10.1073/pnas.1116368109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Goodspeed D, et al. Postharvest circadian entrainment enhances crop pest resistance and phytochemical cycling. Current biology : CB. 2013;23:1235–1241. doi: 10.1016/j.cub.2013.05.034. [DOI] [PubMed] [Google Scholar]
- 92.Zhang C, et al. Crosstalk between the circadian clock and innate immunity in Arabidopsis. PLoS pathogens. 2013;9:e1003370. doi: 10.1371/journal.ppat.1003370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.McClung CR. Beyond Arabidopsis: the circadian clock in non-model plant species. Seminars in cell & developmental biology. 2013;24:430–436. doi: 10.1016/j.semcdb.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 94.Lou P, et al. Preferential retention of circadian clock genes during diploidization following whole genome triplication in Brassica rapa. Plant Cell. 2012;24:2415–2426. doi: 10.1105/tpc.112.099499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shaw LM, et al. The impact of photoperiod insensitive Ppd-1a mutations on the photoperiod pathway across the three genomes of hexaploid wheat (Triticum aestivum) Plant J. 2012;71:71–84. doi: 10.1111/j.1365-313X.2012.04971.x. [DOI] [PubMed] [Google Scholar]
- 96.Turner A, et al. The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science. 2005;310:1031–1034. doi: 10.1126/science.1117619. [DOI] [PubMed] [Google Scholar]
- 97.Yang Y, et al. OsELF3 is involved in circadian clock regulation for promoting flowering under long-day conditions in rice. Mol Plant. 2013;6:202–215. doi: 10.1093/mp/sss062. [DOI] [PubMed] [Google Scholar]
- 98.Zakhrabekova S, et al. Induced mutations in circadian clock regulator Mat-a facilitated short-season adaptation and range extension in cultivated barley. Proc Natl Acad Sci U S A. 2012;109:4326–4331. doi: 10.1073/pnas.1113009109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Faure S, et al. Mutation at the circadian clock gene EARLY MATURITY 8 adapts domesticated barley (Hordeum vulgare) to short growing seasons. Proc Natl Acad Sci U S A. 2012;109:8328–8333. doi: 10.1073/pnas.1120496109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Campoli C, et al. HvLUX1 is a candidate gene underlying the early maturity 10 locus in barley: phylogeny, diversity, and interactions with the circadian clock and photoperiodic pathways. The New phytologist. 2013;199:1045–1059. doi: 10.1111/nph.12346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Murphy RL, et al. Coincident light and clock regulation of pseudoresponse regulator protein 37 (PRR37) controls photoperiodic flowering in sorghum. Proc Natl Acad Sci U S A. 2011;108:16469–16474. doi: 10.1073/pnas.1106212108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Preuss SB, et al. Expression of the Arabidopsis thaliana BBX32 gene in soybean increases grain yield. PloS one. 2012;7:e30717. doi: 10.1371/journal.pone.0030717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Farre EM, et al. Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Current biology : CB. 2005;15:47–54. doi: 10.1016/j.cub.2004.12.067. [DOI] [PubMed] [Google Scholar]
- 104.Perales M, et al. The proteasome-dependent degradation of CKB4 is regulated by the Arabidopsis biological clock. Plant J. 2006;46:849–860. doi: 10.1111/j.1365-313X.2006.02744.x. [DOI] [PubMed] [Google Scholar]
- 105.Sawa M, et al. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science. 2007;318:261–265. doi: 10.1126/science.1146994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kim WY, et al. Release of SOS2 kinase from sequestration with GIGANTEA determines salt tolerance in Arabidopsis. Nature communications. 2013;4:1352. doi: 10.1038/ncomms2357. [DOI] [PubMed] [Google Scholar]
- 107.Kim J, et al. FIONA1 is essential for regulating period length in the Arabidopsis circadian clock. Plant Cell. 2008;20:307–319. doi: 10.1105/tpc.107.055715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jones MA, et al. Jumonji domain protein JMJD5 functions in both the plant and human circadian systems. Proc Natl Acad Sci U S A. 2010;107:21623–21628. doi: 10.1073/pnas.1014204108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lu SX, et al. The Jumonji C domain-containing protein JMJ30 regulates period length in the Arabidopsis circadian clock. Plant Physiol. 2011;155:906–915. doi: 10.1104/pp.110.167015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kevei E, et al. Arabidopsis thaliana Circadian Clock Is Regulated by the Small GTPase LIP1. Current Biology. 2007;17:1456–1464. doi: 10.1016/j.cub.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 111.Wang Y, et al. LIGHT-REGULATED WD1 and PSEUDO-RESPONSE REGULATOR9 form a positive feedback regulatory loop in the Arabidopsis circadian clock. Plant Cell. 2011;23:486–498. doi: 10.1105/tpc.110.081661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wu JF, et al. Two new clock proteins, LWD1 and LWD2, regulate Arabidopsis photoperiodic flowering. Plant Physiol. 2008;148:948–959. doi: 10.1104/pp.108.124917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Panda S, et al. tej defines a role for poly(ADP-ribosyl)ation in establishing period length of the arabidopsis circadian oscillator. Developmental cell. 2002;3:51–61. doi: 10.1016/s1534-5807(02)00200-9. [DOI] [PubMed] [Google Scholar]
- 114.Sanchez-Villarreal A, et al. TIME FOR COFFEE is an essential component in the maintenance of metabolic homeostasis in Arabidopsis thaliana. Plant J. 2013;76:188–200. doi: 10.1111/tpj.12292. [DOI] [PubMed] [Google Scholar]
- 115.Sanchez-Villarreal A, et al. TIME FOR COFFEE is an Essential Component in the Maintenance of Arabidopsis thaliana Metabolic Homeostasis. Plant J. 2013 doi: 10.1111/tpj.12292. [DOI] [PubMed] [Google Scholar]
- 116.Martin-Tryon EL, Harmer SL. XAP5 CIRCADIAN TIMEKEEPER coordinates light signals for proper timing of photomorphogenesis and the circadian clock in Arabidopsis. Plant Cell. 2008;20:1244–1259. doi: 10.1105/tpc.107.056655. [DOI] [PMC free article] [PubMed] [Google Scholar]