ABSTRACT
For nearly 3 decades, listeriologists and immunologists have used mainly three strains of the same serovar (1/2a) to analyze the virulence of the bacterial pathogen Listeria monocytogenes. The genomes of two of these strains, EGD-e and 10403S, were released in 2001 and 2008, respectively. Here we report the genome sequence of the third reference strain, EGD, and extensive genomic and phenotypic comparisons of the three strains. Strikingly, EGD-e is genetically highly distinct from EGD (29,016 single nucleotide polymorphisms [SNPs]) and 10403S (30,296 SNPs), and is more related to serovar 1/2c than 1/2a strains. We also found that while EGD and 10403S strains are genetically very close (317 SNPs), EGD has a point mutation in the transcriptional regulator PrfA (PrfA*), leading to constitutive expression of several major virulence genes. We generated an EGD-e PrfA* mutant and showed that EGD behaves like this strain in vitro, with slower growth in broth and higher invasiveness in human cells than those of EGD-e and 10403S. In contrast, bacterial counts in blood, liver, and spleen during infection in mice revealed that EGD and 10403S are less virulent than EGD-e, which is itself less virulent than EGD-e PrfA*. Thus, constitutive expression of PrfA-regulated virulence genes does not appear to provide a significant advantage to the EGD strain during infection in vivo, highlighting the fact that in vitro invasion assays are not sufficient for evaluating the pathogenic potential of L. monocytogenes strains. Together, our results pave the way for deciphering unexplained differences or discrepancies in experiments using different L. monocytogenes strains.
IMPORTANCE
Over the past 3 decades, Listeria has become a model organism for host-pathogen interactions, leading to critical discoveries in a broad range of fields, including bacterial gene regulation, cell biology, and bacterial pathophysiology. Scientists studying Listeria use primarily three pathogenic strains: EGD, EGD-e, and 10403S. Despite many studies on EGD, it is the only one of the three strains whose genome has not been sequenced. Here we report the sequence of its genome and a series of important genomic and phenotypic differences between the three strains, in particular, a critical mutation in EGD’s PrfA, the main regulator of Listeria virulence. Our results show that the three strains display differences which may play an important role in the virulence differences observed between the strains. Our findings will be of critical relevance to listeriologists and immunologists who have used or may use Listeria as a tool to study the pathophysiology of listeriosis and immune responses.
INTRODUCTION
Listeria monocytogenes is a low-GC-content, Gram-positive, rod-shaped bacterium living in a variety of environments, such as soil and decaying vegetation, and can infect animals and humans by means of contaminated food products. The pathogenic properties of L. monocytogenes rely on its ability to cross three host barriers (the intestinal, placental, and blood-brain barriers) and also its ability to enter, replicate, and survive in wide range of human cell types, such as macrophages, epithelial cells, and endothelial cells, thanks to an arsenal of virulence factors. More than 50 virulence factors have been described (1), and the list continuously expands.
During the last three decades, L. monocytogenes has emerged as a model organism for the study of host-pathogen interactions (2–5), leading to critical discoveries in a broad range of fields, including virulence factor regulation, cell biology, bacterial adaptation to the host cytosol, and bacterial pathophysiology. In addition, since the pioneering studies of Mackaness (6), L. monocytogenes has been widely used as a model to study its interaction with professional phagocytes and host T-cell responses. Remarkably, most of these discoveries have been made using three L. monocytogenes strains. These widely used strains are the 10403S, EGD, and EGD-e L. monocytogenes strains. The genome of the EGD-e strain was sequenced in 2001 (RefSeq accession number NC_003210 [7]). The sequence and annotation of the 10403S genome have recently been released (NC_017544), as have those of several other strains (8–12). Currently, NCBI’s RefSeq database contains 39 L. monocytogenes genomes, and this number will probably continue to grow exponentially in the coming years. In this context, the unknown sequence of the extensively used strain EGD remained a gap to fill.
The EGD strain is from the Trudeau Institute (NCTC7973) and derived from the original strain isolated from guinea pigs by E. G. D. Murray et al. in 1926 (13). The name Listeria monocytogenes was definitively coined by Pirie (14). Strain EGD was brought back to France by Patrick Berche (see reference 15) in 1982 after a stay at the Trudeau Institute with Robert North. Helmuth Hahn also obtained strain EGD from the Trudeau Institute and gave it to Trinad Chakraborty in 1986 (see reference 16). The two strains from the Trudeau Institute used to be passaged through mice to maintain virulence. When the Listeria genome sequencing project was initiated, the European consortium chose to sequence strain EGD, which was retested for its virulence in mice by Trinad Chakraborty and thereafter named EGD-e (where “e” stands for “European” [7]). L. monocytogenes 10403S is a streptomycin-resistant (83) derivative of 10403 reported to be isolated from human skin lesions in Bozeman, MT (17).
The three strains belong to serovar 1/2a. The serotyping scheme, based on somatic (O) and flagellar (H) antigens, is the oldest technique used to differentiate L. monocytogenes strains (18) and has enabled classification of L. monocytogenes in three main lineages (I, II, and III). A subpopulation of lineage III, lineage IIIB, is now called lineage IV (19, 20). Strikingly, a phylogenetic study by multilocus sequence typing (MLST) demonstrated that despite the fact that EGD-e is of serotype 1/2a, it clusters with 1/2c strains and is distantly related to 10403S and EGD (21). Phenotypic differences among the three strains have in the past been observed by listeriologists but not published. However, in different studies that we reported, EGD was used in preference to EGD-e because of its higher invasiveness in human cells (22–27). Nevertheless, until now, no study has been performed to characterize in detail the differences between the three Listeria reference strains.
We report here the sequence and the annotation of the genome of L. monocytogenes EGD and a genomic and phenotypic comparison of the three laboratory model strains, EGD, EGD-e, and 10403S. A comparison of protein-coding genes and noncoding RNAs shows that even if two of the three strains have nearly the same name (EGD-e and EGD), they differ, with EGD being closer to 10403S and EGD-e being more distant. One major difference is a PrfA mutation found in EGD that induces an overexpression of the PrfA-regulated genes (PrfA*), leading to a higher invasiveness in cultured cells and a difference in virulence in animal models.
RESULTS
Resequencing of the EGD-e genome sequence.
Prior to sequencing the L. monocytogenes EGD genome, we resequenced the genome of strain EGD-e using the Illumina technique. Only five differences compared to the published sequence were found (Fig. 1A), confirming the high quality of the first published sequence (7, 28). As shown in Data Set S1 in the supplemental material, four of the five differences are in intergenic regions, where no small RNAs (sRNAs) have been identified so far, and only one difference induces an amino acid change, i.e., a glycine to a valine, in Lmo0247, a hypothetical protein.
FIG 1 .
SNPs, synteny, and sequence type analysis of EGD, EGD-e, and 10403S. (A) SNPs among the EGD, 10403S, and EGD-e reference genomes. Purple indicates synonymous changes, blue indicates nonsynonymous changes, and black indicates intergenic changes. (B) Minimum spanning tree analysis of 360 L. monocytogenes strains based on MLST (multilocus sequence typing) data (adapted from reference 21). The EGD-e, EGD, and 10403S strains are highlighted in red. (C) Linear synteny view of the three strains. Phage BO25 is integrated into EGD in tRNAArg. Phage A118 is integrated inside the comK gene in EGD-e and 10403S. ComK is complete in EGD.
EGD’s genome sequence and its comparison to those of EGD-e and 10403S.
The EGD genome was sequenced by the Illumina technique, assembled, annotated, and deposited in the European Nucleotide Archive (ENA) (accession number HG421741). Strain EGD has one chromosome of 2,907,193 bp and no plasmid. This genome is of approximately the same size as that of strain 10403S (2,903,106 bp). The EGD-e genome (7) is 40 kb larger, with a total size of 2,944,528 bp (Table 1).
TABLE 1 .
General properties of EGD-e, EGD, and 10403S sequences and annotations
| Listeria monocytogenes strain | Sequence |
Annotation |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Genome size (bp) | No. of SNPs | % G+C content | No. of ORFs | No. of rRNAs | No. of tRNAs | No. of sRNAs | No. of internalins | Prophage (attB integration site) | |
| EGD-e | 2,944,528 | 5 between EGD-e (7) and resequenced EGD-e | 39 | 2,846 | 18 | 67 | 154 | 26 | A118 (comK) |
| EGD | 2,907,193 | 29,016 between EGD and EGD-e; 317 between EGD and 10403S | 39 | 2,848 | 18 | 67 | 144 | 25 | B025 (tRNAArg) |
| 10403S | 2,903,106 | 30,296 between 10403S and EGD-e | 39 | 2,814 | 18 | 67 | 145 | 26 | A118 (comK) |
A single nucleotide polymorphism (SNP) search, using MUMmer (29), comparing all three strains to each other revealed 29,016 SNPs (Table 1) between EGD and EGD-e (Fig. 1A and Data Set S1) and 30,296 SNPs between 10403S and EGD-e. In contrast, only 317 SNPs distinguish EGD from 10403S, indicating that EGD and 10403S are genetically very close. This result is consistent with the reported MLST analysis of EGD and 10403S (21), which shows a high similarity (Fig. 1B) in terms of sequence type (ST) between EGD (ST 12) and 10403S (ST 85), with both strains being classified in clonal complex 7 (CC7), whereas EGD-e belongs to CC9.
We found 2,848 open reading frames (ORFs) in EGD, a number close to the 2,846 ORFs predicted for EGD-e (7) and 2,814 ORFs predicted for 10403S (Table 1). To investigate further the differences between these ORFs, we performed a bidirectional best-hit search (threshold E value, <1e−4). As shown in Fig. 2A, more than 95% (2,683) of EGD-e’s ORFs are shared by the three strains. EGD, EGD-e, and 10403S have exactly the same number of rRNAs (18 rRNAs) and tRNAs (67 tRNAs). They also have the same low GC content (39%) at the level of the whole genome (Table 1).
FIG 2 .
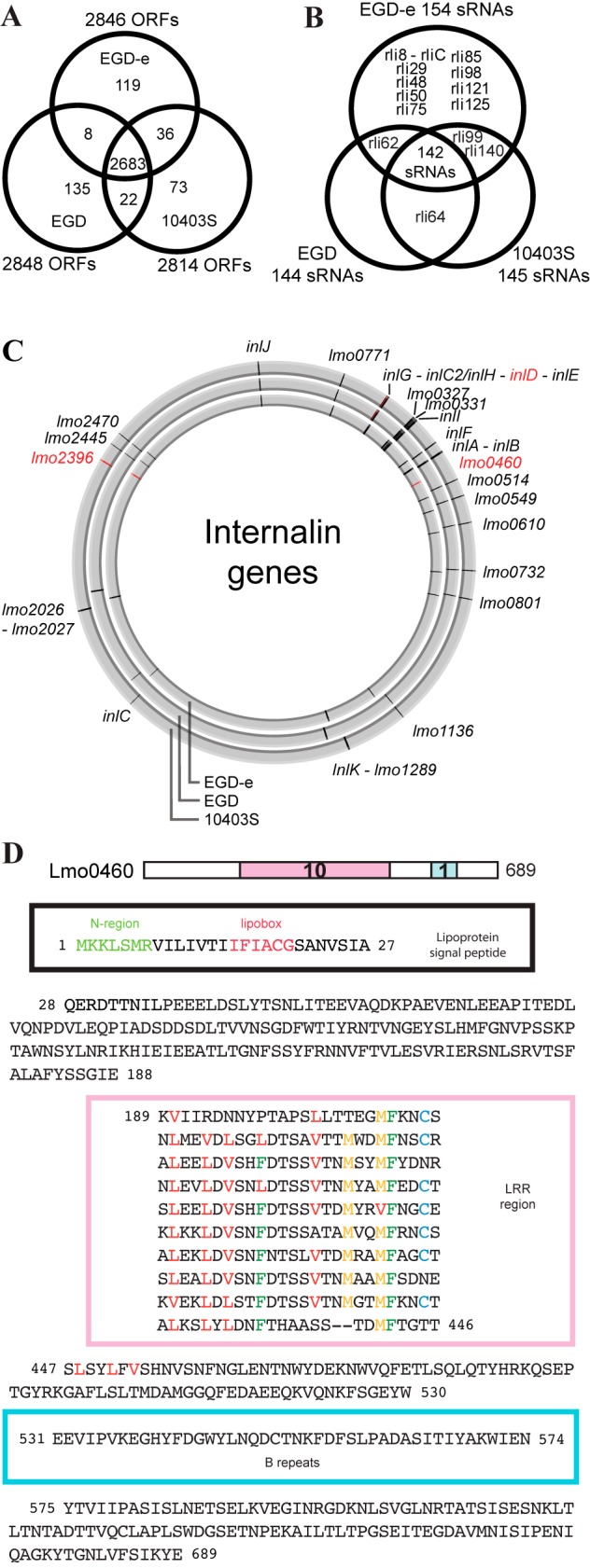
Conservation of ORFs, small RNAs, and internalins in EGD, EGD-e, and 10403S. (A) Venn diagram showing the numbers of ORFs common to the different strains. A bidirectional best-hit search with an E value score lower than 1e−4 was used to determine homologies. (B) Venn diagram of the small RNAs found in the three strains. The percentage of similarity was calculated from BLASTN results. Small RNAs with a percentage lower than 10% were not considered conserved. (C) Genomic locations of 27 internalins in the EGD-e, EGD, and 10403S genomes (using CGView [81]). In red are indicated internalins present only in one or two strains. (D) Lmo0460 amino acid sequence. Lmo0460 is a predicted lipoprotein present only in EGD-e which contains an atypical leucine-rich repeat (LRR) domain.
Of the 393 ORFs not shared by the three strains, only 8 are common to EGD and EGD-e and not present in 10403S (Data Set S2). EGD and 10403S share 22 ORFs that are absent in EGD-e (Data Set S1), and EGD-e and 10403S share 36 ORFs (Fig. 2A) that are not found in EGD. Of these, 30 come from the A118 prophage, which is integrated into both EGD-e and 10403S, as previously described for EGD-e (7, 30). EGD has 135 ORFs which are not found in the two other strains (Fig. 2A and Data Set S2); 52 are phage proteins, and 50 are hypothetical proteins for which RAST automatic annotation software has found no homolog.
EGD has a prophage different from that of EGD-e and 10403S.
Since EGD has 52 specific genes encoding putative phage proteins, we examined whether EGD had an integrated phage. A BLASTN search of each phage gene of the three strains (Fig. S1A) using 8 sequenced Listeria species phage genomes (31) indicated the presence of A118 phage genes in EGD-e and 10403S, integrated into the competence gene comK (32), and the presence of B025 phage genes in EGD (Fig. 1C), integrated into the tRNAArg gene. B025 is found in the first half of the EGD genome (between LMON_1236 and LMON_1299) (Fig. S1B). A118 is integrated in the second half of the EGD-e genome (between lmo2271 and lmo2332) and the 10403S genome (between LMRG_01560 and LMRG_01510).
Conservation of sRNAs.
In the past decade, noncoding RNAs in Listeria have been studied in detail (33–42). One study concerns strain 10403S (36), and all the other studies concern the EGD-e strain. We compiled a list of small noncoding RNAs (sRNAs) from these various publications. Altogether, 305 noncoding RNA elements have now been reported in L. monocytogenes, with 155 sRNAs, 46 cis regulatory elements (cisRegs), and 104 antisense RNAs (asRNAs).
A comparison of EGD-e RNAs by a BLASTN search (−e 0.001 –W 4) in EGD and 10403S showed a very high conservation (Data Set S1) with regard to protein-coding genes. Regarding sRNAs, 142 out of the 155 (92%) are common to the three strains (Fig. 2B); 100% of cisRegs are conserved, as are 97% of the asRNAs. Only 9 sRNAs are found only in EGD-e (Fig. S2A and B). The particular case of Rli38 is interesting, as it seems that the whole region from lmo1097 (which encodes an integrase) up to the 5′ end of the Rli38 gene, has been integrated in EGD-e upstream from lmo1116 (Fig. S2C).
Conservation of internalin genes.
L. monocytogenes encodes a large family of proteins known as internalins, which possess a leucine-rich repeat (LRR)-containing domain. Twenty-five members of this family, including several virulence factors, that have been classified into three types (Fig. S3A) were described for strain EGD-e (43). InlA, the prototype internalin, and InlB promote L. monocytogenes internalization into mammalian cells and were initially identified in EGD (44–46). We found that InlA and InlB show, respectively, 8 and 6 nonsynonymous amino acid differences in EGD and 10403S compared to their counterparts in EGD-e. With a BLASTP search of the different internalins already described in EGD-e, we found 27 internalins present in the different genomes of strains EGD-e, EGD, and 10403S (Fig. 2C and Table 2).
TABLE 2 .
List of 27 internalins found in EGD-e, EGD, and 10403Sc
| Internalin (no. of SNPs) for: |
inl gene namea | ||
|---|---|---|---|
| EGD-e | EGD | 10403S | |
| lmo0171 | LMON_0168 (6) | LMRG_02416 (6) | |
| lmo0262 | LMON_0259 (0) | LMRG_02647 (0) | inlG |
| lmo0263 | LMON_0260 (7) | LMRG_02646 (3) | inlH, inlC2b |
| LMON_0261 | LMRG_02851 | inlD | |
| lmo0264 | LMON_0262 (6) | LMRG_02612 (6) | inlE |
| lmo0327 | LMON_0334 (77) | LMRG_00021 (172) | |
| lmo0331 | LMON_0336 (20) | LMRG_00023 (25) | |
| lmo0333 | LMON_0338 (14) | LMRG_00025 (14) | inlI |
| lmo0409 | LMON_0418 (6) | LMRG_00102 (6) | inlF |
| lmo0433 | LMON_0441 (24) | LMRG_00126 (24) | inlA |
| lmo0434 | LMON_0442 (13) | LMRG_00127 (15) | inlB |
| lmo0460 | |||
| lmo0514 | LMON_0514 (19) | LMRG_00195 (19) | |
| lmo0549 | LMON_0549 (17) | LMRG_00231 (27) | |
| lmo0610 | LMON_0611 (7) | LMRG_00293 (7) | |
| lmo0732 | LMON_0737 (6) | LMRG_00420 (5) | |
| lmo0801 | LMON_0805 (8) | LMRG_02867 (6) | |
| lmo1136 | LMON_1129 (12) | LMRG_00579 (9) | |
| lmo1289 | LMON_1350 (0) | LMRG_00739 (0) | |
| lmo1290 | LMON_1352 (13) | LMRG_00740 (13) | inlK |
| lmo1786 | LMON_1853 (2) | LMRG_02825 (2) | inlC |
| lmo2026 | LMON_2097 (16) | LMRG_01175 (16) | |
| lmo2027 | LMON_2098 (2) | LMRG_01176 (1) | |
| lmo2396 | LMRG_01852 (8) | ||
| lmo2445 | LMON_2456 (0) | LMRG_01803 (0) | |
| lmo2470 | LMON_2481 (5) | LMRG_01778 (0) | |
| lmo2821 | LMON_2840 (5) | LMRG_01877 (5) | inlJ |
Internalin genes extensively studied.
inlH is in EGD-e; inlC2 is in EGD and 10403S.
Boldface indicates internalin genes present in one or two strains.
Our new analysis of the EGD-e genome revealed the presence of a type IV internalin represented by Lmo0460, a predicted lipoprotein (47) containing an atypical LRR domain (Fig. 2D). Whether Lmo0460 is a bona fide lipoprotein remains to be confirmed. The lipobox of Lmo0460 is located at the expected distance from the N terminus and differs only slightly from the consensus lipobox, L − 3-S/A − 2-A/G − 1-C + 1. Lmo0460 displays a novel type of LRR domain, with 10 repeats diverging from the internalin-LRR prototype motif by a longer length (26 instead of 22 amino acids [43]) and the presence of an MFXXCX sequence at the end of most repeats (Fig. 2D). The unusual Lmo0460 LRR repeats with the M-F motif are found in various predicted surface proteins (often lipoproteins) from other species, such as Listeria innocua, Enterococcus faecalis, Lactobacillus plantarum, Mycoplasma mycoides, and Helicobacter hepaticus. The functions of these proteins are still unknown. A BLASTP search did not reveal any homolog of Lmo0460 in EGD and 10403S (Table 2). However, the gene is conserved in many L. monocytogenes strains of different serovars.
As previously reported, inlH from EGD-e comprises the 5′ end of inlC2 and the 3′ end of inlD, both found in EGD and 10403S, and likely results from a recombination event (Fig. S3B). InlH and InlC2 proteins are highly homologous; they have the same LRR domain and C-terminal regions that differ by only 13 amino acids (Fig. S3C).
Presence of a PrfA* mutation in EGD.
Expression of virulence genes at the right time and place during infection is critical for the outcome of the disease and is thus highly regulated. PrfA is a regulator of the major virulence genes (48–50). It belongs to the cyclic AMP (cAMP) receptor protein (Crp)/fumarate nitrate reductase regulator (Fnr) family of bacterial transcription factors. PrfA is itself regulated by an RNA thermosensor allowing PrfA-regulated genes to be expressed at 37°C (41), the temperature of infected hosts. PrfA is also regulated by nutrient availability via a short noncoding RNA generated by a riboswitch (40).
Among the SNPs detected between the different genomes, a remarkable one is present in the prfA gene of strain EGD. We found 2 amino acid changes in PrfA of strain EGD compared to PrfA in EGD-e and 10403S; one glycine is changed into a serine at position 145, and one cysteine is changed into a tyrosine at position 229 (Fig. 3A). While the impact of the latter change, located in the G α-helix, on PrfA function is not known, the former, located in the D α-helix, is well known. It is a PrfA* mutation (51). This Gly145Ser mutation is believed to induce a conformational change in the PrfA protein, leading to a constitutively active protein and overexpression of the virulence locus and of the whole PrfA regulon (52). Strikingly, PrfA is the only protein in the whole virulence locus with an amino acid sequence in EGD that is different from that of 10403S. All the other proteins are similar in the two strains but show some differences in strain EGD-e (Fig. 3B and Data Set S1). This result predicted that EGD might express the PrfA regulon in a way very different from that of EGD-e and 10403S (see below).
FIG 3 .

PrfA* mutation and the overexpression of the PrfA core regulon in EGD. (A) Protein sequence alignment of PrfA in 43 L. monocytogenes strains. The well-known PrfA* mutation G145S (51, 82) is highlighted in red and appears only in the EGD and M7 strains. All other amino acid changes found are drawn showing their positions in the different domains of PrfA (52). HTH, helix turn helix. (B) Schematic representation of the virulence locus synteny in EGD-e, EGD, and 10403S. Amino acid differences from EGD-e’s sequence are displayed. (C) Genome browser view showing tiling array whole-transcriptome coverage of the virulence locus and the inlA-inlB operon in EGD-e, EGD-e PrfA*, and EGD. Each tiled probe indicating expression from the two genomic strands (top for plus strand, bottom for minus strand) is represented as a black dot for EGD-e, an orange dot for EGD-e PrfA*, and a green dot for EGD. (D) Comparison of expression levels of InlA, InlB, and LLO in EGD-e, EGD-e PrfA*, EGD, and L. innocua Clip11262 (used as a nonpathogenic reference bacterium) in whole bacterial lysates or in the cell wall fraction (InlA). (E) Immunofluorescence of InlA and ActA in EGD and EGD-e PrfA* in BHI medium.
It is noteworthy that our analysis of NrdD, a class III anaerobic ribonucleotide reductase (RNR), in EGD showed that a KITPFE motif present in strain EGD (Fig. S1C), as well as in 10403S, is absent in EGD-e (53), revealing a higher capacity for the first two strains to live under anaerobic conditions, including the gastrointestinal tract.
The PrfA core regulon in EGD is overexpressed.
To assess the impact of the PrfA* mutation in EGD, we constructed a PrfA* mutant in EGD-e by generating a Gly145Ser mutation and compared the phenotypes of the two strains (EGD and EGD-e PrfA*) to the EGD-e strain in exponential phase, after growth in brain heart infusion (BHI) at 37°C. We first performed a whole-genome transcriptomic analysis of the resulting EGD-e PrfA* strain using our Affymetrix tiling array (35). As EGD-e and EGD share more than 95% of their ORFs and sRNAs, our tiling arrays could also be used for EGD transcriptomic analysis. We found that in both EGD and EGD-e PrfA*, the core PrfA regulon (54), which contains the whole virulence locus, the inlA-inlB operon, inlC (lmo1786), and hpt (lmo0838), is overexpressed compared to its expression in the reference strain EGD-e (Fig. 3C and Data Set S3), confirming the effect of the Gly145Ser PrfA* mutation on the core PrfA regulon. All these genes have a canonical PrfA box upstream from their start codon, which allows the direct binding of PrfA.
Notably, we found that only 15 genes in the EGD-e PrfA* strain (Fig. S4A) are expressed differently from those in EGD-e when bacteria are grown to an optical density at 600 nm (OD600) of 1.0 at 37°C. This list includes the 11 genes of the PrfA core regulon. Lmo2269 was found to be differently expressed in L. monocytogenes P14prfA* versus P14ΔprfA after growth in BHI (85). The three remaining genes, argC (lmo1591), argG (lmo2090), and lmo0640, have to our knowledge never been described in PrfA regulation studies. In EGD, the number of genes expressed differently (128 genes) compared to EGD-e under reference conditions is much larger (Fig. S4A) than for EGD-e PrfA*, but also includes the core PrfA regulon (54). Overexpression of inlA, inlB, inlC, hly, and lmo0042 (which is similar to the gene for the Escherichia coli DedA protein, an inner membrane protein) was confirmed by quantitative reverse transcription-PCR (qRT-PCR) in the three strains (Fig. S5). The overproduction of the InlA, LLO, and InlB proteins was also confirmed by Western blotting (Fig. 3D). Examination of InlA at the bacterial surface (Fig. 3E) by immunofluorescence assay (55) showed that InlA decorates the bacterial body and accumulates at poles in EGD-e PrfA* and EGD. In contrast, in EGD-e, InlA is detected at the surface as helical dots, in agreement with the results of our previous studies (56). ActA was more highly expressed at the bacterial surface in EGD-e PrfA* and EGD than in EGD-e. Of interest, exposure of ActA on the surface seems to be a bistable process, as only half of the cells express it (Fig. 3E).
We also looked at differently expressed RNAs. Our statistical analysis revealed, in total, 27 sRNAs that were expressed in EGD and EGD-e PrfA* differently from in EGD-e (Fig. S4B and S5 and Data Set S3).
Phenotypic effect of the PrfA* mutation.
The two PrfA* strains EGD-e PrfA* and EGD grow more slowly in broth than strains EGD-e and 10403S. This was confirmed by a colony size analysis showing larger colonies for EGD-e than for EGD-e PrfA* and for 10403S than for EGD on BHI agar plates after 24 h of growth (Fig. 4A). This is in accordance with the defect already observed in 10403S PrfA* strains (57).
FIG 4 .

Differential bacterial invasion phenotypes in vitro. (A) Colony sizes of the three strains after 24 h of growth on solid BHI agar plates reveal that EGD-e PrfA* has smaller colonies than EGD-e and that EGD has smaller colonies than 10403S. (B) Gentamicin assays at 2 h postinfection of HeLa, JEG3, and Raw264 cells by the four different strains, EGD, EGD-e PrfA*, 10403S, and EGD. ns, not significantly different; *, P value of <0.05; **, P value of <0.005; ***, P value of <0.0005.
We performed classical gentamicin invasion assays (58) using strains EGD, 10403S, EGD-e PrfA*, and EGD-e in three different cell lines: HeLa (in which entry is InlB dependent), JEG3 (in which entry is InlA and InlB dependent), and Raw264 (macrophages). In HeLa and JEG3 cells, strains EGD and EGD-e PrfA* were more invasive than EGD-e and 10403S (Fig. 4B). There were also higher bacterial counts in mouse Raw264 macrophages for EGD-e PrfA* than for EGD-e and for EGD than for 10403S.
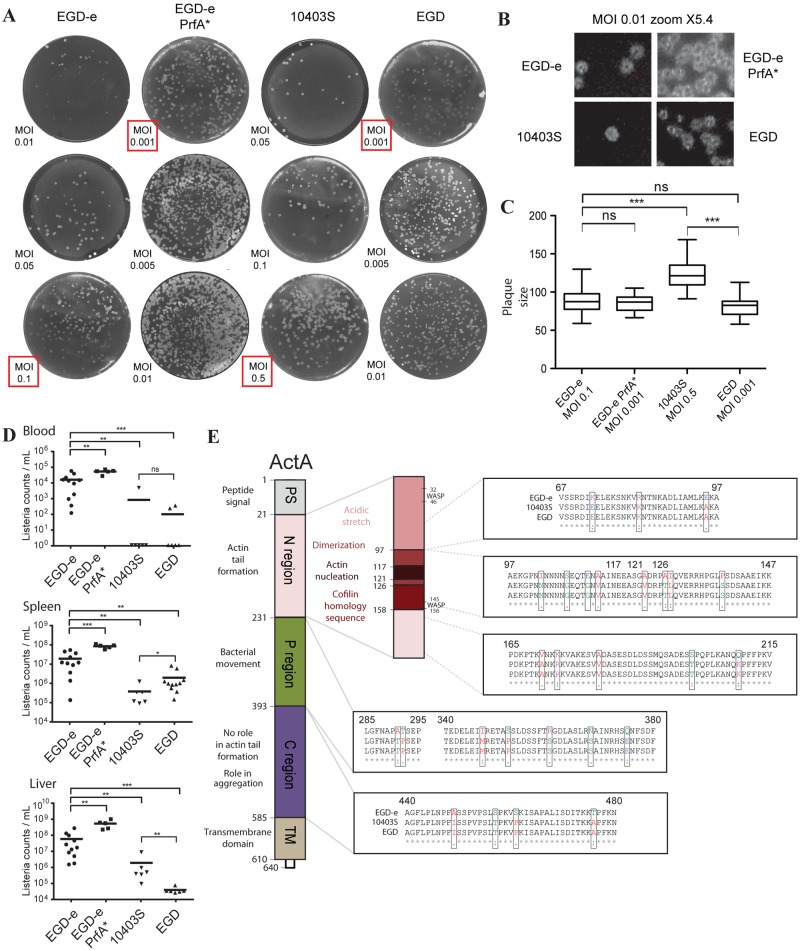
We performed plaque assays in L2 fibroblast cells. We observed a larger number of plaques for each PrfA* strain (Fig. 5A and B). Strikingly, plaque size was larger for the 10403S strain than for the EGD-e, EGD-e PrfA*, and EGD strains (Fig. 5C).
FIG 5 .
Plaque assays, virulence in mice, and ActA amino acid changes. (A) Plaque assays of EGD-e, EGD-e PrfA*, 10403S, and EGD at different MOIs show different sizes depending on the strain. Highlighted in red are the MOIs used for plaque size measurement. (B) Magnifications (×5.4) of the plaques for EGD-e, EGD-e PrfA*, 10403S, and EGD at an MOI of 0.01. (C) Measurement of plaque size (in square pixels) using Icy image analysis software (80). Different MOIs were used for each strain in order to have the same number of plaques in each well. Differences between strains were assessed by unpaired t test. Plaques from 10403S are bigger than the ones from EGD-e and the two PrfA* strains (EGD-e PrfA*, EGD). (D) CFU counts measured 72 h after intravenous infection with EGD-e, EGD-e PrfA*, 10403S, and EGD. Each dot represents the value for one mouse, and asterisks indicate Mann-Whitney statistical test results; results are from two independent experiments. (E) Motifs of the ActA protein (adapted from reference 61) and the different amino acid changes between EGD-e and EGD are shown. ActA has the same amino acid sequence in EGD and 10403S. The two WASP-like sequences of ActA present no differences between EGD-e, EGD, and 10403S.
We then evaluated the virulence of the different strains in mice. Clearly, strains EGD and 10403S are less virulent than EGD-e, as revealed by lower bacterial counts in blood, liver, and spleen (Fig. 5D), showing that many factors control virulence. In addition, the number of EGD-e PrfA* bacteria was higher than the number of wild-type EGD-e bacteria in blood, liver, and spleen, in agreement with the observed overexpression of the PrfA regulon (Fig. 5D). An increased virulence was also observed for 10403S PrfA* (57). Strikingly, despite a clear phenotypic difference in tissue culture cells, bacterial counting in mice spleen and liver 72 h after intravenous infection did not demonstrate clear differences between EGD and 10403S strains.
DISCUSSION
Here we report the genome sequence of L. monocytogenes strain EGD and compare it to the genomes of strains EGD-e and 10403S, the two other Listeria strains widely used by immunologists and listeriologists. Despite the fact that more than 95% of the ORFs are conserved in EGD, 10403S, and EGD-e, we found many critical differences between these strains. Altogether, our study revealed that the EGD strain is closer to 10403S than to EGD-e and that EGD-e is quite different from EGD. We detected a PrfA* mutation in EGD.
We confirmed the effect of the PrfA* mutation on the invasion of cells both by invasion assays and by plaque assays, but we did not observe an increased virulence of EGD in mice. Finally, the plaque size comparison revealed no difference between the EGD-e, EGD-e PrfA*, and EGD strains. However, we detected larger plaques for 10403S. These many discrepancies, which cannot be explained only by the overexpression of PrfA-regulated virulence factors, need more investigation. A first element to decipher is the complete role of ActA in these phenotypes. ActA is known to trigger intra- and intercellular movements (59) and to mediate escape from autophagy (60), and it is also implicated in interbacterial adhesion during intestinal colonization (61). Here were found 27 amino acid changes between EGD-e ActA and the ActA proteins of strains EGD and 10403S (Fig. 5C); it is the most variable protein within the whole virulence locus (Fig. 3B). The highest proportion of amino acid variation is found in the actin nucleation motif of ActA (Fig. 5E). A thorough comparative analysis of the actin tail lengths and intracellular speeds of the different strains may provide insight into the implication of these ActA amino acid changes for plaque size differences.
Altogether, our analysis indicates that PrfA* mutation does not confer an advantage during the whole process of Listeria infection of cells. We performed a comparison of all PrfA protein sequences in 39 published L. monocytogenes genomes. The PrfA* mutation appears only in the EGD and M7 strains (Fig. 3A). (M7 is a nonpathogenic serovar 4a strain isolated from cow’s milk [62].) We conclude that the PrfA* mutation does not provide an advantage. It would otherwise have been found in many more strains. In the specific case of the EGD strain, the PrfA* mutation might have been acquired through years of passage in mice, followed by plating on blood agar.
To understand whether the differences between EGD and EGD-e come from a mislabeling or an accumulation of mutations during evolution, we searched for the phylogenetic strains closest to EGD and EGD-e. According to the NCBI genome database, the strain phylogenetically closest to EGD is strain SLCC5850. It is a serotype 4b strain isolated in 1983 from a man with meningitis, according to the Seeliger collection database (63). Its main feature is the loss of important motifs in its PrfA protein (Fig. 3A). The loss of these motifs was also found in the nonhemolytic SLCC53 strain when PrfA was sequenced in 1991 (49). SLCC53 is the type strain of Listeria (64) and thus originates from the rabbit strain isolated by E. G. D. Murray in 1924 (see reference 13). The strains phylogenetically closest to EGD-e are SLCC2372, a serotype 1/2c strain isolated from human in 1935, and SLCC2479, a serotype 3c strain which is of unknown origin, isolated in 1966 (12). By comparing the close phylogenetic neighbors of each strain, it seems more likely that EGD is closer to the original type strain and that EGD-e has been mislabeled and exchanged for another strain. However, a complete answer cannot be given until a phylogenomics analysis of all Listeria strains available is performed. It would be the only solution to characterize the relationship between strains. However, we face almost a century of Listeria strain isolation and cultures, and it clearly seems impossible to decipher completely the many events which might have occurred to create what seems to be a mislabeling of strains.
Since the pioneer work of Mackaness, L. monocytogenes has been used and is still widely used as a tool to study the induction of a T-cell response as well as to analyze the response to infection in knockout mice (65–68). In these studies, infections are performed with a variety of L. monocytogenes strains, including the three strains EGD, EGD-e, and 10403S. However, strain-specific differences are not taken into account except when using mutants, such as the nonhemolytic mutant or the ActA mutant strains. We consider that many factors in addition to LLO and ActA can affect survival in the host. It is thus of the utmost importance in any report to precisely indicate which strain has been used. It is to be noticed that Listeria has recently been engineered as a promising live-vaccine strain against viral infection and cancer (69–74). In most cases, strain 10403S is the original strain used. Given the results reported here, it will be important to use the same original strain in future constructions and vaccine trials. In conclusion, our results highlight strain-specific genomic differences with important consequences for the interpretation of results in both infection biology and immunology. We hope the genomic comparisons that we provided here will help listeriologists to go further in their investigations and strongly recommend that authors always indicate the names and origins of the Listeria strains used in their studies.
MATERIALS AND METHODS
For more information on materials and methods, see Text S4 in the supplemental material.
Listeria monocytogenes EGD and EGD-e sequencing and annotation.
Briefly, genomic DNA was prepared as described in reference 75. Library preparation was achieved using NEBNext DNA sample prep master mix set 1 with the multiplexing sample preparation oligonucleotide according to the manufacturer’s recommendations. Libraries were then sequenced on a HiSeq 2000 sequencer in 100-base single-end reads. Sequence files were generated using Illumina Analysis Pipeline version 1.7 (CASAVA). After quality filtering, 25,827,948 reads were aligned with the Listeria monocytogenes 10403S genome sequence (GenBank accession number CP002002) using CLC Genomics Workbench (version 3.20), and more than 98.4% of reads mapped successfully. The remaining 407,195 reads were then used to sequentially fill gaps in the final sequence. The overall final coverage was 875×, with only 47,125 unmapped reads. EGD-e was sequenced using the same protocol, with a total of 13,414,584 reads.
The consensus sequence of EGD has been exported and annotated using RAST annotation software (76). Automatic annotation provided by RAST was curated using homology to proteins in Listeria monocytogenes EGD-e (7) and Listeria monocytogenes 10403S, and the sequence was submitted to the ENA database (accession number HG421741). Interactive visualization of the syntenic organization of Listeria genomes is available with the Flash-based SynTView (77) software available at http://genopole.pasteur.fr/SynTView/flash/Listeria_monocytogenes/SynWebEGD_final.html.
Transcriptomic analysis.
Bacterial overnight cultures were diluted in BHI, and bacteria were grown to an OD of 1. RNA was extracted and samples for each chip were prepared as previously described (35). The tiling chip works with two types of arrays: the gene expression array (link E-MTAB-1676; https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-1676/) and tiling array (link E-MTAB-1677; https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-1677/). Genes having an average false-discovery rate (Benjamini and Yekutieli method [84]) (FDRBY) under 0.05 and an absolute log fold change (|logFC|) value of >1.5 were selected as potential differentially expressed genes. For small RNAs, we applied the cutoff t test P value of <0.05 and an |logFC| value of >1.5; after manual curation, we obtained a potential list of differentially expressed sRNAs. Genes and sRNAs of interest were then studied using the real-time PCR system ABI PRISM 7900HT (Applied Biosystems), normalized to expression of the gyrase (lmo0007) gene, and values were compared by an unpaired t test.
Listeria strains used.
For every experiment in this paper, we used the following strains: EGD (BUG600), 10403S (BUG1361), EGD-e (BUG1600), EGD-e PrfA* (BUG3057), and L. innocua Clip11262 (BUG499). BUG numbers are identification numbers of the Unité des Interactions Bactéries Cellules laboratory's Listeria strain collection.
Bacterial lysis, cell wall extraction, and protein detection.
For preparation of whole bacterial lysates, 1 × 109 bacteria of overnight cultures were washed 3 times in phosphate-buffered saline (PBS), lysed in 200 µl Laemmli buffer containing 10% dithiothreitol (DTT), boiled for 10 min, and sonicated. Cell wall extraction and Western blotting of InlA, InlB, LLO, and EF-Tu were performed as previously described (78).
Gentamicin invasion assay and in vivo studies.
We performed classical gentamicin invasion assays as described in reference 58. Cells were plated in 24-well plates the day before infection with the indicated strains at a multiplicity of infection (MOI) between 1 and 25 depending on the host cell type. Bacteria on BHI agar plates for the inoculum and output after gentamicin treatment were counted. Invasion was quantified as a percentage of the inoculum.
All experiments involving mice were handled in accordance with the Pasteur Institute’s guidelines for animal welfare. Eight-week-old BALB/c mice (Charles River) were injected intravenously with 104 CFU of Listeria monocytogenes per mouse. Liver, spleen, and blood samples were recovered 72 h after infection. The organs were disrupted in 2 ml of PBS. Serial dilutions of organ homogenates and of the mouse blood were plated on BHI agar plates and numbers of CFU determined.
Plaque assay.
The plaque assay procedure was adapted from the work of Kuhn et al. (79). L2 from cells were grown in Ham’s F-12K medium (GIBCO, Life Technologies). Before the infection, monolayers were infected at different MOIs. Infected cells were subsequently incubated at 37°C for 1 h and then washed several times with medium. Following a 48-h incubation at 37°C, cells were fixed with paraformaldehyde (4% in PBS for 20 min) and stained with crystal violet.
In order to measure the plaque size, we needed to have the same number of plaques on each well for each bacterium. We thus selected the following MOIs: 0.1 for EGD-e, 0.001 for EGD-e PrfA*, 0.5 for 10403S, and 0.001 for EGD. Directly on a picture of the plaques, we measured plaque size (in square pixels) using the interior value of the region of interest (ROI) manually defined by Icy software (80). Almost 30 plaque sizes were measured for each bacterium. An unpaired t test was used to assess plaque size differences between strains.
Nucleotide sequence accession number.
The sequence of EGD was submitted to the ENA database under accession number HG421741.
SUPPLEMENTAL MATERIAL
Extended materials and methods. We describe more precisely the different methods used, such as the EGD-e PrfA* strain construction. Download
List of SNPs among the three strains. (First spreadsheet) Numbers of SNPs for each EGD-e gene in a comparison of the EGD and 10403S genome sequences. The annotation information for each EGD-e gene is indicated. CLC Genomic Workbench software was used to determine the number of SNPs. (Second spreadsheet) 10403S versus EGD. Number of SNPs for each 10403S gene compared with the EGD genome sequence. Annotation information for each 10403S gene is indicated. CLC Genomic Workbench software was used to determine the number of SNPs. (Third spreadsheet) EGD-e resequencing. Position of each SNP when the EGD-e genome sequence from 2001 (7) is compared to the new sequence obtained in our analysis. (Fourth spreadsheet) Virulence locus amino acid (aa) changes. Numbers of amino acid changes found for each gene of the virulence locus (lmo0200 to lmo02007) and the InlA-InlB locus (lmo0433 and lmo0434). The amino acid changes between EGD-e and EGD and between EGD-e and 10403S are indicated. Download
Conservation of genes and sRNAs among the EGD-e, EGD, and 10403S strains. Several tables showing genes conserved in the three strains, genes conserved in only two of the strains, and genes found in only one of the strains are presented. The results of the BLASTP search performed between strain proteins are shown. Finally, DNA similarity indexes of all three types of noncoding RNAs (sRNAs, cis-regulatory RNAs, asRNAs) are shown, showing the conservation of these noncoding RNAs in the EGD-e, EGD, and 10403S strains. Download
Differently expressed genes and sRNAs in a comparison of the EGD and EGD-e PrfA* strains grown in BHI under the reference conditions (EGD-e grown in BHI). (First spreadsheet) Genes. Only genes from the EGD-e strain found differently expressed in a comparison of EGD with EGD-e or EGD-e PrfA* versus EGD-e are shown. For gene expression and tiling arrays, the log of the fold change (logFC) and statistical values (Local-pooled-error test [LPE], t test, FDRBY) are displayed in different columns, along with the conservation in the EGD strain and annotation information. (Second spreadsheet) sRNAs. Only sRNAs from the EGD-e strain found differently expressed in a comparison of EGD with EGD-e or EGD-e PrfA* versus EGD-e are shown. Logs of the fold change (logFC) and t test values are displayed, along with the DNA identity values in the EGD strain, PCR validation information, and annotation data. One asterisk in the PCR validation column means validation of one of the differential expressions predicted by tiling chip assays; two asterisks indicate PCR validation of the two differential expressions predicted by tiling chip assays. Download
Phages and NrdD in EGD, EGD-e, and 10403S. (A) Genes of 8 Listeria phages, B054 (NC_009813), B025 (NC_009812), PSA (NC_003291), A500 (NC_009810), A118 (NC_003216), A006 (NC_009815), P35 (NC_009814), and P40 (NC_011308), were identified with BLASTN (default parameters) in EGD-e, EGD, and 10403S. The results show that B025 is an EGD prophage but that A118 is an EGD-e and 10403S prophage. (B) BLASTP results of B025 and A118 phage proteins against the EGD-e, EGD, and 10403S genomes are displayed in a circular view. Listeria proteins are color coded by amino acid similarity to phage proteins. (Inset) comK locus in EGD-e and 10403S, in which comK is split into two parts, with the A118 prophage integrated in between. The B025 attB integration site in EGD is a tRNAArg. (C) Multisequence alignment of NrdD (Lmo0279) an anaerobic ribonucleoside triphosphate reductase in the three strains shows (in red) a deletion of the KITPFE motif in EGD-e (adapted from reference 53), indicating a higher capacity for EGD and 10403S to live in a low-oxygen environment. Download
Small RNA conservation in EGD, EGD-e, and 10403S. (A) One hundred fifty-four known small RNAs (sRNAs) previously described in the EGD-e genome (34–38) are displayed using CGView (81) according to their position in the genome. In red are the sRNAs whose DNA sequences are conserved at <10% in the EGD and 10403S strains. (B) DNA similarity heat map showing sRNAs with a similarity lower than 95% in one of the strains. (C) The Rli38 region in the three strains shows that two loci, one from lmo1097 (an integrase) to lmo1115 and the other from lmo1118 to lmo1119, including the genes for Rli8/RliC, Rli85, and Rli125, are present only in EGD-e. In EGD (and 10403S), only the 3′ end of the small RNA is present, with only 3 nucleotide changes, located between a GMP synthase (LMON_1106) and an AraC family transcriptional regulator (LMON_1107). Rli38 is an interesting case, as it is overexpressed in blood under low oxygen, and an Rli38-deleted EGD-e mutant is attenuated in a murine model of infection (35). The whole region from lmo1097 (which encodes an integrase) up to the 5′ end of the Rli38 gene has been integrated into EGD-e upstream from lmo1116. In addition, Rli8/RliC, Rli85, Rli125, Lmo1118, and Lmo1119, which are absent in EGD and 10403S, might have been integrated together between Lmo1117 and Lmo1120. Download
Internalins in EGD-e, EGD, and 10403S. (A) Schematic domain organization of the four families of EGD-e internalins (adapted from reference 43). (B) inlH represents a fusion of the 5′ end of inlC2, encoding the internalin domain, with the 3′ end of inlD, encoding the B repeats and the sorting signal (adapted from reference 43). (C) Amino acid differences between InlH and InlC2. Download
Transcriptomic analysis of EGD-e, EGD-e prfA*, and EGD. (A) Fold changes of EGD-e, EGD-e PrfA*, and EGD from the reference condition (EGD-e). Indicated with an asterisk are EGD-e genes showing no homolog (BDBH value, lower than 10e−4) in EGD. (B) Small RNAs differentially expressed in EGD-e prfA* and EGD are shown in a log fold change table. One asterisk indicates sRNAs validated using qRT-PCR, and two asterisks indicate sRNAs for which qRT-PCR was not conclusive. The fourth column of the table indicates the DNA similarity of each sRNA in EGD. Download
Validation of differentially expressed genes and sRNAs by qRT-PCR. (A) Four genes, inlA, inlB, inlC, and hly, are shown to be overexpressed in EGD and EGD-e PrfA*. One gene, lmo0048, is down-regulated in EGD and shows no differential expression in EGD-e PrfA*. (B) Fourteen sRNAs are shown to be differently expressed in EGD. Three sRNAs were not validated in EGD. Only rli51, which is part of the virulence locus, was found overexpressed in EGD-e PrfA*. Expression has been normalized to expression of the gyrase (lmo0007) gene. A two-tailed unpaired t test was used in three independent experiments. Download
ACKNOWLEDGMENTS
This work received financial support from the European Research Council (advanced grant 233348), the French Agence Nationale de la Recherche (grants BACNET 10-BINF-02-01, IBEID ANR-10-LABX-62-01, and ERA-NET ANR-2010-PATH), the Institut Pasteur, the Institut National de la Santé et de la Recherche Médicale, and the Institut National de la Recherche Agronomique. A.K. is a recipient of a scholarship from the Pasteur-Paris University International Doctoral Program/Institut Carnot Maladies Infectieuses.
We thank Rich Calendar and Patrick Berche for important discussions, Nina Sesto for the help in the construction of the PrfA* mutant, Marie-Anne Nahori for the help in performing in vivo studies, and Guillaume Soubigou for the help in performing the tiling array experiments.
P.C. conceived and designed the experiments. C. Bécavin, C. Bouchier, C.A., Z.W., A.K., M.G.P., J.P.-C., and H.B. performed the experiments. C. Bécavin, C. Bouchier, P.L., C.A., S.C., F.G.-D.P., I.M., and H.B. analyzed the data. S.B. and I.M. provided analysis tools. C. Bécavin, T.H., D.A.P., T.C., M.L., H.B., and P.C. wrote the manuscript.
Footnotes
Citation Bécavin C, Bouchier C, Lechat P, Archambaud C, Creno S, Gouin E, Wu Z, Kühbacher A, Brisse S, Pucciarelli MG, García-del Portillo F, Hain T, Portnoy DA, Chakraborty T, Lecuit M, Pizarro-Cerdá J, Moszer I, Bierne H, Cossart P. 2014. Comparison of widely used Listeria monocytogenes strains EGD, 10403S, and EGD-e highlights genomic differences underlying variations in pathogenicity. mBio 5(2):e00969-14. doi:10.1128/mBio.00969-14.
REFERENCES
- 1. Camejo A, Carvalho F, Reis O, Leitão E, Sousa S, Cabanes D. 2011. The arsenal of virulence factors deployed by Listeria monocytogenes to promote its cell infection cycle. Virulence 2:379–394. 10.4161/viru.2.5.17703 [DOI] [PubMed] [Google Scholar]
- 2. Hamon M, Bierne H, Cossart P. 2006. Listeria monocytogenes: a multifaceted model. Nat. Rev. Microbiol. 4:423–434. 10.1038/nrmicro1413 [DOI] [PubMed] [Google Scholar]
- 3. Lecuit M. 2007. Human listeriosis and animal models. Microbes Infect. 9:1216–1225. 10.1016/j.micinf.2007.05.009 [DOI] [PubMed] [Google Scholar]
- 4. Cossart P, Toledo-Arana A. 2008. Listeria monocytogenes, a unique model in infection biology: an overview. Microbes Infect. 10:1041–1050. 10.1016/j.micinf.2008.07.043 [DOI] [PubMed] [Google Scholar]
- 5. Cossart P. 2011. Illuminating the landscape of host-pathogen interactions with the bacterium Listeria monocytogenes. Proc. Natl. Acad. Sci. U. S. A. 108:1984–1991. 10.1073/pnas.1112371108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mackaness GB. 1964. The immunological basis of acquired cellular resistance. J. Exp. Med. 120:105–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Glaser P, Frangeul L, Buchrieser C, Rusniok C, Amend A, Baquero F, Berche P, Bloecker H, Brandt P, Chakraborty T, Charbit A, Chetouani F, Couvé E, de Daruvar A, Dehoux P, Domann E, Domínguez-Bernal G, Duchaud E, Durant L, Dussurget O, Entian KD, Fsihi H, García-del Portillo F, Garrido P, Gautier L, Goebel W, Gómez-López N, Hain T, Hauf J, Jackson D, Jones LM, Kaerst U, Kreft J, Kuhn M, Kunst F, Kurapkat G, Madueno E, Maitournam A, Vicente JM, Ng E, Nedjari H, Nordsiek G, Novella S, de Pablos B, Pérez-Diaz JC, Purcell R, Remmel B, Rose M, Schlueter T, Simoes N, Tierrez A, Vázquez-Boland Ja, Voss H, Wehland J, Cossart P. 2001. Comparative genomics of Listeria species. Science 294:849–852. 10.1126/science.1063447 [DOI] [PubMed] [Google Scholar]
- 8. Nelson KE, Fouts DE, Mongodin EF, Ravel J, DeBoy RT, Kolonay JF, Rasko DA, Angiuoli SV, Gill SR, Paulsen IT, Peterson J, White O, Nelson WC, Nierman W, Beanan MJ, Brinkac LM, Daugherty SC, Dodson RJ, Durkin AS, Madupu R, Haft DH, Selengut J, Van Aken S, Khouri H, Fedorova N, Forberger H, Tran B, Kathariou S, Wonderling LD, Uhlich GA, Bayles DO, Luchansky JB, Fraser CM. 2004. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res. 32:2386–2395. 10.1093/nar/gkh562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Doumith M, Cazalet C, Simoes N, Frangeul L, Jacquet C, Kunst F, Martin P, Cossart P, Glaser P, Buchrieser C. 2004. New aspects regarding evolution and virulence of Listeria monocytogenes revealed by comparative genomics and DNA arrays. Infect. Immun. 72:1072–1083. 10.1128/IAI.72.2.1072-1083.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gilmour MW, Graham M, Van Domselaar G, Tyler S, Kent H, Trout-Yakel KM, Larios O, Allen V, Lee B, Nadon C. 2010. High-throughput genome sequencing of two Listeria monocytogenes clinical isolates during a large foodborne outbreak. BMC Genomics 11:120. 10.1186/1471-2164-11-120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Orsi RH, den Bakker HC, Wiedmann M. 2011. Listeria monocytogenes lineages: genomics, evolution, ecology, and phenotypic characteristics. Int. J. Med. Microbiol. 301:79–96. 10.1016/j.ijmm.2010.05.002 [DOI] [PubMed] [Google Scholar]
- 12. Kuenne C, Billion A, Mraheil MA, Strittmatter A, Daniel R, Goesmann A, Barbuddhe S, Hain T, Chakraborty T. 2013. Reassessment of the Listeria monocytogenes pan-genome reveals dynamic integration hotspots and mobile genetic elements as major components of the accessory genome. BMC Genomics 14:47. 10.1186/1471-2164-14-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Murray EGD, Webb RA, Swann MBR. 1926. A disease of rabbits characterised by a large mononuclear leucocytosis, caused by a hitherto undescribed bacillus Bacterium monocytogenes (n. sp.). J. Pathol. Bacteriol. 29:407–439. 10.1002/path.1700290409 [DOI] [Google Scholar]
- 14. Pirie JHH. 1940. Listeria: change of name for a genus bacteria. Nature 145:264. 10.1038/145264a0 [DOI] [Google Scholar]
- 15. Gaillard JL, Berche P, Sansonetti P. 1986. Transposon mutagenesis as a tool to study the role of hemolysin in the virulence of Listeria monocytogenes. Infect. Immun. 52:50–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Näher H, Sperling U, Hahn H. 1984. Developmental interrelationship of specific Lyt 123 and Lyt 1 cell sets in expression of antibacterial immunity to Listeria monocytogenes. Infect. Immun. 44:252–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Edman DC, Pollock MB, Hall ER. 1968. Listeria monocytogenes L forms. I. Induction maintenance, and biological characteristics. J. Bacteriol. 96:352–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Seeliger H, Hohne K. 1979. Serotyping of Listeria monocytogenes and related species. Methods Microbiol. 13:31–49 [Google Scholar]
- 19. Den Bakker HC, Bowen BM, Rodriguez-Rivera LD, Wiedmann M. 2012. FSL J1-208, a virulent uncommon phylogenetic lineage IV Listeria monocytogenes strain with a small chromosome size and a putative virulence plasmid carrying internalin-like genes. Appl. Environ. Microbiol. 78:1876–1889. 10.1128/AEM.06969-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Roberts A, Nightingale K, Jeffers G, Fortes E, Kongo JM, Wiedmann M. 2006. Genetic and phenotypic characterization of Listeria monocytogenes lineage III. Microbiology 152:685–693. 10.1099/mic.0.28503-0 [DOI] [PubMed] [Google Scholar]
- 21. Ragon M, Wirth T, Hollandt F, Lavenir R, Lecuit M, Le Monnier A, Brisse S. 2008. A new perspective on Listeria monocytogenes evolution. PLoS Pathog. 4:e1000146. 10.1371/journal.ppat.1000146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pizarro-Cerdá J, Payrastre B, Wang YJ, Veiga E, Yin HL, Cossart P. 2007. Type II phosphatidylinositol 4-kinases promote Listeria monocytogenes entry into target cells. Cell. Microbiol. 9:2381–2390. 10.1111/j.1462-5822.2007.00967.x [DOI] [PubMed] [Google Scholar]
- 23. Bonazzi M, Kühbacher A, Toledo-Arana A, Mallet A, Vasudevan L, Pizarro-Cerdá J, Brodsky FM, Cossart P. 2012. A common clathrin-mediated machinery co-ordinates cell-cell adhesion and bacterial internalization. Traffic 13:1653–1666. 10.1111/tra.12009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Le Monnier A, Autret N, Join-Lambert OF, Jaubert F, Charbit A, Berche P, Kayal S. 2007. ActA is required for crossing of the fetoplacental barrier by Listeria monocytogenes. Infect. Immun. 75:950–957. 10.1128/IAI.01570-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lecuit M, Dramsi S, Gottardi C, Fedor-Chaiken M, Gumbiner B, Cossart P. 1999. A single amino acid in E-cadherin responsible for host specificity towards the human pathogen Listeria monocytogenes. EMBO J. 18:3956–3963. 10.1093/emboj/18.14.3956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lecuit M, Vandormael-Pournin S, Lefort J, Huerre M, Gounon P, Dupuy C, Babinet C, Cossart P. 2001. A transgenic model for listeriosis: role of internalin in crossing the intestinal barrier. Science 292:1722–1725. 10.1126/science.1059852 [DOI] [PubMed] [Google Scholar]
- 27. Nikitas G, Deschamps C, Disson O, Niault T, Cossart P, Lecuit M. 2011. Transcytosis of Listeria monocytogenes across the intestinal barrier upon specific targeting of goblet cell accessible E-cadherin. J. Exp. Med. 208:2263–2277. 10.1084/jem.20110560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Quereda JJ, Pucciarelli MG, Botello-Morte L, Calvo E, Carvalho F, Bouchier C, Vieira A, Mariscotti JF, Chakraborty T, Cossart P, Hain T, Cabanes D, García-Del Portillo F. 2013. Occurrence of mutations impairing sigma factor B (SigB) function upon inactivation of Listeria monocytogenes genes encoding surface proteins. Microbiology 159:1328–1339. 10.1099/mic.0.067744-0 [DOI] [PubMed] [Google Scholar]
- 29. Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5:R12. 10.1186/gb-2004-5-6-p12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Verghese B, Lok M, Wen J, Alessandria V, Chen Y, Kathariou S, Knabel S. 2011. comK prophage junction fragments as markers for Listeria monocytogenes genotypes unique to individual meat and poultry processing plants and a model for rapid niche-specific adaptation, biofilm formation, and persistence. Appl. Environ. Microbiol. 77:3279–3292. 10.1128/AEM.00546-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dorscht J, Klumpp J, Bielmann R, Born Y, Zimmer M, Loessner MJ, Schmelcher M, Calendar R. 2009. Comparative genome analysis of Listeria bacteriophages reveals extensive mosaicism, programmed translational frameshifting, and a novel prophage insertion site. J. Bacteriol. 191:7206–7215. 10.1128/JB.01041-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Loessner MJ, Inman RB, Lauer P, Calendar R. 2000. Complete nucleotide sequence, molecular analysis and genome structure of bacteriophage A118 of Listeria monocytogenes: implications for phage evolution. Mol. Microbiol. 35:324–340. 10.1046/j.1365-2958.2000.01720.x [DOI] [PubMed] [Google Scholar]
- 33. Christiansen JK, Nielsen JS, Ebersbach T, Valentin-hansen P, Søgaard-Andersen L, Kallipolitis BH. 2006. Identification of small Hfq-binding RNAs in Listeria monocytogenes. RNA 12:1383–1396. 10.1261/rna.49706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mandin P, Repoila F, Vergassola M, Geissmann T, Cossart P. 2007. Identification of new noncoding RNAs in Listeria monocytogenes and prediction of mRNA targets. Nucleic Acids Res. 35:962–974. 10.1093/nar/gkl1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, Loh E, Gripenland J, Tiensuu T, Vaitkevicius K, Barthelemy M, Vergassola M, Nahori MA, Soubigou G, Régnault B, Coppée JY, Lecuit M, Johansson J, Cossart P. 2009. The Listeria transcriptional landscape from saprophytism to virulence. Nature 459:950–956. 10.1038/nature08080 [DOI] [PubMed] [Google Scholar]
- 36. Oliver HF, Orsi RH, Ponnala L, Keich U, Wang W, Sun Q, Cartinhour SW, Filiatrault MJ, Wiedmann M, Boor KJ. 2009. Deep RNA sequencing of L. monocytogenes reveals overlapping and extensive stationary phase and sigma B-dependent transcriptomes, including multiple highly transcribed noncoding RNAs. BMC Genomics 10:641. 10.1186/1471-2164-10-641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mraheil MA, Billion A, Mohamed W, Mukherjee K, Kuenne C, Pischimarov J, Krawitz C, Retey J, Hartsch T, Chakraborty T, Hain T. 2011. The intracellular sRNA transcriptome of Listeria monocytogenes during growth in macrophages. Nucleic Acids Res. 39:1–14. 10.1093/nar/gkq742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wurtzel O, Sesto N, Mellin JR, Karunker I, Edelheit S, Bécavin C, Archambaud C, Cossart P, Sorek R. 2012. Comparative transcriptomics of pathogenic and non-pathogenic Listeria species. Mol. Syst. Biol. 8:1–14. 10.1038/msb.2012.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mellin JR, Cossart P. 2012. The non-coding RNA world of the bacterial pathogen Listeria monocytogenes. RNA Biol. 9:372–378. 10.4161/rna.19235 [DOI] [PubMed] [Google Scholar]
- 40. Loh E, Dussurget O, Gripenland J, Vaitkevicius K, Tiensuu T, Mandin P, Repoila F, Buchrieser C, Cossart P, Johansson J. 2009. A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell 139:770–779. 10.1016/j.cell.2009.08.046 [DOI] [PubMed] [Google Scholar]
- 41. Johansson J, Mandin P, Renzoni A, Chiaruttini C, Springer M, Cossart P. 2002. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell 110:551–561. 10.1016/S0092-8674(02)00905-4 [DOI] [PubMed] [Google Scholar]
- 42. Sesto N, Touchon M, Andrade JM, Kondo J, Rocha EPC, Arraiano CM, Archambaud C, Westhof E, Romby P, Cossart P. 2014. A PNPase dependent CRISPR system in Listeria. PLoS Genet. 10:e1004065. 10.1371/journal.pgen.1004065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bierne H, Sabet C, Personnic N, Cossart P. 2007. Internalins: a complex family of leucine-rich repeat-containing proteins in Listeria monocytogenes. Microbes Infect. 9:1156–1166. 10.1016/j.micinf.2007.05.003 [DOI] [PubMed] [Google Scholar]
- 44. Gaillard JL, Berche P, Frehel C, Gouin E, Cossart P. 1991. Entry of L. monocytogenes into cells is mediated by internalin, a repeat protein reminiscent of surface antigens from gram-positive cocci. Cell 65:1127–1141. 10.1016/0092-8674(91)90009-N [DOI] [PubMed] [Google Scholar]
- 45. Pizarro-Cerdá J, Kühbacher A, Cossart P. 2012. Entry of Listeria monocytogenes in mammalian epithelial cells: an updated view. Cold Spring Harb. Perspect. Med. 2(11):pii:a010009. 10.1101/cshperspect.a010009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dramsi S, Biswas I, Maguin E, Braun L, Mastroeni P, Cossart P. 1995. Entry of Listeria monocytogenes into hepatocytes requires expression of inlB, a surface protein of the internalin multigene family. Mol. Microbiol. 16:251–261. 10.1111/j.1365-2958.1995.tb02297.x [DOI] [PubMed] [Google Scholar]
- 47. Baumgärtner M, Kärst U, Gerstel B, Loessner M, Wehland J, Jänsch L. 2007. Inactivation of Lgt allows systematic characterization of lipoproteins from Listeria monocytogenes. J. Bacteriol. 189:313–324. 10.1128/JB.00976-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Leimeister-Wächter M, Haffner C, Domann E, Goebel W, Chakraborty T. 1990. Identification of a gene that positively regulates expression of listeriolysin, the major virulence factor of Listeria monocytogenes. Proc. Natl. Acad. Sci. U. S. A. 87:8336–8340. 10.1073/pnas.87.21.8336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mengaud J, Dramsi S, Gouin E, Vazquez-Boland JA, Milon G, Cossart P. 1991. Pleiotropic control of Listeria monocytogenes virulence factors by a gene that is autoregulated. Mol. Microbiol. 5:2273–2283. 10.1111/j.1365-2958.1991.tb02158.x [DOI] [PubMed] [Google Scholar]
- 50. De las Heras A, Cain RJ, Bielecka MK, Vázquez-Boland JA. 2011. Regulation of Listeria virulence: PrfA master and commander. Curr. Opin. Microbiol. 14:118–127. 10.1016/j.mib.2011.01.005 [DOI] [PubMed] [Google Scholar]
- 51. Ripio MT, Domínguez-Bernal G, Lara M, Suárez M, Vazquez-Boland JA. 1997. A Gly145Ser substitution in the transcriptional activator PrfA causes constitutive overexpression of virulence factors in Listeria monocytogenes. J. Bacteriol. 179:1533–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vega Y, Rauch M, Banfield MJ, Ermolaeva S, Scortti M, Goebel W, Vázquez-Boland JA. 2004. New Listeria monocytogenes prfA* mutants, transcriptional properties of PrfA* proteins and structure-function of the virulence regulator PrfA. Mol. Microbiol. 52:1553–1565. 10.1111/j.1365-2958.2004.04052.x [DOI] [PubMed] [Google Scholar]
- 53. Ofer A, Kreft J, Logan DT, Cohen G, Borovok I, Aharonowitz Y. 2011. Implications of the inability of Listeria monocytogenes EGD-e to grow anaerobically due to a deletion in the class III NrdD ribonucleotide reductase for its use as a model laboratory strain. J. Bacteriol. 193:2931–2940. 10.1128/JB.01405-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Scortti M, Monzó HJ, Lacharme-Lora L, Lewis DA, Vázquez-Boland JA. 2007. The PrfA virulence regulon. Microbes Infect. 9:1196–1207. 10.1016/j.micinf.2007.05.007 [DOI] [PubMed] [Google Scholar]
- 55. Bierne H, Mazmanian SK, Trost M, Pucciarelli MG, Liu G, Dehoux P, Genome L, Jänsch L, Portillo FG, Schneewind O, Cossart P, European, Listeria Genome Consortium 2002. Inactivation of the srtA gene in Listeria monocytogenes inhibits anchoring of surface proteins and affects virulence. Mol. Microbiol. 43:869–881. 10.1046/j.1365-2958.2002.02798.x [DOI] [PubMed] [Google Scholar]
- 56. Bruck S, Personnic N, Prevost MC, Cossart P, Bierne H. 2011. Regulated shift from helical to polar localization of Listeria monocytogenes cell wall-anchored proteins. J. Bacteriol. 193:4425–4437. 10.1128/JB.01154-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bruno JC, Freitag NE. 2010. Constitutive activation of PrfA tilts the balance of Listeria monocytogenes fitness towards life within the host versus environmental survival. PLoS One 5:e15138. 10.1371/journal.pone.0015138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pizarro-Cerdá J, Jonquières R, Gouin E, Vandekerckhove J, Garin J, Cossart P. 2002. Distinct protein patterns associated with Listeria monocytogenes InlA- or InlB-phagosomes. Cell. Microbiol. 4:101–115. 10.1046/j.1462-5822.2002.00169.x [DOI] [PubMed] [Google Scholar]
- 59. Kocks C, Gouin E, Tabouret M, Berche P, Ohayon H, Cossart P. 1992. L. monocytogenes-induced actin assembly requires the actA gene product, a surface protein. Cell 68:521–531. 10.1016/0092-8674(92)90188-I [DOI] [PubMed] [Google Scholar]
- 60. Yoshikawa Y, Ogawa M, Hain T, Yoshida M, Fukumatsu M, Kim M, Mimuro H, Nakagawa I, Yanagawa T, Ishii T, Kakizuka A, Sztul E, Chakraborty T, Sasakawa C. 2009. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nat. Cell Biol. 11:1233–1240. 10.1038/ncb1967 [DOI] [PubMed] [Google Scholar]
- 61. Travier L, Guadagnini S, Gouin E, Dufour A, Chenal-Francisque V, Cossart P, Olivo-Marin J-C, Ghigo J-M, Disson O, Lecuit M. 2013. ActA promotes Listeria monocytogenes aggregation, intestinal colonization and carriage. PLoS Pathog. 9:e1003131. 10.1371/journal.ppat.1003131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chen J, Xia Y, Cheng C, Fang C, Shan Y, Jin G, Fang W. 2011. Genome sequence of the nonpathogenic Listeria monocytogenes serovar 4a strain M7. J. Bacteriol. 193:5019–5020. 10.1128/JB.05501-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Haase JK, Murphy RA, Choudhury KR, Achtman M. 2011. Revival of Seeliger’s historical “Special Listeria Culture Collection.” Environ. Microbiol. 13:3163–3171. 10.1111/j.1462-2920.2011.02610.x [DOI] [PubMed] [Google Scholar]
- 64. Kathariou S, Pine L. 1991. The type strain(s) of Listeria monocytogenes: a source of continuing difficulties. Int. J. Syst. Bacteriol. 41:328–330. 10.1099/00207713-41-2-328 [DOI] [PubMed] [Google Scholar]
- 65. Kaufmann S, Weber L, Hahn H. 1975. Macrophage inhibiting activity in serum and central lymph of Listeria-immune mice. Eur. J. Immunol. 5:799–800. 10.1002/eji.1830051114 [DOI] [PubMed] [Google Scholar]
- 66. North RJ, Berche PA, Newborg MF. 1981. Immunologic consequences of antibiotic-induced abridgement of bacterial infection: effect on generation and loss of protective T cells and level of immunologic memory. J. Immunol. 127:342–346 [PubMed] [Google Scholar]
- 67. Kobayashi K, Inohara N, Hernandez LD, Galán JE, Núñez G, Janeway CA, Medzhitov R, Flavell RA. 2002. RICK/Rip2/CARDIAK mediates signalling for receptors of the innate and adaptive immune systems. Nature 416:194–199. 10.1038/416194a [DOI] [PubMed] [Google Scholar]
- 68. Reynders A, Yessaad N, Vu Manh TP, Dalod M, Fenis A, Aubry C, Nikitas G, Escalière B, Renauld JC, Dussurget O, Cossart P, Lecuit M, Vivier E, Tomasello E. 2011. Identity, regulation and in vivo function of gut NKp46+RORγt+ and NKp46+RORγt− lymphoid cells. EMBO J. 30:2934–2947. 10.1038/emboj.2011.201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Goossens PL, Milon G, Cossart P, Saron MF. 1995. Attenuated Listeria monocytogenes as a live vector for induction of CD8+ T cells in vivo: a study with the nucleoprotein of the lymphocytic choriomeningitis virus. Int. Immunol. 7:797–805. 10.1093/intimm/7.5.797 [DOI] [PubMed] [Google Scholar]
- 70. Shen H, Slifka MK, Matloubian M, Jensen ER, Ahmed R, Miller JF. 1995. Recombinant Listeria monocytogenes as a live vaccine vehicle for the induction of protective anti-viral cell-mediated immunity. Proc. Natl. Acad. Sci. U. S. A. 92:3987–3991. 10.1073/pnas.92.9.3987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Brockstedt DG, Giedlin MA, Leong ML, Bahjat KS, Gao Y, Luckett W, Liu W, Cook DN, Portnoy DA, Dubensky TW. 2004. Listeria-based cancer vaccines that segregate immunogenicity from toxicity. Proc. Natl. Acad. Sci. U. S. A. 101:13832–13837. 10.1073/pnas.0406035101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kim SH, Castro F, Paterson Y, Gravekamp C. 2009. High efficacy of a Listeria-based vaccine against metastatic breast cancer reveals a dual mode of action. Cancer Res. 69:5860–5866. 10.1158/0008-5472.CAN-08-4855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Guirnalda P, Wood L, Paterson Y. 2012. Listeria monocytogenes and its products as agents for cancer immunotherapy. Adv. Immunol. 113:81–118. 10.1016/B978-0-12-394590-7.00004-X [DOI] [PubMed] [Google Scholar]
- 74. Rothman J, Paterson Y. 2013. Live-attenuated Listeria-based immunotherapy. Expert Rev. Vaccines 12:493–504. 10.1586/erv.13.34 [DOI] [PubMed] [Google Scholar]
- 75. Wilson K. 2001. Preparation of genomic DNA from bacteria. Curr. Protoc. Mol. Biol. Chapter 2:Unit 2.4. 10.1002/0471142727.mb0204s56 [DOI] [PubMed] [Google Scholar]
- 76. Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Lechat P, Souche E, Moszer I. 2013. SynTView—an interactive multi-view genome browser for next-generation comparative microorganism genomics. BMC Bioinformatics 14:277. 10.1186/1471-2105-14-277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Archambaud C, Gouin E, Pizarro-Cerda J, Cossart P, Dussurget O. 2005. Translation elongation factor EF-Tu is a target for Stp, a serine-threonine phosphatase involved in virulence of Listeria monocytogenes. Mol. Microbiol. 56:383–396. 10.1111/j.1365-2958.2005.04551.x [DOI] [PubMed] [Google Scholar]
- 79. Kuhn M, Prevost M, Mounier J, Sansonetti PJ. 1990. A nonvirulent mutant of Listeria monocytogenes does not move intracellularly but still induces polymerization of actin. Infect. Immun. 58:3477–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. De Chaumont F, Dallongeville S, Chenouard N, Hervé N, Pop S, Provoost T, Meas-Yedid V, Pankajakshan P, Lecomte T, Le Montagner Y, Lagache T, Dufour A, Olivo-Marin JC. 2012. Icy: an open BioImage informatics platform for extended reproducible research. Nat. Methods 9:690–696. 10.1038/nmeth.2075 [DOI] [PubMed] [Google Scholar]
- 81. Stothard P, Wishart DS. 2005. Circular genome visualization and exploration using CGView. Bioinformatics 21:537–539. 10.1093/bioinformatics/bti054 [DOI] [PubMed] [Google Scholar]
- 82. Eiting M, Hagelüken G, Schubert WD, Heinz DW. 2005. The mutation G145S in PrfA, a key virulence regulator of Listeria monocytogenes, increases DNA-binding affinity by stabilizing the HTH motif. Mol. Microbiol. 56:433–446. 10.1111/j.1365-2958.2005.04561.x [DOI] [PubMed] [Google Scholar]
- 83. Bishop DK, Hinrichs DJJ. 1987. Adoptive transfer of immunity to Listeria monocytogenes. The influence of in vitro stimulation on lymphocyte subset requirements. J. Immunol. 139:2005–2009 [PubMed] [Google Scholar]
- 84. Reiner A, Yekutieli D, Benjamini Y. 2003. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 19(3):368–375 [DOI] [PubMed] [Google Scholar]
- 85. Milohanic E, Glaser P, Coppée JY, Frangeul L, Vega Y, Vázquez-Boland JA, Kunst F, Cossart P, Buchrieser C. 2003. Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol. Microbiol. 47(6):1613–1625 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Extended materials and methods. We describe more precisely the different methods used, such as the EGD-e PrfA* strain construction. Download
List of SNPs among the three strains. (First spreadsheet) Numbers of SNPs for each EGD-e gene in a comparison of the EGD and 10403S genome sequences. The annotation information for each EGD-e gene is indicated. CLC Genomic Workbench software was used to determine the number of SNPs. (Second spreadsheet) 10403S versus EGD. Number of SNPs for each 10403S gene compared with the EGD genome sequence. Annotation information for each 10403S gene is indicated. CLC Genomic Workbench software was used to determine the number of SNPs. (Third spreadsheet) EGD-e resequencing. Position of each SNP when the EGD-e genome sequence from 2001 (7) is compared to the new sequence obtained in our analysis. (Fourth spreadsheet) Virulence locus amino acid (aa) changes. Numbers of amino acid changes found for each gene of the virulence locus (lmo0200 to lmo02007) and the InlA-InlB locus (lmo0433 and lmo0434). The amino acid changes between EGD-e and EGD and between EGD-e and 10403S are indicated. Download
Conservation of genes and sRNAs among the EGD-e, EGD, and 10403S strains. Several tables showing genes conserved in the three strains, genes conserved in only two of the strains, and genes found in only one of the strains are presented. The results of the BLASTP search performed between strain proteins are shown. Finally, DNA similarity indexes of all three types of noncoding RNAs (sRNAs, cis-regulatory RNAs, asRNAs) are shown, showing the conservation of these noncoding RNAs in the EGD-e, EGD, and 10403S strains. Download
Differently expressed genes and sRNAs in a comparison of the EGD and EGD-e PrfA* strains grown in BHI under the reference conditions (EGD-e grown in BHI). (First spreadsheet) Genes. Only genes from the EGD-e strain found differently expressed in a comparison of EGD with EGD-e or EGD-e PrfA* versus EGD-e are shown. For gene expression and tiling arrays, the log of the fold change (logFC) and statistical values (Local-pooled-error test [LPE], t test, FDRBY) are displayed in different columns, along with the conservation in the EGD strain and annotation information. (Second spreadsheet) sRNAs. Only sRNAs from the EGD-e strain found differently expressed in a comparison of EGD with EGD-e or EGD-e PrfA* versus EGD-e are shown. Logs of the fold change (logFC) and t test values are displayed, along with the DNA identity values in the EGD strain, PCR validation information, and annotation data. One asterisk in the PCR validation column means validation of one of the differential expressions predicted by tiling chip assays; two asterisks indicate PCR validation of the two differential expressions predicted by tiling chip assays. Download
Phages and NrdD in EGD, EGD-e, and 10403S. (A) Genes of 8 Listeria phages, B054 (NC_009813), B025 (NC_009812), PSA (NC_003291), A500 (NC_009810), A118 (NC_003216), A006 (NC_009815), P35 (NC_009814), and P40 (NC_011308), were identified with BLASTN (default parameters) in EGD-e, EGD, and 10403S. The results show that B025 is an EGD prophage but that A118 is an EGD-e and 10403S prophage. (B) BLASTP results of B025 and A118 phage proteins against the EGD-e, EGD, and 10403S genomes are displayed in a circular view. Listeria proteins are color coded by amino acid similarity to phage proteins. (Inset) comK locus in EGD-e and 10403S, in which comK is split into two parts, with the A118 prophage integrated in between. The B025 attB integration site in EGD is a tRNAArg. (C) Multisequence alignment of NrdD (Lmo0279) an anaerobic ribonucleoside triphosphate reductase in the three strains shows (in red) a deletion of the KITPFE motif in EGD-e (adapted from reference 53), indicating a higher capacity for EGD and 10403S to live in a low-oxygen environment. Download
Small RNA conservation in EGD, EGD-e, and 10403S. (A) One hundred fifty-four known small RNAs (sRNAs) previously described in the EGD-e genome (34–38) are displayed using CGView (81) according to their position in the genome. In red are the sRNAs whose DNA sequences are conserved at <10% in the EGD and 10403S strains. (B) DNA similarity heat map showing sRNAs with a similarity lower than 95% in one of the strains. (C) The Rli38 region in the three strains shows that two loci, one from lmo1097 (an integrase) to lmo1115 and the other from lmo1118 to lmo1119, including the genes for Rli8/RliC, Rli85, and Rli125, are present only in EGD-e. In EGD (and 10403S), only the 3′ end of the small RNA is present, with only 3 nucleotide changes, located between a GMP synthase (LMON_1106) and an AraC family transcriptional regulator (LMON_1107). Rli38 is an interesting case, as it is overexpressed in blood under low oxygen, and an Rli38-deleted EGD-e mutant is attenuated in a murine model of infection (35). The whole region from lmo1097 (which encodes an integrase) up to the 5′ end of the Rli38 gene has been integrated into EGD-e upstream from lmo1116. In addition, Rli8/RliC, Rli85, Rli125, Lmo1118, and Lmo1119, which are absent in EGD and 10403S, might have been integrated together between Lmo1117 and Lmo1120. Download
Internalins in EGD-e, EGD, and 10403S. (A) Schematic domain organization of the four families of EGD-e internalins (adapted from reference 43). (B) inlH represents a fusion of the 5′ end of inlC2, encoding the internalin domain, with the 3′ end of inlD, encoding the B repeats and the sorting signal (adapted from reference 43). (C) Amino acid differences between InlH and InlC2. Download
Transcriptomic analysis of EGD-e, EGD-e prfA*, and EGD. (A) Fold changes of EGD-e, EGD-e PrfA*, and EGD from the reference condition (EGD-e). Indicated with an asterisk are EGD-e genes showing no homolog (BDBH value, lower than 10e−4) in EGD. (B) Small RNAs differentially expressed in EGD-e prfA* and EGD are shown in a log fold change table. One asterisk indicates sRNAs validated using qRT-PCR, and two asterisks indicate sRNAs for which qRT-PCR was not conclusive. The fourth column of the table indicates the DNA similarity of each sRNA in EGD. Download
Validation of differentially expressed genes and sRNAs by qRT-PCR. (A) Four genes, inlA, inlB, inlC, and hly, are shown to be overexpressed in EGD and EGD-e PrfA*. One gene, lmo0048, is down-regulated in EGD and shows no differential expression in EGD-e PrfA*. (B) Fourteen sRNAs are shown to be differently expressed in EGD. Three sRNAs were not validated in EGD. Only rli51, which is part of the virulence locus, was found overexpressed in EGD-e PrfA*. Expression has been normalized to expression of the gyrase (lmo0007) gene. A two-tailed unpaired t test was used in three independent experiments. Download