Abstract
Background
Ambient particulate matter (PM) has been associated with mortality and morbidity for cardiovascular disease (CVD). MicroRNAs control gene expression at a post-transcriptional level. Altered microRNA expression has been reported in processes related to CVD and PM exposure, e.g. systemic inflammation, endothelial dysfunction and atherosclerosis. Polymorphisms in microRNA-related genes could influence response to PM.
Methods
We investigated the association of exposure to ambient particles in several time windows (4-hours to 28-days moving averages) and blood-leukocyte expression changes in fourteen candidate microRNAs, in 153 elderly males from the Normative Aging Study (examined 2005–2009). Potential effect modification by six single nucleotide polymorphisms (SNPs) in three microRNA-related genes was investigated. Fine PM (PM2.5), black carbon, organic carbon and sulfates were measured at a stationary ambient monitoring site. Linear regression models, adjusted for potential confounders, were used to assess effects of particles and SNP-by-pollutant interaction. An in silico pathways analysis was performed on target genes of miRNAs associated with the pollutants.
Results
We found a negative association for pollutants in all moving averages and miR-1, -126, -135a, -146a, -155, -21, -222 and -9. The strongest associations were observed with the 7-day moving averages for PM2.5 and black carbon and with the 48-hour moving averages for organic carbon. The association with sulfates was stable across the moving averages. The in silico pathway analysis identified 18 pathways related to immune response shared by at least two miRNAs; in particular, the “HMGB1/RAGE signaling pathway” was shared by miR-126, -146a, -155, -21 and -222.
No important associations were observed for miR-125a-5p, -125b, -128, -147, -218 and -96. We found significant SNP-by-pollutant interactions for rs7813, rs910925 and rs1062923 in GEMIN4 and black carbon and PM2.5 for miR-1, -126, -146a, -222 and -9, and for rs1640299 in DGCR8 and SO42− for miR-1 and -135a.
Conclusions
Exposure to ambient particles could cause a downregulation of microRNAs involved in processes related to PM exposure. Polymorphisms in GEMIN4 and DGCR8 could modify these associations.
Exposure to ambient particulate matter (PM) has been associated with increased mortality and morbidity for cardiovascular disease (CVD).1 Although some biological mechanisms have been identified (including systemic inflammation, endothelial dysfunction and atherosclerosis2), the underlying mechanisms for ambient particles toxicity are not completely understood. Moreover, particles are a complex mixture of primary particles (e.g. black carbon) as well as secondary particles (e.g. various organic carbon particles and sulfates [SO42−]) that may act through different mechanisms.
MicroRNAs (miRNAs) are small endogenous 20 to 23 nucleotide non-coding RNAs that can pair to sites in specific messenger RNAs (mRNAs) of protein-coding genes and control gene expression at a post-transcriptional level by degrading or repressing mRNAs.3 Altered expression of several miRNAs have been reported in processes related to inflammation (e.g. miR-1, -128, -135a, -146a, -147, -155, -21, and -94–8), endothelial dysfunction (e.g. miR-126 and -2189,10), and atherosclerosis (e.g. miR-125a-5p, -125b, -155, -222, -9611–14).
Few studies have investigated changes in miRNAs expression in response to environmental stressors, including PM.15 A dysregulation of miRNAs has been found associated with exposure to PM, diesel exhaust particles and carbon black nanoparticles in vitro16,17 and in animal studies.18,19 Expression changes in miRNAs related to inflammation and oxidative stress following exposure to metal-rich PM in foundry workers has been reported.20,21
Several genes are involved in miRNAs biogenesis and processing, including Gem-associated protein 4 (GEMIN4) and DiGeorge critical region-8 (DGCR8) genes.22 Polymorphisms in these genes may affect miRNA expression. Our group recently observed a modification of pollutant effects on health outcomes by a number of single nucleotide polymorphisms (SNPs) in miRNA processing genes,23,24 indicating that miRNA expression may represent a biological mechanism linked to PM effects.
In the present study, we investigated whether exposure to overall fine particulate matter (PM2.5), as well as particles from mobile sources (black carbon) and secondary transported particles (organic carbon and sulfates) in several time windows, was associated with expression changes in selected candidate miRNAs in blood leukocytes. Furthermore, we investigated whether the effects were modified by SNPs in a selection of miRNA-related genes previously shown to modify particles effects.
Methods
Study population
Our study participants were members of the Veterans Normative Aging Study. This cohort, established in 1963, enrolled men age 21–80 years from the Greater Boston area who were free of known chronic medical conditions.25 Participants were reevaluated every 3–5 years using on-site comprehensive clinical examinations. Residency was verified during each visit. Buffy coat for miRNAs measurement were collected in 166 participants between December 2005 and May 2009. Collection of blood samples for genetic analysis began in the late 1990s; 149 participants provided blood samples for some or all miRNA-related SNPs. Participants reported to the study center on the morning of their scheduled examinations. Lifestyle data were collected using a questionnaire. Written informed consent from all participants and approval from the Institutional Review Boards of all participant institutions were obtained.
Ambient air pollutants measurement
Measurements of pollutants were obtained from a stationary ambient monitoring site, < 1 km from the examination site where the participant visit took place. The median distance from participants residence to monitor site was 23.9 (25th–75th percentiles= 11.2 – 58.3) km. Ambient black carbon was measured hourly using an aethalometer (Magee Scientific, Berkley, CA), and PM2.5 was measured continuously using a tapered element oscillating microbalance (model 1400A, Rupprecht&Pataschnick Co, Albany, NY). Hourly averages for PM2.5 were calculated based on the continuous measurements. Daily average of both black carbon and PM2.5 were calculated in the 24-hr (0800 to 0800 hours) period preceding examination. When data were missing for either black carbon or PM2.5, levels were imputed through a linear regression model.26 Twenty-four-hour integrated sulfur was measured on daily particulate filter samples using X-ray fluorescence spectroscopy and was multiplied by 3 to compute SO42− levels. After 2007 a continuous sulfate monitor was available to fill in occasional missing values from X-ray fluorescence analyses. Twenty-four-hour integrated organic carbon was measured using the Partisol model 2300 sequential sampler (Rupprecht&Pataschnick Co). Particle-health associations have been reported for a range of averaging times ranging from hours to a month or longer. To assess the most relevant exposure windows for the associations with miRNAs, we evaluated several time windows of exposure, using as the exposure index the average pollutant concentration taken from time periods of 4-hour to 28-day moving averages before the time of blood draw.
MiRNAs selection and analysis
We selected 14 miRNAs that have been previously reported as involved in processes related to inflammation,4–8 endothelial dysfunction,9,10 and atherosclerosis.11–14 Total RNA was extracted from stored frozen buffy coat of whole blood, and RNA purity and concentration were determined (for further details see eAppendix).
We used stem-loop quantitative real-time RT-PCR to detect and quantify 14 miRNAs. In the reverse transcription step, we used TaqMan® MicroRNA Reverse Transcription Kit (Applied Biosystems) and custom 96 well plates pre-spotted with specific miRNA stem-loop primers (Applied Biosystems). Real-time polymerase chain reaction (PCR) was performed using TaqMan® custom 384 well plates (Applied Biosystems) and TaqMan® Universal PCR Master Mix (Applied Biosystems). All PCR runs were performed in triplicate on a 7900HT Fast Real-Time PCR System (Applied Biosystems) (see eAppendix). The relative gene expression was calculated via a 2−ΔΔCt method. Blood samples for miRNAs were analyzed in two batches (20 and 146 participants respectively). Data are presented as the relative quantity of target miRNA, normalized to endogenous control miRNAs (i.e. RNU24 and RNU48) and a calibrator built as a pool of random samples (see eAppendix). DataAssistTM Software (Applied Biosystems) was employed to provide relative quantification analysis of miRNAs expression.
SNPs selection and genotyping
SNPs were selected based on previously published works investigating modification of effects of pollutants on health outcomes.23,24 Genotyping was performed using multiplex PCR assays designed with Sequenom Spectro DESIGNER software (Sequenom, Inc., San Diego, CA). The extension product was then spotted onto a 384-well spectroCHIP before analysis in the MALDITOF mass spectrometer (Sequenom, Inc.). Duplication was performed on 5% of the samples. The 6 SNPs analyzed for this study were all successfully genotyped. After genotyping, we excluded those SNPs for which fewer than 3 participants were homozygous variant carriers (rs13078 in DICER, rs197414 in GEMIN3), leaving a total of 4 SNPs in 2 genes (rs7813, rs910925 and rs1062923 in GEMIN4, rs1640299 in DGCR8).
Statistical methods
We investigated the effect of exposure to PM2.5 on miRNAs levels within the population. The effects of black carbon, organic carbon and SO42− were also assessed to determine whether certain sources/types of particle pollution such as traffic and coal combustion produced different effects. We also examined gene-by-environment interactions between pollutants and selected SNPs in miRNA-related genes. We tested for nonlinearity using penalized splines in generalized linear models. Linear regression multivariate models were constructed to estimate the effects of each air pollutant. MiRNAs measurements were natural-log-transformed to improve normality. The following adjusting variables were selected a priori, based on previous work investigating associations between miRNAs and particles in foundry workers20: age, body mass index (BMI), cigarette smoking (never, former, current), and pack-years. We adjusted for percent of granulocytes (to control for possible shifts in leukocyte differential count), date, and seasonality (using sine and cosine). We first determined for each pollutant the most representative time window to be used in our main analysis to examine the association of investigated pollutants with 14 miRNAs. We then examined SNP-by-pollutant cross-product terms for the selected pollutants’ time window to assess gene-by-environment interactions, only for those miRNAs that we found associated with pollutants. To reduce the number of tested associations we examined only the recessive model of inheritance.
All statistical analyses were carried out using R 2.12.1 (R Foundation for Statistical Computing, Vienna, Austria). All effect estimates (β) and their 95% confidence intervals (CI) are presented as percent changes per-interquartile-range (IQR) change of pollutant at each averaging time period, using the formula (eβ*IQR−1) × 100.
Sensitivity analyses
Because blood samples for SNPs genotyping was available in a subset of participants, we restricted our analysis of the association between miRNA and pollutants to participants with some or all miRNA-related SNPs. Since blood samples for miRNAs were analyzed in two batches, results from the two groups may differ. We therefore controlled for batch in joint analyses and performed a sensitivity analysis restricting to the group with more participants. To account for potential population stratification, we ran a sensitivity analysis by restricting to persons of the most represented race based on self-report. Moreover, to take into account differences in the distance from the monitoring site to the participant’s residence, we performed a sensitivity analysis by restricting to the 95% of people living closest to the monitoring site.
In silico pathway analysis
MiRNAs that were significantly associated with the pollutants were further investigated, together with their target genes, with GeneGO pathways enrichment analysis (see eAppendix) in MetaCore™ v6.9 (GeneGo Inc.), a web-based computational platform for multiple applications in systems biology. MetaCore analyses are based on MetaBase, a proprietary database of mammalian biology that contains over 6 million manually curated experimental findings on protein-protein, protein-DNA and protein-compound interactions, metabolic and signaling pathways, supported by proprietary ontologies and controlled vocabulary.
Experimentally validated target genes were obtained from miRTarBase v3.5, a database of miRNA-target interactions that are collected by surveying pertinent literature, as well as on predicted targets from TargetScan v6.2. TargetScan uses an algorithm to calculate the Total Context Score for each predicted miRNA hit as a measure of targeting efficacy (see eAppendix). To reduce the false-positive rate, predicted targets with a Total Context Score < −0.2 were considered biologically relevant and acceptable for analysis.
MiRNA-target expression correlation
Since only small portions of validated target messenger RNAs (mRNAs) are known for each miRNA and the prediction algorithm can provide false-positive results,27 we also tested the expression correlation of miRNAs and predicted targets included in the pathways analysis, using the data available in the MirGator v3.0, a public database that collects deep-sequencing human data (see eAppendix). We considered inversely correlated miRNA/mRNA couples with Spearman’s r < −0.5.
Results
All participants made one study visit between December 2005 and May 2009. MiRNA data were available for 156 out of the 166 participants who provided a blood sample. Complete covariate data were available on 153 participants. Organic carbon measurements were available for a subset of 132 participants, with clinical characteristics similar to those with all data available (results not shown). The clinical characteristics of the participants are described in Table 1. Participants were non-Hispanic white (95%), elderly males, most of whom were never (30%) or former (69%) smokers. eTable 1 summarizes daily pollutants levels during the study period. PM2.5 was highly correlated with all other pollutants, in particular SO42− (Spearman’s correlation coefficient [ρ] = 0.93). Daily concentrations of black carbon and organic carbon were moderately correlated (ρ = 0.69) (eTable 1).
Table 1.
Characteristicsa of Study Participants (n = 153).
| Characteristics | |
|---|---|
| Age (years) | 77.4 (5.9) |
| BMI (kg/m2) | 28.7 (4.7) |
| miR-1b | 0.9 (1.6) |
| miR-125a-5pb | 1.9 (2.3) |
| miR-125bb | 1.4 (1.7) |
| miR-126b | 1.4 (1.4) |
| miR-128b | 2.1 (4.1) |
| miR-135ab | 0.8 (0.8) |
| miR-146ab | 1.5 (1.7) |
| miR-147b,c | 5.8 (20.9) |
| miR-155b | 2.4 (2.1) |
| miR-21b | 1.7 (2.1) |
| miR-218b | 2.4 (7.4) |
| miR-222b | 1.6 (1.7) |
| miR-9b | 1 (0.8) |
| miR-96b | 2 (2.8) |
| Smoking status; no. (%) | |
| Never | 46 (30.1) |
| Current | 2 (1.3) |
| Former | 105 (68.6) |
| No. packs years of smoking | 18.1 (21.8) |
| Race; no. (%) | |
| Non-Hispanic white | 145 (94.8) |
| Hispanic white | 4 (2.6) |
| African American | 4 (2.6) |
Mean (SD), unless otherwise specified.
Expressed as 2−ΔΔCt.
n = 152.
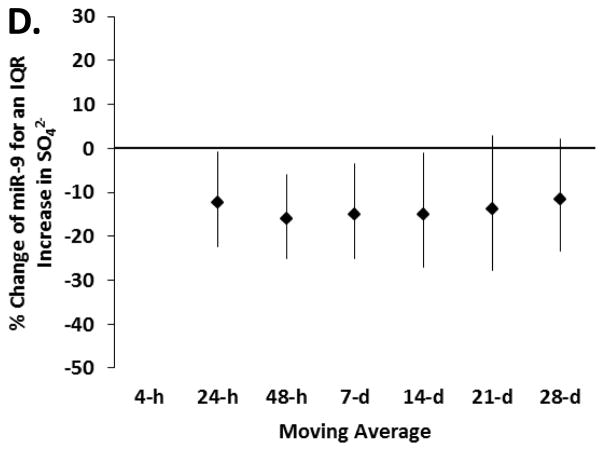
We first examined the association between exposure to particles for several moving averages (4-hour, 24-hour, 48-hour, 7-day, 14-day and 28-day) and changes in miRNAs, to determine for each pollutant the representative time window for our main analysis. We excluded a non-linear relationship between metals and miRNA using penalized splines in generalized additive models (eFigure 1). In our models adjusted for age, BMI, cigarette smoking (never, former, current), pack-years of smoking, granulocytes (%), date, seasonality and miRNA batch, we found an overall association in the negative direction for pollutants in all the exposure windows and eight miRNAs (i.e. miR-1, -126, -135a, -146a, -155, -21, -222 and -9) (eTable 2). No important associations were observed for the miR-125a-5p, -125-b, -128, -147, -218 and -96 (eTable 2). The trend of associations through the time-windows was similar for all the eight miRNAs; an example is reported in Figure 1, showing results for miR-9.
Figure 1.
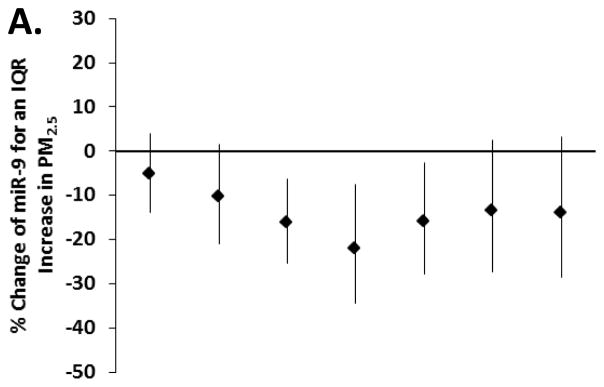
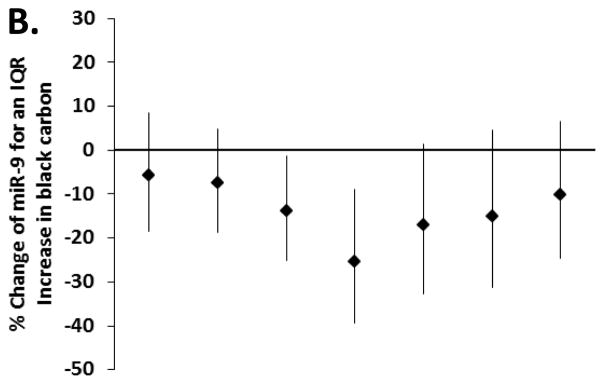

Estimated percentage change and 95% CI in miR-9 for an interquartile range increase in (A) PM2.5, (B) black carbon, (C) organic carbon and (D) sulfates, in several exposure windows (n = 153, except n=132 for organic carbon). Model adjusted for age, BMI, cigarette smoking (never, former, current), pack-years, granulocytes (%), date, seasonality (using sine and cosine of day) and miRNA batch.
For each pollutant we then selected a representative moving average, on the basis of this trend. In general, the observed decrease in miRNAs levels in relation to PM2.5 and black carbon was strongest for the 7-day moving average. The negative association between organic carbon and miRNAs was generally strongest for the 48-hour moving average. The negative association between sulfates and miRNAs was mostly stable across the moving averages (eTable 2). In Table 2 we report the percentage change in miRNAs for an IQR increase in pollutant in the selected exposure window for each pollutant, i.e. 7-days for PM2.5 (IQR= 3.83 μg/m3) and black carbon (IQR= 0.26 μg/m3), 48-hours for organic carbon (IQR= 1.56 μg/m3) and 48-hours for SO42− (IQR= 1.47μg/m3). The strongest associations in relation to higher PM2.5 and black carbon were for miR-21 and -146a. An IQR increase in the 7-day moving average of PM2.5 was associated with 34% (95% CI = −48% to −17%) and 35% (−48% to −18%) lower miR-146a and miR-21, respectively. Similarly, an IQR increase in the 7-day moving average of black carbon was associated with a 28% (−45% to −4%) and 35% (−51% to −15%) lower miR-146a and miR-21, respectively. MiR-1, -135a and -21 showed the strongest associations in relation to organic carbon. An IQR increase in the 48-hour moving average of organic carbon was associated with a 34% (−54% to −6%), 28% (−45% to −6%) and 26% (−45% to −1%) lower miR-1, miR-135a and miR-21, respectively. The strongest associations with the 48-hour moving average of SO42− were for miR-1 (−25% association [−38% to −8%]), miR-146a (−24% [−35% to −11%]) and miR-21 (−23% [−34% to −10%]).
Table 2.
Estimated Percentage Changea in MiRNAs for an Interquartile Range Increase in the Selected Exposure Window for Each Air Pollutant (n = 153, Except n=132 for Organic Carbon).
| Pollutant (IQR; moving average) | MicroRNA | % change | (95% CI) |
|---|---|---|---|
| PM2.5 (3.83μg/m3, 7-d) | |||
| miR-1 | −33 | (−50.2 to −10.2) | |
| miR-126 | −25 | (−38.1 to −8.1) | |
| miR-135a | −29 | (−42.9 to −12.4) | |
| miR-146a | −34 | (−47.6 to −17.4) | |
| miR-155 | −21 | (−32.8 to −6.7) | |
| miR-21 | −35 | (−48 to −18.1) | |
| miR-222 | −20 | (−32.9 to −4.3) | |
| miR-9 | −22 | (−34.4 to −7.6) | |
| Black carbon (0.26μg/m3, 7-d) | |||
| miR-1 | −20 | (−44 to 14.8) | |
| miR-126 | −24 | (−39.8 to −3) | |
| miR-135a | −26 | (−43.1 to −4.4) | |
| miR-146a | −28 | (−45.2 to −4.2) | |
| miR-155 | −13 | (−28.8 to 6.1) | |
| miR-21 | −35 | (−50.8 to −14.8) | |
| miR-222 | −17 | (−32.9 to 2.9) | |
| miR-9 | −26 | (−39.4 to −8.9) | |
| Organic carbon (1.56μg/m3, 48-h)b | |||
| miR-1 | −34 | (−53.5 to −5.7) | |
| miR-126 | −24 | (−41 to −3) | |
| miR-135a | −28 | (−45.4 to −6.1) | |
| miR-146a | −14 | (−35.2 to 14) | |
| miR-155 | −15 | (−30.7 to 5.3) | |
| miR-21 | −26 | (−44.5 to −0.7) | |
| miR-222 | −5 | (−23.9 to 18.4) | |
| miR-9 | −22 | (−36.5 to −4.6) | |
| SO42− (1.47 μg/m3, 48-h) | |||
| miR-1 | −25 | (−38.2 to −8.4) | |
| miR-126 | −17 | (−27.5 to −5.5) | |
| miR-135a | −18 | (−29 to −5.2) | |
| miR-146a | −24 | (−34.6 to −11.1) | |
| miR-155 | −15 | (−23.4 to −4.5) | |
| miR-21 | −23 | (−33.7 to −9.9) | |
| miR-222 | −13 | (−23 to −2.3) | |
| miR-9 | −16 | (−25.1 to −5.9) | |
Adjusted for age, BMI, cigarette smoking (never, former, current), pack-years, granulocytes (%), date, seasonality (using sine and cosine of day) and miRNA batch.
The complete list of the 4 SNPs analyzed is described in Table 3. All or some of the miRNA-related genotyping were available for 141 of 153 participants. Using the selected exposure window for each pollutant we examined the gene-by-environment interactions between pollutants and these four SNPs. We found significant SNP-by-pollutant interactions for black carbon and PM2.5 in relation to SNPs in GEMIN4 for five microRNAs (i.e. miR-1, -126, -146a, -222 and -9), and for SO42− and rs1640299 in DGCR8 for miR-135a and -21 (Table 4). There were no significant SNP-by-pollutant interaction for organic carbon and miR-155 (results not shown). For wild-type persons and heterozygotes carriers of rs1062923, an IQR change in the 7-day moving average of black carbon was associated with a 32% decrease (−48% to −9%) in miR-146a levels, whereas in homozygous variant carriers (n = 6) we observed a 768% increase (6% to 7043%) (P= 0.021 for interaction term). Rs1062923 also modified in a similar fashion the effect of PM2.5 and black carbon on miR-1 and miR-126, and the effect of black carbon on miR-9. In homozygous variant carriers for rs7813 and rs910925, we observed lower miR-146a and miR-222 in relation with higher levels of black carbon, compared with wild-type and heterozygotes. For wild-type and heterozygotes for rs1640299, an IQR change in the 48-hour moving average of SO42− was associated with a 38% decrease (−52% to −19%) in miR-1 levels, whereas in homozygous variant carriers no significant association was observed (test for interaction, P = 0.032). Rs1640299 also modified in a similar fashion the effect of SO42− on miR-135a (Table 4). We performed a number of sensitivity analyses to assess the robustness of our results. Restricting to participants with some or all miRNA-related SNPs (n = 141) did not change our results. Since blood samples for miRNAs were analyzed in two batches, results from the two groups may differ; we then performed sensitivity analysis restricting to the group with more participants with all covariates (n = 135), with no changes in findings. We ran a sensitivity analysis by restricting our analyses to non-Hispanic white men (n = 145), to account for potential population stratification, with no changes in the results. Restricting to the 95% of participants living closest to the monitoring site did not have important effects on our results. We included in our model multiple variables a priori based on previous work,20 we then ran a sensitivity analysis by removing the non-significant covariates in the subset of models showing the strongest association in relation to higher PM2.5 (i.e. those investigating miR-21 and miR-146a [eTable 3]), and we found no important changes in our results.
Table 3.
SNPs in MiRNA Processing Genes Included in Analysis.
| Genea | RS number | SNP position | Alleles | Role | Amino acid change | Minor allele frequencyb |
|---|---|---|---|---|---|---|
| Gem-associated protein 4 (GEMIN4) | rs7813 | chromosome17:594936 | cytosine>thymine | Coding exon | arginine>cytosine | 0.31 |
| rs910925 | chromosome17:596297 | cytosine>guanosine | Coding exon | alanine>glycine | 0.31 | |
| rs1062923 | chromosome17:595817 | cytosine>thymine | Coding exon | isoleucine>threonine | 0.08 | |
| DiGeorge syndrome critical region gene 8 (DGCR8) | rs1640299 | chromosone22:18478359 | guanosine>thymine | 3′ untranslated region | n/a | 0.39 |
n/a indicates not applicable.
Gene name (abbreviation).
As reported in dbSNP database
Table 4.
Effect Modification of the Association Between an IQR Increase in PM2.5, Black Carbon and SO42− Concentrations and MiRNAs by Gene Variants Related to GEMIN4 and DGCR8 Under Recessive Models.
| Pollutant (IQR; moving averange) | MiRNA | Gene/SNP | Wild-type and heterozygotes | Homozygous variant carriers | Test for interaction | ||
|---|---|---|---|---|---|---|---|
|
| |||||||
| No. | % change (95% CI) | No. | % change (95% CI) | ||||
| PM2.5 (3.83 μg/m3; 7-d) | |||||||
| miR-1 | GEMIN4/rs1062923 | 134 | −33 (−50 to −10) | 6 | 3241 (−15.1 to 131304.4) | P=0.040 | |
| miR-126 | GEMIN4/rs1062923 | 134 | −27 (−40.2 to −11.5) | 6 | 777 (−24 to 10023.1) | P=0.049 | |
| BC (0.26 μg/m3; 7-d) | |||||||
| miR-1 | GEMIN4/rs1062923 | 134 | −23 (−46.2 to 10.4) | 6 | 1336 (−1.3 to 20782.2) | P=0.036 | |
| miR-126 | GEMIN4/rs1062923 | 134 | −30 (−44.9 to −11.6) | 6 | 466 (−2.1 to 3175) | P=0.022 | |
| miR-146a | GEMIN4/rs1062923 | 134 | −32 (−48.4 to −9.1) | 6 | 768 (5.6 to 7042.7) | P=0.021 | |
| GEMIN4/rs910925 | 119 | −16 (−37.9 to 14.9) | 19 | −66 (−83.6 to −29.4) | P=0.024 | ||
| GEMIN4/rs7813 | 121 | −17 (−38.6 to 13.4) | 19 | −66 (−83.7 to −29.4) | P=0.026 | ||
| miR-222 | GEMIN4/rs910925 | 119 | −5 (−24.2 to 20.2) | 19 | −58 (−75.7 to −27.3) | P=0.007 | |
| GEMIN4/rs7813 | 121 | −7 (−25.8 to 17.8) | 19 | −58 (−75.9 to −27.3) | P=0.009 | ||
| miR-9 | GEMIN4/rs1062923 | 134 | −30 (−42.8 to −13.4) | 6 | 243 (−26.9 to 1513.7) | P=0.049 | |
| SO42− (1.47 μg/m3; 48-h) | |||||||
| miR-1 | DGCR8/rs1640299 | 102 | −38 (−52.4 to −19.1) | 37 | −4 (−28.8 to 28.4) | P=0.032 | |
| miR-135a | DGCR8/rs1640299 | 102 | −29 (−41.5, −13.3) | 37 | 2 (−18.3, 26.4) | 0.018 | |
To explore the functional significance of the eight miRNAs associated with pollutants, we performed an in silico pathway analysis on the validated and predicted target genes identified for miR-1, -126, -135a, -146a, -155, -21, -222, and -9. We first determined the putative downstreams (i.e. gene targets) for each miRNA. We found 1575 miRNA-target interactions either validated or predicted with a Total Context Score < −0.20 (Table 5). The total number of gene targets considered for analysis ranged from 33 (miR-126) to 407 (miR-1). We ran a pathway analysis for each miRNA. In addition to those related to general functions, pathways related to immune response represented between 12% and 39% of the noteworthy pathways (false discovery rate <0.05) in six out of eight miRNAs (namely, miR-1, -126, -146a, -155, -21 and -222) (Table 6; for a complete pathway list, see eTables 5–12). We therefore identified those pathways shared by two or more miRNAs, to investigate the potential cross-talk among the miRNAs in regulating pathways related to immune response. 18 out of 54 of these pathways related to immune response were shared by at least two miRNAs (Table 7), in particular the “High-mobility group protein B1 (HMGB1)/Advanced glycosylation end product-specific receptor (RAGE) signaling pathway” was shared by mir-126, -146a, -155, -21 and -222 (eFigure 2).
Table 5.
MiRNA Experimentally Validated Gene Targets and Predicted Gene Targets.
| MiRNA | Gene Targets
|
||||
|---|---|---|---|---|---|
| No. Validated | Predicted
|
No. Analyzedb | |||
| No. Conserved targets | No. Sites (no. conserved) | Total Context Score < −0.2 (% inversely correlateda) | |||
| miR-1 | 190 | 790 | 1080 (860) | 269 (41%) | 407 |
| miR-126 | 18 | 25 | 25 (25) | 20 (37%) | 33 |
| miR-135a | 3 | 718 | 990 (786) | 269 (48%) | 269 |
| miR-146a | 23 | 224 | 321 (233) | 79 (25%) | 99 |
| miR-155 | 161 | 440 | 626 (472) | 135 (34%) | 273 |
| miR-21 | 67 | 307 | 412 (329) | 121 (37%) | 175 |
| miR-222 | 19 | 446 | 619 (465) | 217 (38%) | 232 |
| miR-9 | 15 | 1237 | 1753 (1408) | 232 (55%) | 244 |
Percentage of predicted targets with total context score <−0.2 inversely correlated (r<−0.5) with miRNA in at least one deep sequencing dataset in MirGator.
Total analyzed targets for each miRNA (experimentally validated and predicted with total context score < −0.2).
Table 6.
GeneGo Pathways Significantly Enriched in Target Genes of the Investigated MiRNAs.
| miRNA | No. Pathways
|
|
|---|---|---|
| Total | False Discovery Rate < 0.05 Total (Immune response-related) |
|
| miR-1 | 348 | 6 (1) |
| miR-126 | 184 | 60 (7) |
| miR-135a | 334 | 16 (0) |
| miR-146a | 296 | 52 (20) |
| miR-155 | 427 | 95 (17) |
| miR-21 | 295 | 48 (9) |
| miR-222 | 387 | 82 (25) |
| miR-9 | 304 | 15 (0) |
| Total | 613 | 243 (54) |
Table 7.
Immune-Response-Related GeneGo Pathways Enriched in Target Genes of at Least Two out of Eight MiRNAs Associated With Pollutants (FDR < 0.05).
| GeneGo Pathway | miRNA | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| miR-126 | miR-146a | miR-155 | miR-21 | miR-222 | ||||||
| ratioa | −log(p) | ratioa | −log(p) | ratioa | −log(p) | ratioa | −log(p) | ratioa | −log(p) | |
|
|
||||||||||
| C5a signaling | n/a | n/a | 3/50 | 2.71 | 4/50 | 2.71 | n/a | n/a | n/a | n/a |
| CCL2 signaling | 2/54 | 2.55 | n/a | n/a | 6/54 | 4.65 | n/a | n/a | 7/54 | 6.48 |
| CD40 signaling | n/a | n/a | 6/65 | 6.12 | n/a | n/a | n/a | n/a | n/a | n/a |
| ETV3 affect on CSF1-promoted macrophage differentiation | n/a | n/a | n/a | n/a | 3/31 | 2.36 | n/a | n/a | 3/31 | 2.64 |
| Gastrin in inflammatory response | 2/69 | 2.35 | 4/69 | 3.42 | n/a | n/a | n/a | n/a | n/a | n/a |
| HMGB1/RAGE signaling pathway | 2/53 | 2.57 | 3/53 | 2.63 | 5/53 | 3.61 | 4/53 | 3.18 | 3/53 | 1.98 |
| HSP60 and HSP70/TLR signaling pathway | n/a | n/a | 7/54 | 8.13 | n/a | n/a | 4/54 | 3.15 | n/a | n/a |
| IL-17 signaling pathways | n/a | n/a | 3/60 | 2.48 | 6/60 | 4.39 | n/a | n/a | 4/60 | 2.77 |
| IL-22 signaling pathway | n/a | n/a | n/a | n/a | n/a | n/a | 3/34 | 2.68 | 4/34 | 3.72 |
| IL-3 activation and signaling pathway | n/a | n/a | n/a | n/a | 3/31 | 2.36 | n/a | n/a | 3/31 | 2.64 |
| IL-5 signaling | n/a | n/a | n/a | n/a | n/a | n/a | 4/44 | 3.5 | 4/44 | 3.28 |
| IL-9 signaling pathway | 2/36 | 2.9 | n/a | n/a | n/a | n/a | n/a | n/a | 3/36 | 2.45 |
| Innate immune response to RNA viral infection | n/a | n/a | 3/28 | 3.45 | 3/28 | 2.49 | n/a | n/a | n/a | n/a |
| MIF in innate immunity response | n/a | n/a | 3/40 | 2.99 | 3/40 | 2.05 | n/a | n/a | n/a | n/a |
| MIF-mediated glucocorticoid regulation | n/a | n/a | 2/22 | 2.27 | 4/22 | 4.1 | n/a | n/a | 4/22 | 4.49 |
| PIP3 signaling in B lymphocytes | n/a | n/a | n/a | n/a | 3/42 | 1.99 | n/a | n/a | 3/42 | 2.26 |
| Role of HMGB1 in dendritic cell maturation and migration | n/a | n/a | 4/27 | 5.05 | n/a | n/a | 3/27 | 2.98 | n/a | n/a |
| Signaling pathway mediated by IL-6 and IL-1 | n/a | n/a | 3/30 | 3.36 | 4/30 | 3.56 | n/a | n/a | n/a | n/a |
Abbreviations: C5a, complement component 5a; CCL2, C-C motif chemokine 2; CSF1, colony stimulating factor 1; ETV3, ets variant 3; HMGB1, High mobility group protein B1; HSP, heat shock protein; IL, interleukin; MIF, macrophage migration inhibitory factor; PIP3, Phosphatidylinositol (3,4,5)-triphosphate; RAGE, Advanced glycosylation end product-specific receptor; TLR, toll-like receptor; n/a, not applicable.
miRN/A target genes/total genes in the pathway.
Since prediction algorithms can provide false-positives results,27 we also tested the expression correlation of miRNAs and predicted targets included in the pathways analysis, using data available in MirGator. The percentage of predicted targets that were inversely correlated (Spearman’s r < −0.5) with miRNA in at least one deep-sequencing dataset in MirGator ranged from 25% (miR-146a) to 55% (miR-9) (Table 5; for complete results see eTable 4).
Discussion
In this study of elderly men, we investigated the effects of several exposure windows to PM2.5, black carbon, organic carbon and SO42− on the expression of fourteen candidate miRNAs involved in processes related to PM exposure (i.e. inflammation, endothelial dysfunction and atherosclerosis).4,5,9,11–14 We observed an overall negative association between the investigated pollutants in all exposure windows (4 hours to 28 days) and miR-1, -126, -135a, -146a, -155, -21, -222, and -9, with similar trends of the associations through the time windows in all miRNAs. Since higher levels of these miRNAs tend to be associated with less expression of their target mRNAs, this negative association would be expected to result in increased inflammation, endothelial dysfunction, and atherosclerosis. The strongest effect for PM2.5 and black carbon was observed for seven-day exposure. The negative association between organic carbon and miRNAs were generally strongest for the 48-hour moving average. The association with sulfates was mostly stable across the moving averages. Seven-day moving averages of PM2.5 and of black carbon (an indicator of traffic-related particles) have also been reported to be relevant for several outcomes in the Normative Aging Study study, including DNA methylation, another mechanism of epigenetic regulation of gene expression.28 For PM2.5 the associations emerged as early as the 4-hour exposure measure. We did not find meaningful associations for the miR-125a-5p, -125b, -128, -147, -218, and -96. Moreover, we found three SNPs in GEMIN4 that modified the observed association with PM2.5 and black carbon for miR-1, miR-126, miR-146a, miR-222, and miR-9, and one SNP in DGCR8 that modified the observed association with SO42− for miR-1 and miR-135a.
Few studies have investigated changes in miRNAs expression in response to environmental stressors, and even fewer have addressed exposure to PM.15,29,30197 of 313 miRNAs were up- or down-regulated ≥ 1.5-fold in human bronchial epithelial cells after treatment with diesel exhaust particles.16 MiR-375 was found upregulated following diesel-exhaust-particle exposure and ambient fine PM exposure in primary human bronchial epithelial cells.17 A decrease of several miRNAs was demonstrated in the myocardium of PM exposed rats.18 In a study of pulmonary global miRNA responses to carbon black nanoparticles in mice, the authors found a marked increases in miR-135b and subtle changes in miR-21 and miR-146b.19 In a study of foundry workers, an up-regulation of selected miRNA, measured by real-time PCR (i.e. miR-222 and -21, but not miR-146a) was observed after short-term occupational exposure (3 days) to metal-rich PM.20 In the same study, the authors performed a microarray analysis of 847 human miRNAs and identified four differentially expressed miRNAs post-exposure (namely, miR-421, -146a, -29a, and let-7g).21
Our study investigated the association in humans between ambient particles in different time windows and in vivo expression of candidate miRNAs, and investigated the role of polymorphisms in miRNA processing genes as effects modifiers in these associations. None of the fourteen miRNAs investigated in our study was upregulated. This is in keeping with results in in vitro and in in vivo studies showing mainly downregulation of miRNAs in the myocardium of PM-exposed rats18 and in cigarette-smoke exposed human airway epithelial cells31 and murine lungs.32
We found miR-21, miR-146a, and miR-222 to be negatively associated with exposure to pollutants. Two of these miRNAs (i.e. miR-146a and -21) have also been found to be decreased in the myocardium of rats exposed to PM.18 In contrast, the expression of miR-21 was increased in a study on pulmonary global miRNA responses to carbon black nanoparticles in mice.19 Moreover, the expression of these miRNAs, measured by real-time PCR, was increased (miR-21 and -222) or not changed (miR-146a) in peripheral blood leukocytes of foundry workers after three days of work, compared with baseline measurements.20 The authors did not find any association with particle mass (PM10 and PM1), but an association of miR-222 and miR-146a with specific metal components of PM.20 In the same study, the authors performed a microarray analysis of 847 human miRNAs and identified four differentially expressed miRNAs post-exposure (fold change > |2|; p < 0.05), including miR-146a (fold change = 2.62; p = 0.007).21 The opposite results we obtained could be due to differences in particle composition and in the characteristics of studied participants.
In addition to databases of manually curated experimental findings on miRNA-target gene interactions, (e.g. miRTarBase), bioinformatic strategies are now available to predict potential miRNA targets, such as those implemented in TargetScan. We explored both experimentally validated and predicted targets (with the understanding that the latter are speculative) for those miRNAs associated with pollutants (namely, miR-1, -126, 135a, -146a, -155, -21, -222, and -9). We examined whether these genes are overrepresented in GeneGo pathways annotated in the MetaCore database. We understand that the predicted targets are speculative, and we therefore investigated the expression correlation of miRNAs and predicted targets included in the pathways analysis. We found the percentage of negatively correlated miRNA/mRNA couples to be lower than expected based on the estimated false-response rate for TargetScan.27 This might be due partially to the type of tissues/cells included in MirGator (i.e. mostly cancer tissues and immortalized cell lines).
We identified several pathways involved in the immune response for all the miRNAs except miR-135a and miR-9. In particular, the “HMGB1/RAGE signaling pathway” was shared by mir-126, -146a, -155, -21, and -222. HMGB1 - a highly conserved, ubiquitous protein present in the nuclei and cytoplasm of nearly different cell types, including activated macrophages monocytes and dendritic cells - is a necessary and sufficient mediator of inflammation during sterile and infection-associated responses.33 HMGB1 activates cells through the differential engagement of multiple surface receptors including RAGE.34 HMGB1-induced intracellular signaling through RAGE can activate ERK1/2 and stress-activated mitogen-activated protein kinases p38 mitogen-activated protein kinase (p38 MAPK) and c-Jun N-terminal kinases (JNK), leading to activation of transcription factors such as NF-kappa-B (NF-kB), Cyclic AMP-responsive element-binding protein 1 (CREB1), SP1, and Transcription factor AP-1 (c-Jun).35,36
The activation of these transcription factors leads to the enhanced expression of cytokines and molecules thought to play a role in particle effects,2 including proinflammatory cytokines (such as tumor necrosis factor alpha, interleukin [IL] 1-beta, IL-6 and IL-8), adhesion molecules (i.e. vascular cell adhesion molecule 1 and intercellular adhesion molecule 1), and coagulation factors (i.e. plasminogen activator inhibitor-1, tissue-type plasminogen activator and tissue factor).37–40 Moreover, several miRNAs have been reported to directly or indirectly inhibit the transcription factor NF-kB, including miR-146-a, -155, -21 and -9,4 and NF-kB can in turn regulate the expression of several miRNA (e.g. miR-146a, -21).4 RAGE and HMGB1 mRNAs were found to be increased in rats exposed to ozone plus diesel exhaust particulate,41 suggesting a role in PM mechanisms. Also, cellular signaling through RAGE has been suggested to have a role in diesel-exhaust-particle-induced NF-kB-activation and chemokine responses in a type-I-like epithelial cell line (R3/1).42 Our results suggest that the “HMGB1/RAGE signaling pathway” may also be involved in mechanisms related to PM2.5 and the investigated components, such as inflammation, coagulation and endothelial dysfunction, and that miR-126, -146a, -155, -21, and -222 can play a role in the regulation of this pathway in response to particles.
Expression of miRNAs can be controlled by several mechanisms, including epigenetic mechanisms (e.g. DNA methylation), and SNPs in both the promoter region of miRNA or in miRNA processing genes. Several findings suggest miRNA gene silencing by CpG island methylation in the miRNA promoter region.43,44 On the other hand, SNPs in the promoter region of some miRNA (e.g. miR-146a45) have been shown to regulate miRNA expression. Moreover, SNPs in genes involved in miRNA biogenesis and processing may affect miRNA expression. We focused on SNPs in two genes involved in miRNA processing - namely GEMIN4 and DGCR8, which we recently found to modify the effect of black carbon and PM2.5 on blood pressure and adhesion molecules.23,24 According to the current model for miRNAs transcription and processing,22 miRNAs originate from longer precursor RNAs called primary miRNAs (pri-miRNAs). Pri-miRNAs are cleaved in the nucleus into a 70- to 100-nucleotide hairpin-shaped precursor miRNA (pre-miRNA) by an enzymatic complex consisting of the RNase III enzyme Drosha and its binding partner DGCR8. Pre-miRNAs are transported from the nucleus and further processed into a 19- to 25-nucleotide double-stranded duplex. The double-strand duplex is then separated by a helicase into the functional guide strand and the passenger strand. Multiple helicases have been linked to the miRNA pathway, including GEMIN4.46 The functional strand of the mature miRNA interact with argonaute-2 to form a ribonucleoprotein, which guides the miRNA into the RNA-induced silencing complex, where the miRNA strand anneals to the 3′ untranslated regions of target mRNAs, promoting translational repression or mRNA degradation.22
We found a modification of effect of black carbon and PM2.5 for SNPs in GEMIN4 for some but not all the miRNA that were found associated with pollutants. We can speculate that helicases other than GEMIN4 could be more relevant for some of the investigated miRNAs. We found that two SNPs in GEMIN4 (rs7813 and rs910925) that have previously been found to be in high linkage disequilibrium,23 similarly predicted miR-146a and -222, with the homozygous variant carrier showing lower miRNAs expression for exposure to black carbon. Conversely, rs1062923 in GEMIN4 wild-type persons and heterozygous variant carriers showed lower expression of all five miRNAs except for miR-222 in response to black carbon and miR-1 and -126 in response to PM2.5. For rs1062923 there were only 6 homozygous variant carriers, and we cannot exclude that the observed results were due to chance. The opposite direction in effect modification by these SNPs is consistent with the results from a previous study by our group.23 We also found that rs1640299 in DGCR8 modified the association between SO42− and miR-1 and -135a. The downregulation of DGCR8 in murine myoblast cells overexpressing heme-oxigenases (a cytoprotective enzyme) was associated with lower expression of 50% of the investigated miRNA (including miR-1) compared with control cells,47 suggesting that DGCR8 may be involved in the biogenesis of specific miRNAs.
Our results are subject to a number of limitations. Generalizability of results to other populations is limited, because we investigated predominantly non-Hispanic white elderly males and because air pollutant composition and concentration of particles in the Greater Boston area may differ from those in other urban environments.
There may be some misclassification of exposure in our analysis. We have utilized stationary measures of air pollution to represent personal exposures. Prior research indicates that when examining longitudinal exposures to air pollution, most error is of the Berkson type. Simulation studies indicate that this exposure misclassification may lead to an underestimation of the health effects of air pollution.48 In addition, several studies, including one conducted in the Greater Boston area, have found that longitudinal measures of ambient particulate concentrations are representative of longitudinal variation in personal exposures.49
Since expression levels of key target genes were not measured in this study, we were not able to demonstrate that observed changes in the expression of miRNAs corresponded to changes in the expression of their targets. Moreover, using data available in a public database we found the percentage of negatively correlated miRNA/mRNA couples to be lower than expected based on the estimated false-response rate for the software we used,27 although this might be due in part to the type of tissues/cells included in the database (i.e. mostly cancer tissues and immortalized cell lines).
MiRNAs are tissue-specific, and tissue other than peripheral blood leukocytes could be more representative for the investigated mechanisms; however, blood is an easily obtainable biological medium. Whereas blood leukocytes have been linked to PM-related inflammatory and coagulatory responses, our findings cannot be directly compared to experimental models using other tissues. We cannot exclude the possibility that other miRNAs could be more relevant for PM-related disease mechanisms.
We tested several pollutants and miRNAs, as well as several SNP-by-pollutant interactions, and thus the possibility of chance findings due to multiple testing should be considered. However, if one in 20 tests at the 95% confidence level are expected to be significant due to chance, 26 significant findings out of 56 tests in our main analysis (4 pollutants * 14 miRNAs) and 12 significant findings out of 128 tests in the SNP-by-pollutant analysis (4 pollutants * 8 miRNA * 4 SNPs) exceeds this.
In conclusion, our results suggests that exposure to ambient air particles, in particular traffic-related particles, causes a downregulation of candidate microRNAs involved in processed related to PM exposure (such as inflammation, endothelial dysfunction and coagulation) in elderly men. Polymorphisms in two microRNA-related genes, GEMIN4 and DGCR8, could modify the observed associations. Further research is needed to replicate our findings in different and larger cohorts, considering both a wider number of candidate miRNAs and the expression of candidate target genes.
Supplementary Material
Acknowledgments
Source of Funding
This publication was made possible by: NIEHS 1-RO1 ES015172, 2-RO1 ES015172, ES014663, ES00002, ES009825, ES020010, USEPA R832416, RD 83479801 and P42 ES016454. The VA Normative Aging Study, a component of the Massachusetts Veterans Epidemiology Research and Information Center, Boston, Massachusetts, is supported by the Cooperative Studies Program/Epidemiology Research and Information Center of the U.S. Department of Veterans Affairs.
Footnotes
Conflict of Interest
The authors declare they have no actual or potential competing financial interests.
References
- 1.Zanobetti A, Schwartz J. The effect of fine and coarse particulate air pollution on mortality: a national analysis. Environmental health perspectives. 2009;117(6):898–903. doi: 10.1289/ehp.0800108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brook RD, Rajagopalan S, Pope CA, et al. Particulate matter air pollution and cardiovascular disease: An update to the scientific statement from the American Heart Association. Circulation. 2010;121(21):2331–78. doi: 10.1161/CIR.0b013e3181dbece1. [DOI] [PubMed] [Google Scholar]
- 3.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nature reviews. Genetics. 2004;5(7):522–31. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 4.Ma X, Becker Buscaglia LE, Barker JR, Li Y. MicroRNAs in NF-{kappa}B signaling. Journal of molecular cell biology. 2011;3(3):159–66. doi: 10.1093/jmcb/mjr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guerau-de-Arellano M, Smith KM, Godlewski J, et al. Micro-RNA dysregulation in multiple sclerosis favours pro-inflammatory T-cell-mediated autoimmunity. Brain_: a journal of neurology. 2011;134(Pt 12):3578–89. doi: 10.1093/brain/awr262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gonsalves CS, Kalra VK. Hypoxia-mediated expression of 5-lipoxygenase-activating protein involves HIF-1alpha and NF-kappaB and microRNAs 135a and 199a-5p. Journal of immunology (Baltimore, Md: 1950) 2010;184(7):3878–88. doi: 10.4049/jimmunol.0902594. [DOI] [PubMed] [Google Scholar]
- 7.Lindberg RLP, Hoffmann F, Mehling M, Kuhle J, Kappos L. Altered expression of miR-17-5p in CD4+ lymphocytes of relapsing-remitting multiple sclerosis patients. European journal of immunology. 2010;40(3):888–98. doi: 10.1002/eji.200940032. [DOI] [PubMed] [Google Scholar]
- 8.Liu G, Friggeri A, Yang Y, Park Y-J, Tsuruta Y, Abraham E. miR-147, a microRNA that is induced upon Toll-like receptor stimulation, regulates murine macrophage inflammatory responses. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(37):15819–24. doi: 10.1073/pnas.0901216106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris Ta, Yamakuchi M, Ferlito M, Mendell JT, Lowenstein CJ. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(5):1516–21. doi: 10.1073/pnas.0707493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Small EM, Sutherland LB, Rajagopalan KN, Wang S, Olson EN. MicroRNA-218 regulates vascular patterning by modulation of Slit-Robo signaling. Circulation research. 2010;107(11):1336–44. doi: 10.1161/CIRCRESAHA.110.227926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yao R, Ma Y, Du Y, et al. The altered expression of inflammation-related microRNAs with microRNA-155 expression correlates with Th17 differentiation in patients with acute coronary syndrome. Cellular & molecular immunology. 2011;8(6):486–95. doi: 10.1038/cmi.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen T, Huang Z, Wang L, et al. MicroRNA-125a-5p partly regulates the inflammatory response, lipid uptake, and ORP9 expression in oxLDL-stimulated monocyte/macrophages. Cardiovascular research. 2009;83(1):131–9. doi: 10.1093/cvr/cvp121. [DOI] [PubMed] [Google Scholar]
- 13.Rink C, Khanna S. MicroRNA in ischemic stroke etiology and pathology. Physiological genomics. 2011;43(10):521–8. doi: 10.1152/physiolgenomics.00158.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kondkar AA, Bray MS, Leal SM, et al. VAMP8/endobrevin is overexpressed in hyperreactive human platelets: suggested role for platelet microRNA. Journal of thrombosis and haemostasis_: JTH. 2010;8(2):369–78. doi: 10.1111/j.1538-7836.2009.03700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hou L, Wang D, Baccarelli A. Environmental chemicals and microRNAs. Mutation research. 2011;714(1–2):105–112. doi: 10.1016/j.mrfmmm.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jardim MJ, Fry RC, Jaspers I, Dailey L, Diaz-Sanchez D. Disruption of microRNA expression in human airway cells by diesel exhaust particles is linked to tumorigenesis-associated pathways. Environmental health perspectives. 2009;117(11):1745–51. doi: 10.1289/ehp.0900756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bleck B, Grunig G, Chiu A, et al. MicroRNA-375 Regulation of Thymic Stromal Lymphopoietin by Diesel Exhaust Particles and Ambient Particulate Matter in Human Bronchial Epithelial Cells. Journal of immunology (Baltimore, Md: 1950) 2013;190(7):3757–63. doi: 10.4049/jimmunol.1201165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farraj AK, Hazari MS, Haykal-Coates N, et al. ST depression, arrhythmia, vagal dominance, and reduced cardiac micro-RNA in particulate-exposed rats. American journal of respiratory cell and molecular biology. 2011;44(2):185–96. doi: 10.1165/rcmb.2009-0456OC. [DOI] [PubMed] [Google Scholar]
- 19.Bourdon JA, Saber AT, Halappanavar S, et al. Carbon black nanoparticle intratracheal installation results in large and sustained changes in the expression of miR-135b in mouse lung. Environmental and molecular mutagenesis. 2012;53(6):462–8. doi: 10.1002/em.21706. [DOI] [PubMed] [Google Scholar]
- 20.Bollati V, Marinelli B, Apostoli P, et al. Exposure to metal-rich particulate matter modifies the expression of candidate microRNAs in peripheral blood leukocytes. Environmental health perspectives. 2010;118(6):763–8. doi: 10.1289/ehp.0901300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Motta V, Angelici L, Nordio F, et al. Integrative Analysis of miRNA and Inflammatory Gene Expression After Acute Particulate Matter Exposure. Toxicological sciences_: an official journal of the Society of Toxicology. 2013;132(2):307–16. doi: 10.1093/toxsci/kft013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nature cell biology. 2009;11(3):228–34. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 23.Wilker EH, Baccarelli A, Suh H, Vokonas P, Wright RO, Schwartz J. Black carbon exposures, blood pressure, and interactions with single nucleotide polymorphisms in MicroRNA processing genes. Environmental health perspectives. 2010;118(7):943–8. doi: 10.1289/ehp.0901440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilker EH, Alexeeff SE, Suh H, Vokonas PS, Baccarelli A, Schwartz J. Ambient pollutants, polymorphisms associated with microRNA processing and adhesion molecules: the Normative Aging Study. Environmental health_: a global access science source. 2011;10:45. doi: 10.1186/1476-069X-10-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bell B, Rose CL, Damon A. The Veterans Administration longitudinal study of healthy aging. [Accessed April 25, 2012];The Gerontologist. 1966 6(4):179–84. doi: 10.1093/geront/6.4.179. Available at: http://www.ncbi.nlm.nih.gov/pubmed/5342911. [DOI] [PubMed] [Google Scholar]
- 26.Zanobetti A, Schwartz J. Air pollution and emergency admissions in Boston, MA. Journal of epidemiology and community health. 2006;60(10):890–5. doi: 10.1136/jech.2005.039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Min H, Yoon S. Got target? Computational methods for microRNA target prediction and their extension. Experimental & molecular medicine. 2010;42(4):233–44. doi: 10.3858/emm.2010.42.4.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baccarelli A, Wright RO, Bollati V, et al. Rapid DNA methylation changes after exposure to traffic particles. American journal of respiratory and critical care medicine. 2009;179(7):572–8. doi: 10.1164/rccm.200807-1097OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sonkoly E, Pivarcsi A. MicroRNAs in inflammation and response to injuries induced by environmental pollution. Mutation research. 2011;717(1–2):46–53. doi: 10.1016/j.mrfmmm.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 30.Jardim MJ. microRNAs: Implications for air pollution research. Mutation research. 2011;717(1–2):38–45. doi: 10.1016/j.mrfmmm.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 31.Schembri F, Sridhar S, Perdomo C, et al. MicroRNAs as modulators of smoking-induced gene expression changes in human airway epithelium. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(7):2319–24. doi: 10.1073/pnas.0806383106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Izzotti A, Calin Ga, Arrigo P, Steele VE, Croce CM, De Flora S. Downregulation of microRNA expression in the lungs of rats exposed to cigarette smoke. The FASEB journal_: official publication of the Federation of American Societies for Experimental Biology. 2009;23(3):806–12. doi: 10.1096/fj.08-121384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang H, Tracey KJ. Targeting HMGB1 in inflammation. Biochimica et biophysica acta. 1799(1–2):149–56. doi: 10.1016/j.bbagrm.2009.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sims GP, Rowe DC, Rietdijk ST, Herbst R, Coyle AJ. HMGB1 and RAGE in inflammation and cancer. Annual review of immunology. 2010;28:367–88. doi: 10.1146/annurev.immunol.021908.132603. [DOI] [PubMed] [Google Scholar]
- 35.Andersson U, Erlandsson-Harris H, Yang H, Tracey KJ. HMGB1 as a DNA-binding cytokine. [Accessed April 26, 2013];Journal of leukocyte biology. 2002 72(6):1084–91. Available at: http://www.ncbi.nlm.nih.gov/pubmed/12488489. [PubMed] [Google Scholar]
- 36.Yang H, Wang H, Czura CJ, Tracey KJ. The cytokine activity of HMGB1. Journal of leukocyte biology. 2005;78(1):1–8. doi: 10.1189/jlb.1104648. [DOI] [PubMed] [Google Scholar]
- 37.Taniguchi N, Kawahara K, Yone K, et al. High mobility group box chromosomal protein 1 plays a role in the pathogenesis of rheumatoid arthritis as a novel cytokine. Arthritis and rheumatism. 2003;48(4):971–81. doi: 10.1002/art.10859. [DOI] [PubMed] [Google Scholar]
- 38.Kohka Takahashi H, Sadamori H, Liu K, et al. Role of cell-cell interactions in high mobility group box 1 cytokine activity in human peripheral blood mononuclear cells and mouse splenocytes. European journal of pharmacology. 2013;701(1–3):194–202. doi: 10.1016/j.ejphar.2012.11.058. [DOI] [PubMed] [Google Scholar]
- 39.Treutiger CJ, Mullins GE, Johansson A-SM, et al. High mobility group 1 B-box mediates activation of human endothelium. [Accessed April 26, 2013];Journal of internal medicine. 2003 254(4):375–85. doi: 10.1046/j.1365-2796.2003.01204.x. Available at: http://www.ncbi.nlm.nih.gov/pubmed/12974876. [DOI] [PubMed] [Google Scholar]
- 40.Fiuza C, Bustin M, Talwar S, et al. Inflammation-promoting activity of HMGB1 on human microvascular endothelial cells. Blood. 2003;101(7):2652–60. doi: 10.1182/blood-2002-05-1300. [DOI] [PubMed] [Google Scholar]
- 41.Kodavanti UP, Thomas R, Ledbetter AD, et al. Vascular and cardiac impairments in rats inhaling ozone and diesel exhaust particles. Environmental health perspectives. 2011;119(3):312–8. doi: 10.1289/ehp.1002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reynolds PR, Wasley KM, Allison CH. Diesel particulate matter induces receptor for advanced glycation end-products (RAGE) expression in pulmonary epithelial cells, and RAGE signaling influences NF-κB-mediated inflammation. Environmental health perspectives. 2011;119(3):332–6. doi: 10.1289/ehp.1002520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han L, Witmer PDW, Casey E, Valle D, Sukumar S. DNA methylation regulates microRNA expression. Cancer Biology & Therapy. 2007;6(8):1290–1294. doi: 10.4161/cbt.6.8.4486. [DOI] [PubMed] [Google Scholar]
- 44.Saito Y, Liang G, Egger G, et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer cell. 2006;9(6):435–43. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 45.Luo X, Yang W, Ye D-Q, et al. A functional variant in microRNA-146a promoter modulates its expression and confers disease risk for systemic lupus erythematosus. PLoS genetics. 2011;7(6):e1002128. doi: 10.1371/journal.pgen.1002128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mourelatos Z, Dostie J, Paushkin S, et al. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes & development. 2002;16(6):720–8. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kozakowska M, Ciesla M, Stefanska A, et al. Heme oxygenase-1 inhibits myoblast differentiation by targeting myomirs. Antioxidants & redox signaling. 2012;16(2):113–27. doi: 10.1089/ars.2011.3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeger SL, Thomas D, Dominici F, et al. Exposure measurement error in time-series studies of air pollution: concepts and consequences. [Accessed March 11, 2013];Environmental health perspectives. 2000 108(5):419–26. doi: 10.1289/ehp.00108419. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1638034&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rojas-Bracho L, Suh HH, Koutrakis P. Relationships among personal, indoor, and outdoor fine and coarse particle concentrations for individuals with COPD. [Accessed March 11, 2013];Journal of exposure analysis and environmental epidemiology. 10(3):294–306. doi: 10.1038/sj.jea.7500092. Available at: http://www.ncbi.nlm.nih.gov/pubmed/10910121. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.