Abstract
Although digital detection of nucleic acids has been achieved by amplification of single templates in uniform microfluidic droplets and widely used for genetic analysis, droplet-based digital detection of proteins has rarely been reported, largely due to the lack of an efficient target amplification method for protein in droplets. Here, we report a key step towards digital detection of proteins using a highly parallel microfluidic droplet approach for single enzyme molecule detection in picoliter droplets via enzyme catalyzed signal amplification. An integrated microfluidic chip was designed for high throughput uniform droplet generation, monolayer droplet collection, incubation, detection, and release. Single β-galatosidase (β-Gal) molecules and the fluorogenic substrate fluorescein di-β-D-galactopyranoside were injected from two separated inlets to form uniform 20 μm droplets in fluorinated oil at a frequency of 6.6 kHz. About 200 000 droplets were captured as a monolayer in a capture well on-chip for subsequent imaging detection. A series of β-Gal solutions at different concentrations were analyzed at the single-molecule level. With no enzyme present, no droplets were found to fluoresce, while brightly fluorescent droplets were observed under single-enzyme molecule conditions. Droplet fluorescence intensity distribution analysis showed that the distribution of enzyme molecules under single-molecule conditions matched well with theoretical prediction, further proving the feasibility of detecting single enzyme molecules in emulsion droplets. Moreover, the population of fluorescent droplets increased as the β-Gal concentration increased. Based on a digital counting method, the measured concentrations of the enzyme were found to match well with input enzyme concentration, establishing the accuracy of the digital detection method for the quantification of β-Gal enzyme molecules. The capability of highly parallel detection of single enzyme molecules in uniform picoliter droplets paves the way to microdroplet based digital detection of proteins.
INTRODUCTION
Because of their small volume, low cost, and high throughput, microdroplets have attracted wide attention in recent years and have provided numerous opportunities in chemical and biological research.1, 2, 3, 4 By compartmentalizing reactions into discrete droplets, ultrahigh throughput reactions can be performed in parallel, allowing simultaneous observation of a large number of events at the single-molecule or single-cell level.5 Highly parallel detection of single molecules in uniform droplets facilitates digital detection, one of the most promising applications for microdroplets.6, 7, 8, 9 In digital detection, signal outputs are distinguished by “0,” or “1,” where “0” represents the absence of the molecule while “1” stands for the presence of the molecule in the droplet. Microfluidic droplet-based digital detection methods have been reported for DNA10, 11, 12 and RNA13, 14 detection, rare mutation analysis,15, 16, 17 and directed evolution.18
In contrast to nucleic acid analysis, droplet-based digital detection of proteins has rarely been reported, largely due to the lack of an efficient target amplification method for protein in droplets. As proteins can be sensitively detected by enzyme-linked immunosorbent assay (ELISA) using enzyme-based signal amplification, one key step toward single protein molecule detection in microfluidic droplets is the realization of single enzyme molecule detection in emulsion droplets. The first single enzyme molecule study in emulsion droplets was pioneered by Rotman et al., who compartmentalized single β-galactosidase molecules into droplets generated by a spray method to study the properties of a single enzyme in a droplet.19 Although they were able to determine the molecular weight and turnover number of β-Gal, the broad distribution of droplet diameters generated by the spray method requires tedious and time-consuming selection of appropriate droplets from a large number of individual droplets, wasting precious biochemical reagents. More importantly, the heterogeneous distribution of droplet sizes and the limited number of effective droplets created preclude accurate quantitation of enzyme molecules.
Over the years, great effort has been spent on creating uniform reactors, including fabricated microarrays20, 21, 22, 23, 24, 25 and microfluidic droplets,26, 27, 28 in a high throughput manner for single enzyme analysis. Compared with a microarray, microdroplets have the advantages of ultrahigh throughput, low cost, flexible manipulation, and low non-specific adsorption to channel/chamber wall by oil phase isolation, offering attractive alternative reactors for protein quantification. For example, very recently, Shim et al. fabricated a microfluidic device capable of generating and trapping 200 000 highly uniform femtoliter droplets for the measurement of chemical reactions of single-enzyme molecules and the application to a single-molecule-counting immunoassay.26 Takeuchi et al. developed a method that utilizes an immiscible liquid phase to instantly enclose thousands of uniform femtoliter droplets within microchambers built in microchannel walls to study enzymatic activities at single-molecule levels.27 In these two examples, droplets were stably trapped for imaging analysis, with the former relying on a complicated valve and post design and the latter using a non-scalable microchamber format. Arayanarakool et al. fabricated a nano- and micro-fluidic device capable of generating a large number of droplets with diameter around 3 μm for parallel single-enzyme kinetic analysis.28 However, no digital detection was achieved in this work. More importantly, in their system, the enzyme reaction time was reported to be more than 20 h, which precludes the use for practical digital enzyme detection.
Herein, we report the fabrication a simple microfluidic chip integrated with a capture well for high throughput uniform droplet generation, monolayer droplet collection, incubation, detection, and release to realize digital quantitation of enzyme molecules, a key step towards digital protein detection. Single β-galatosidase (β-Gal) molecules and the fluorogenic substrate fluorescein di-β-D-galactopyranoside (FDG) were injected from two separated inlets to form uniform 20 μm droplets in fluorinated oil at a frequency of 6.6 kHz. About 200 000 droplets were captured as a monolayer in the on-chip capture well for subsequent imaging detection. The signal from the fluorescent product of the hydrolysis reaction catalysed by single enzyme molecules in uniform droplets was easily detected under a fluorescence microscope within a few hours. More importantly, the fluorescence intensity distribution of droplets under single enzyme molecule conditions was found to match well with the theoretical prediction, proving the feasibility of detecting single enzyme molecules in emulsion droplets. Based on a digital counting method, the detected concentrations of the enzyme were found to match well with the input enzyme concentration, establishing the accuracy of the digital detection method for quantification of enzyme molecules. By integrating with single-molecule ELISA in droplets, a general digital detection method for single protein molecules could be easily achieved.
EXPERIMENTAL SECTION
Materials and reagents
The SU-8 2015 was purchased from MicroChem (Newton, MA, U.S.A.). β-galactosidase from Escherichia coli overproducer was purchased from Roche Diagnostics (Shanghai, China). The purity of β-galactosidase in the product was analysed by three repeats of denatured 20% polyacrylamide gel electrophoresis (PAGE) with each band identified by proteolytic digestion and MALDI-MS. The final concentration of β-galactosidase was determined as 0.480.14 mg/ml (0.890.26 μM). Fluorescein di-β-D-galactopyranoside was purchased from Sigma-Aldrich (St. Louis, MO, USA). Polydimethylsiloxane (PDMS, Sylgard 184) and curing agent were obtained from Dow Corning (Shanghai, China). Film photomask was produced by Qingyi Precision Maskmaking Co. Ltd. (Shenzhen, China). 1H, 1H, 2H, 2H-perfluorooctyl-dimethylchlorosilane was obtained from Alfa Aesar (Tianjin, China). Bovine serum albumin (BSA) was purchased from New England Biolabs (Beijing, China). Fluorinated oil GH135 was obtained from Lianqun Co. Ltd. (Shenzhen, China). HFE-7100 was purchased from 3M (Shanghai, China), Poly-DMAP was purchased from Sigma-Aldrich (St. Louis, MO, USA), and 157 FSH was obtained from DuPont (Shanghai, China). PEG-600 was provided by Biomatrik Inc. (Jiaxing, China). Phosphate-buffered saline (PBS) buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4) was prepared in the lab with all reagents obtained from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China).
Synthesis of surfactant E2K0660
Fluorosurfactant E2K0660 was synthesized according to a literature method.29 Briefly, oxalyl chloride (0.617 g, 4.86 mmol) was added to a solution of PFPE-COOH (0.729 g, 0.0972 mmol) in 50 ml HFE7100. The mixture was stirred and refluxed overnight under nitrogen. After cooling to room temperature, the mixture was concentrated on a rotary evaporator to remove excess oxalyl chloride. The resulting PFPE-COCl was then dissolved in 50 ml HFE7100 under nitrogen atmosphere with poly-DMAP (0.1 g, 3 mmol/g), using cross-linked polystyrene with pendant 4-aminopyridine groups to scavenge HCl generated during the reaction. Then, a solution of PEG-diamine (32.0 mg, 0.0535 mmol) in 50 ml CHCl3 was added to the above solution. After refluxing overnight, Poly-DMAP was removed by filtration. The mixture was then concentrated on a rotary evaporator, and the final product was vacuum dried at room temperature.
Device fabrication
The SU-8 pattern was fabricated by a conventional photolithographic method. In brief, SU-8 2015 photoresist was coated on a silica wafer at a spin rate of 2200 rpm to produce a film with a thickness of 20 μm. The photomask pattern was transferred onto SU-8 photoresist via UV exposure. PDMS precursor solution was poured onto the silica wafer and spun at 1000 rpm for 20 s to obtain a thin patterned PDMS layer with the thickness of about 100 μm. The patterned PDMS layer was peeled from SU-8 patterned silica wafer, and a droplet capture well was excised using a scalpel along the well outline. The patterned layer was then bonded to a flat PDMS covering layer, which was peeled from another polished glass slide. The module was punched with three inlets and one outlet before bonding onto a flat bottom PDMS layer to seal the channels and well. Finally, the entire PDMS module was bonded to a glass slide to provide high rigidity and low air permeability. Bonding of PDMS to PDMS and PDMS to glass were carried out immediately after oxygen plasma treatment. After bonding, 2% 1H, 1H, 2H, 2H-perfluorooctyldimethyl-chlorosilane/GH-135 (v/v) solution was injected into the channels and incubated at 65 °C for 1 day to produce fluorinated surfaces.
Generation, capture, and release of droplets
FDG and β-Gal solutions were injected from two separate inlets at an equal flow rate of 0.05 ml/h, while the flow rate of the oil phase, 98% (w/w) GH135 and 2% (w/w) fluorinated surfactant E2K0660, was set at 1 ml/h during droplet generation. Droplets were generated at the cross section of the chip by the hydrodynamic flow-focusing method and trapped in the capture well. After a large number of droplets had gathered in the well, the aqueous phases were stopped, and the flow rate of the oil phase was decreased to 0.1 ml/h. The slow oil flow was used to sweep away lower layers of droplets for the generation of monolayer droplets. After incubation and observation, the chip was flipped over to release the droplets with 1 ml/h oil flow. Images of droplet generation, sweeping, and releasing were photographed under the microscope.
Single β-Gal molecule assay
A 1 cm × 1 cm square capture well was designed to capture droplets to achieve high throughput single-molecule analysis (total 200 000 reactions for the optimized 20 μm diameter droplets). β-Gal solution was diluted with PBS buffer to single- molecule concentration of 0, 0.1, 0.25, 0.5, 0.75, 1, 2, or 3 copies per droplet (cpd) on average and injected into the chip from one water phase inlet. In each experiment, 50 ng/ml BSA was added to the solution to reduce nonspecific absorption. The 100 μM FDG was injected from the other water phase inlet. As a result, the concentration of FDG in the final droplets was 50 μM. After having collected enough droplets for detection, inlets and the outlet were sealed by a hot glue gun, followed by bathing in water at 37 °C for 4 h. After capturing by fluorescence microscopy (Nikon Eclipse Ti-U, Tokyo, Japan), the images of droplets under fluorescence filed were input into NIH ImageJ software. Bright droplets above threshold value were automatically counted by the software. About 4000 droplets (including bright and dark droplets) for each dilution repeat were counted.
For real-time recording experiments, the inlets and outlet were not sealed. Instead, the oil flow was kept to support a fluidic shear to confirm the relative location of droplets. A close-packed single layer of droplets was finally observed by fluorescence microscopy. The statistics of fluorescent droplet distribution was analysed using NIH ImageJ software.
RESULTS AND DISCUSSION
Principle of digital enzyme detection using microfluidic droplet approach
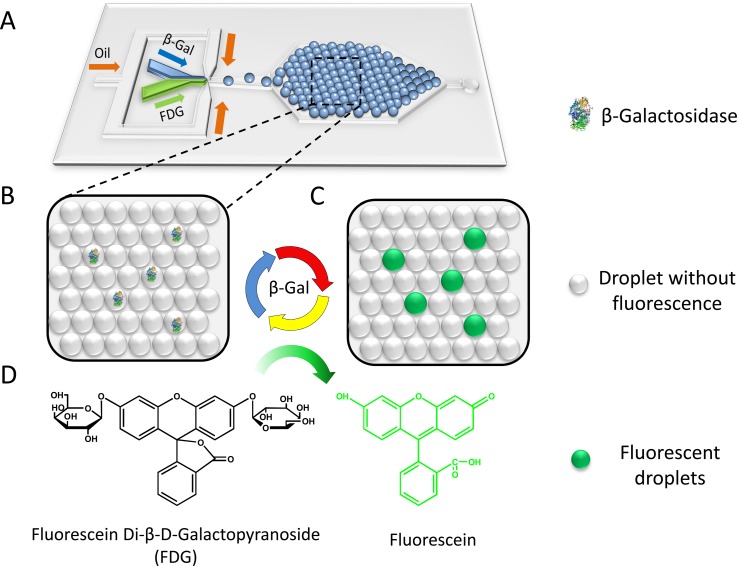
The working principle of the microdroplet-based digital enzyme detection method is schematically shown in Fig. 1. FDG, a non-fluorescent substrate that contains two β-galactoside linkages, can be hydrolysed by β-Gal to produce the highly fluorescent fluorescein molecule (Fig. 1d). Single β-Gal molecules are encapsulated in monodisperse droplets generated on-chip (Fig. 1a). The PDMS chip has three inlets, including a protein solution inlet, an FDG solution inlet, and an oil phase inlet. This two-liquid-phase-inlet design avoids premixing of the reaction components, thus decreasing the fluorescence background. The two aqueous phases are injected at the same flow rate in parallel and dispersed together into droplets with equal volume. Droplets are then trapped in the capture well, where they self-organize into a close-packed monolayer by fluid shear (Fig. 1b). Upon incubation at 37 °C, hydrolysis of FDG takes place inside droplets that contain β-Gal molecules, producing brightly fluorescent droplets as a result (Fig. 1c). For droplets containing no enzyme, no hydrolysis occurs, yielding nonfluorescent droplets. After enzymatic reaction, fluorescent images of the droplets are obtained under the fluorescence microscope and analysed using ImageJ software to obtain the percentage of fluorescent droplets and their fluorescence intensity statistics. Under single-molecule conditions, different enzyme concentrations lead to different percentages of fluorescent droplets. Using Poisson statistics, the concentration of enzyme in the sample can be simply calculated from the fraction of fluorescent or non-fluorescent droplets and the droplet volume.
Figure 1.
Schematic of digital enzyme detection using microfluidic droplet approach. (A) Layout of the microfluidic device for droplet generation and trapping. (B) An enlarged diagrammatic view of monolayer close-packed droplet array. (C) Diagrammatic drawing of droplets after incubation. (D) Hydrolysis of non-fluorescent FDG to fluorescent fluorescein catalysed by β-Gal.
The choice of fluorinated oil GH-135 and surfactant E2K0660
In recent years, fluorinated oils and fluorinated surfactants have been widely used in microdroplet applications because of their favorable biocompatibility.29, 30 In addition, compared to most other oils, such as silicone oil and mineral oil, fluorinated oil has lower viscosity, which can greatly improve the flow rate in chip and droplet generation frequency, while decreasing the frictional force and fluid pressure. Although HFE-7500 and FC 40 are the two most widely used fluorinated oils in literature reports, GH-135 was found to have lower volatility and higher thermal stability compared to the former two types of oil in our experiments. Fluorinated surfactant E2K0660 has been used for polymerase chain reaction (PCR)31 and cell analysis32 with excellent stability and biocompatibility. Therefore, fluorinated oil GH-135 and surfactant E2K0660 were chosen in our work to generate thermostable droplets in the microfluidic chip device.
Since the enzyme reactions take place in water-in-oil (W/O) droplets, to guarantee success of the experiment, we had to verify that the chosen fluorinated oil and surfactant have limited influence on enzyme activity. The enzyme activities of β-Gal in water phase and in W/O droplets were compared. Three groups of samples were prepared: a negative control (bulk water solution without β-Gal), a positive control (bulk PBS buffer solution with 2.25 ng/ml β-Gal), and a droplet group where water phase (with 2.25 ng/ml β-Gal) was shaken into droplets in the same volume of oil containing 2% E2K0660. All three groups contained 50 μM FDG and 50 ng/ml BSA in the final reaction solution. The fluorescent intensity of each group was recorded in real-time by a quantitative PCR instrument (StepOne Real-Time PCR System, Applied Biosystems, USA) at 2 min intervals. There was no fluorescence intensity increase observed in the negative control sample; while in the positive control and droplet group, the fluorescence intensity increased linearly with time (Fig. S1 (Ref. 33)). A small decrease in fluorescent intensity was observed for the droplet group, as compared to the positive control, possibly due to turbidity of the W/O emulsion phase. The result suggested that the oil and surfactant used in the experiment do not have significant negative influence on the enzyme activity.
Droplet generation, capture, and release
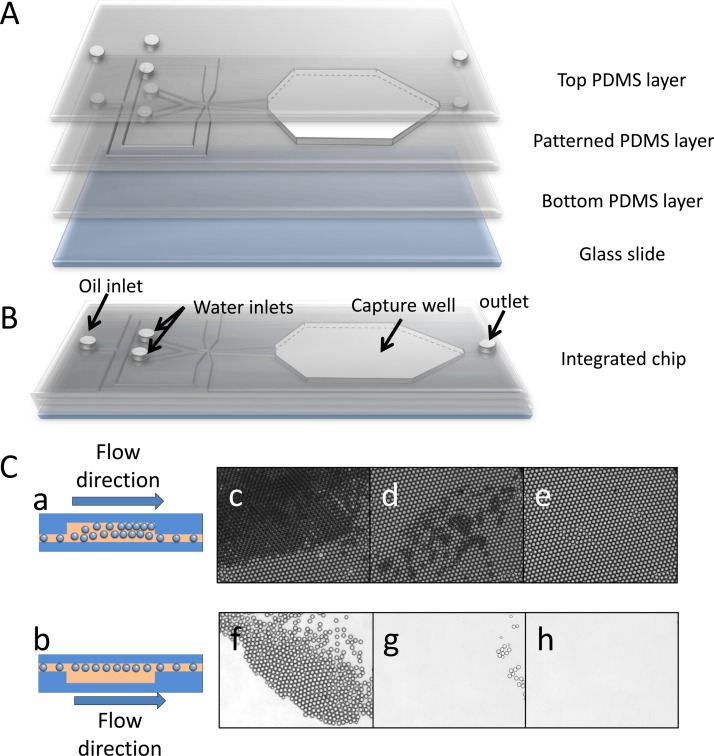
An integrated PDMS microfluidic chip was designed and fabricated for high throughput uniform droplet generation, monolayer droplet collection, incubation, detection, and release. A schematic of chip structure is illustrated in Fig. 2a. The PDMS chip includes a protein solution inlet, an FDG solution inlet, an oil phase inlet, a droplet capture well, and an outlet (Fig. 2b). Droplets are generated at the cross section of the chip by a hydrodynamic flow-focusing method. Due to the significant density difference between water (1 g/ml) and fluorinated oil GH-135 (1.68 g/ml), when droplets are generated, they tend to float along the roof of the channel. This property makes it possible to capture the droplets downstream by an up concave well. As shown in Fig. 2, we designed the chip with an uplifted well right after the droplet generation section. The well is connected with the outlet by a channel at the bottom with width of 200 μm and depth of 20 μm to exhaust waste oil. Droplets jetted into the capture well after generation rise to the roof of the well (Fig. 2). When a large number of droplets are gathered in the capture well, they self-assemble into close-packed layers (Fig. 2) until they occupy most of the space and the surplus droplets start draining away. Dynamic balance between accumulation and release of droplets is finally formed with continuous generation of droplets, but is interrupted when both of the water phases are stopped. Then the bottom layers of droplets are swept away by the relatively low oil flow (0.1 ml/h), whereas the top layer stably trapped in the capture well because of the up-concave structure (Figs. 2 and 2). In this way, a monolayer close-packed droplet array (Fig. 2) is obtained. The final channel depth is 20 μm and the capture well height is about 100 μm. After incubation and observation, the chip is flipped over, inducing the droplets to rise to the outlet channel side of chip and flow out of the well along with the oil phase (Fig. 2). All of the droplets are released after several minutes by oil rinse at 1 ml/h (Figs. 2–2) and the chip can be reused. Compared with other droplet trapping methods,27, 28 our design offers the advantages of simple device fabrication, scalable droplet generation, facile droplet trapping, and rapid device regeneration.
Figure 2.
(A) Three-dimensional illustration of a droplet capture chip, including a top PDMS layer, a patterned PDMS layer with capture well excised, a flat bottom PDMS layer and a glass slide. (B) Schematic diagram of integrated chip. (C) Schematic of capture well when capturing (a) and releasing (b) droplets. (c) Close-packed droplets layers are generated when droplets gather in the capture well. (d) Lower layers of droplets are swept away when the water phases are stopped with the oil flow rate set at 0.1 ml/h. (e) Mono-layer of close-packing droplets is generated after sweeping for several minutes. Release of droplets after flipping-over the microfluidic chip for 1 min (f), 2 min (g), and 4 min (h).
Optimization of the droplet diameter
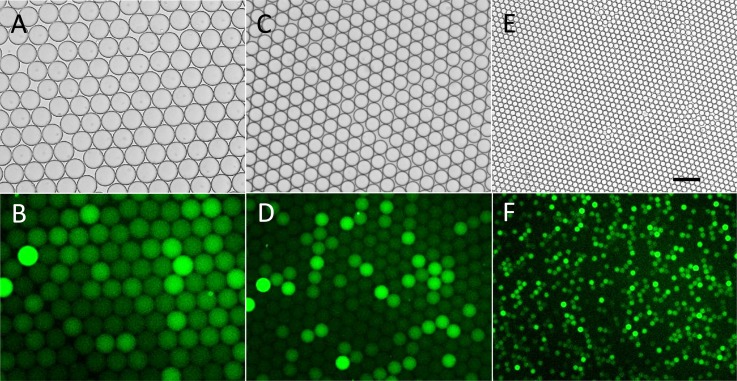
In order to study the influence of droplet diameter on fluorescent intensity and signal-background ratio (SBR, the average fluorescent intensity ratio of fluorescent droplets to non-fluorescent droplets), three kinds of droplets with diameters of about 80 μm, 55 μm, and 20 μm were generated using microfluidic chips with different channel parameters and flow rates. A chip with dimensions of 50 μm × 50 μm × 20 μm (width × width × height) in the flow-focusing section was used to generate droplets with ∼80 μm and ∼55 μm diameter by adjusting the flow rates of the oil and the water phase. A 20 μm × 20 μm × 20 μm chip was used to generate ∼20 μm droplets. The diameters of the droplets generated were measured to be 80.9 ± 1.3 μm, 56.1 ± 0.6 μm, and 20.0 ± 0.2 μm. The relative standard deviation (RSD) of droplet size was found to be smaller than 2%, showing narrow distribution and excellent monodispersity. To provide excess substrate, the concentration of FDG was set as 50 μM in the generated droplet and the concentration of β-Gal was adjusted according to the diameter to guarantee that about 10% of the droplets would be fluorescent. These droplets were collected in chip and incubated at 37 °C and the results were recorded under the fluorescence microscope.
Images of three kinds of droplets are shown in Fig. 3. For SBR calculation, 100 fluorescent droplets and 300 non-fluorescent droplets were measured. The SBR of the smallest (20.0 μm) droplets was found to be the highest (SBR = 2.1 ± 0.9, Fig. 3f) compared to that of 80.9 μm droplets (SBR indeterminable because of the tiny fluorescence intensity difference between droplets, Fig. 3b) and 56.1 μm droplets (SBR = 1.6 ± 0.8, Fig. 3d), even though only 4 h was required for the former reaction, while the latter two groups were allowed to react for 20 h. The improvement of SBR for smaller droplets is reasonable. Within droplets, the enzymatic hydrolysis product fluorescein cannot spread into the oil phase because of the hydrophobicity of the fluorinated oil. Because the substrate was in large excess, the catalytic rate of each enzyme molecule and the amount of catalytic product can be regarded as the same in the three types of droplets within a limited incubation time. This resulted in an increase of fluorescent catalytic product concentration in the smallest droplets.19 To obtain high SBR, all subsequent experiments were conducted based on 20 μm droplets. In addition, although background fluorescence of droplets without β-Gal was also observed, possibly due to partial hydrolysis of FDG during storage, the fluorescent droplets could still be clearly discriminated obviously when the diameter was decreased to 20 μm.
Figure 3.
Fluorescent and bright field images of three different sizes of droplets after single-enzyme reaction. (a)-(b) 80.9 ± 1.3 μm droplets incubated for 20 h. (c)-(d) 56.1 ± 0.62 μm droplets incubated for 20 h. (e)-(f) 20 ± 0.2 μm droplets incubated for 4 h. Horizontal scale bar is 100 μm.
Highly parallel observation of single-enzyme reaction kinetics in droplets
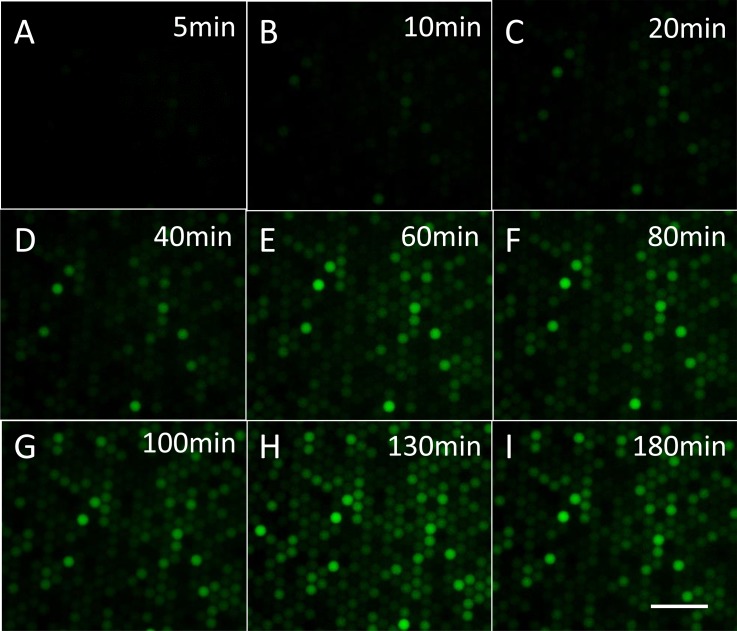
The excellent stability of the monolayer droplet array formed in the capture well offers the ability to perform real-time highly parallel single-enzyme reaction kinetic studies inside droplets. The relative positions of droplets in the capture well barely changed even after several hours' incubation, when the flow rate of the oil phase was set as low as 0.01 ml/h to ensure close-packing of droplets. The fluorescence of about 1000 droplets in a visual field was recorded during incubation. In Fig. 4, fluorescence intensity images of droplets are shown according to increasing incubation time. When β-Gal solution with 2 cpd concentration was injected into the chip, it was found that the fluorescent intensity of some droplets increased as incubation time increased, while the rest had no significant variation during the 3 h incubation. The intensity variation of droplets with different rates of fluorescence increase is shown in Fig. S2.33 The variation is possibly due to the change in the number of enzyme molecules in each droplet and the heterogeneous nature of single-enzyme catalysis. The results clearly suggest that our design can be used to perform highly parallel single-enzyme kinetic analysis.
Figure 4.
Fluorescence images of droplets after incubation for 5 min to 180 min: (a) 5 min, (b) 10 min, (c) 20 min, (d) 40 min, (e) 60 min, (f) 80 min, (g) 100 min, (h) 130 min, and (i) 180 min. Horizontal scale bar is 100 μm.
Poisson distribution of single molecules in droplets
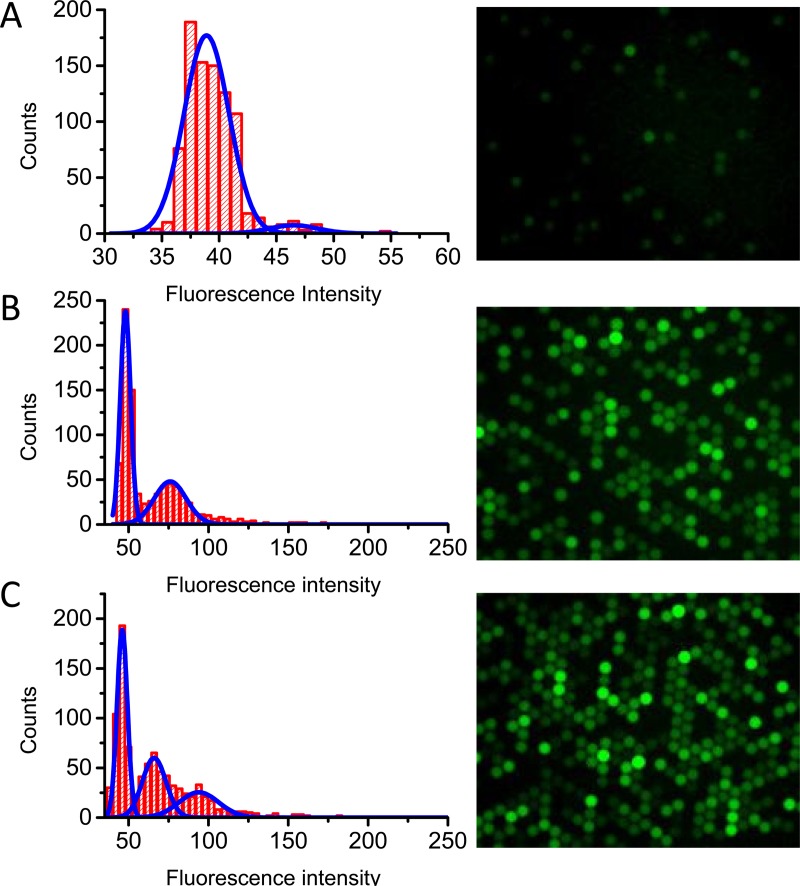
The feasibility of observing single-enzyme reaction in droplets was further verified by analysing the fluorescent intensity distribution of droplets under single-enzyme molecule conditions (0.1, 0.5, and 1 cpd). The fitting results of the fluorescent intensity distribution and fraction of droplets in each distribution are shown in Fig. 5 and Table S1.33 The measured population of 0-molecule, 1-molecule-, and 2-molecule droplets matched with the theoretical value of the Poisson distribution. For example, at a concentration of 0.1 cpd (Fig. 5a), it was expected that about 9.5% droplets would be fluorescent, while about 8.0% droplets were found. In the case of 0.5 cpd (Fig. 5b), while it was expected that 60.6% droplets contained 0 enzyme molecules, 30.3% contained 1 molecule, and 9.1% contained more than 2 molecules, and fluorescent intensity distributions suggested the distribution to be 58.6%, 32.1%, and 9.3%, respectively. A χ2 (chi-square) test was conducted to confirm whether the measured cpd matched well with the theoretical value of the Poisson distribution. For both group droplets with 0.1 cpd and 0.5 cpd, P values higher than 0.05 was observed, strongly suggesting that the measured distribution matched well with the theoretical distribution. For droplets with 1.0 cpd, p was smaller than 0.05. The deviation of the measured value from the theoretical value could possibly be due to the heterogeneous nature of enzyme activity that resulted in the slight overlap of neighboring populations (see Fig. 5c). This deviation causes little effect on the precision of digital detection result, because in digital detection, signal outputs are only distinguished by “0” or “1,” where “0” represents the absence of the molecule and “1” stands for the presence of the molecule in the droplet. As shown in Table S2,33 the observed distributions of bright (“1”) and dark (“0”) droplets for all three groups fit well with the Poisson distribution, as confirmed by chi-square test. The observed fit of experimental data with Poisson statistics strongly confirms the feasibility of observing single-enzyme molecules in emulsion droplets via enzymatic reaction.
Figure 5.
Fluorescence intensity distribution of droplets with concentration 0.1 cpd (a), 0.5 cpd (b), and 1 cpd (c). At the right side are the fluorescent images respective to each cpds.
Digital detection of β-Gal molecules
After confirming the capability of performing single-molecule enzymatic reactions in droplets, we further tested the feasibility of digital quantitation of the enzyme concentration by simply counting the fluorescent droplets. As shown above, the distribution of enzyme molecules in the droplets obeys Poisson distribution
| (1) |
which describes the probability of events occurring randomly in a fixed time or certain space at an average rate λ with a discrete stochastic variable x. First, the probabilities of non-fluorescent droplets (Px = 0) in experiments with different enzyme concentrations at the single molecule level were collected, and the measured average cpd (λmes) was calculated using the following expression derived from the Poisson distribution:
| (2) |
The measured enzyme concentration in droplets (Cmes) was then calculated according to
| (3) |
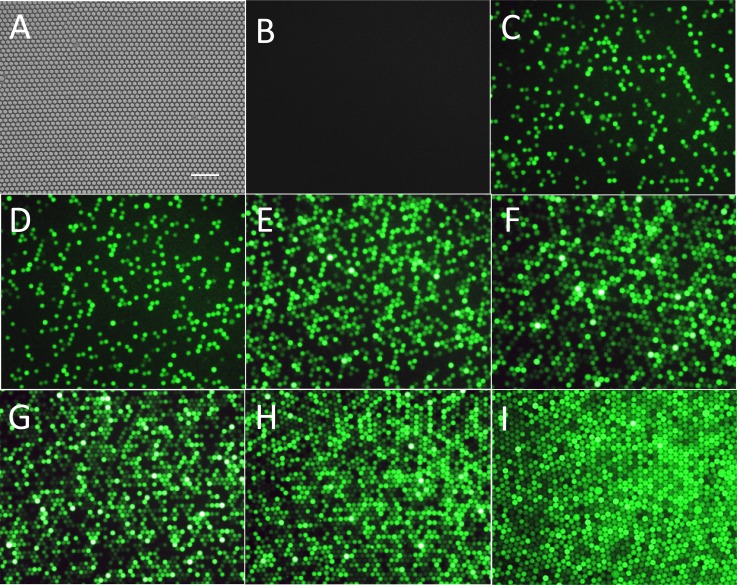
where NA is Avogadro's number (6.02 × 1023), and V is the droplet volume (4.2 pl) calculated from the droplet diameter. To check the reliability of the single-molecule quantification method, different concentrations of β-Gal were injected into the chip. A close-packed single layer droplet array was generated in the capture well (Fig. 6a). As shown in Fig. 6b, no fluorescent droplets were found in the negative control without β-Gal, and the percentage of fluorescent droplets increased as the feeding enzyme concentration increased (Fig. 6). For example, the fraction of fluorescent droplets to total droplets was 0.634 for the 1 cpd sample and increased to 0.882 for the 2 cpd sample. The increase was not linear because the probability of multiple enzyme molecules appearing in the same droplet increased as the concentration increased.
Figure 6.
(a) Typical image of a close-packed single layer droplet array. (b)-(i) Fluorescence images of droplets with different molecule numbers of β-Gal: negative control with no enzyme (b), 0.1 cpd (c), 0.25 cpd (d), 0.5 cpd (e), 0.75 cpd (f), 1 cpd (g), 2 cpd (h), and 3 cpd (i). All the droplets have reacted for 4 h and were single layer close packed. Horizontal scale bar is 100 μm.
As shown in Table TABLE I., to absolutely quantitate the enzyme concentration, the fraction of non-fluorescent droplets was collected and Cmes was calculated using Eqs. 2, 3. Each Cmes for different enzyme concentrations shown in Table TABLE I. was an average of three experiment repeats, with RSD displayed. Almost all of the RSDs were smaller than 5%, which was identified as a random error, except for the two most dilute groups that may have exhibited non-specific adsorption. It is worth noting that consistent values of measured sample concentration were obtained across all diluted enzyme samples (Table TABLE I.) with an average value of 0.880.02 μM. This strongly demonstrates that the digital counting method can be used to determine sample concentrations by single molecule counting. The measured concentration (0.880.02 μM) matches well with the original sample concentration (0.890.26 μM). This result confirmed the feasibility and accuracy of our microfluidic droplet-based single-molecule counting method for digital detection of β-Gal enzyme.
TABLE I.
Digital detection results based on single-molecule counting with different β-Gal concentrations.
| Sample concentration (μM) | Dilution factor | Input concentration (fM) | Fraction of none-fluorescent droplets | λmes | Measured enzyme concentration in droplet (Cmes, fM) | RSD % | Measured sample concentration μM) | Average of measured sample concentration (μM) | |
|---|---|---|---|---|---|---|---|---|---|
| 0.89 0.26 | 4.50 × 10−8 | 40 | 0.899 | 0.106 | 42.0 | 6.8 | 0.93 | 0.880.02 | |
| 1.13 × 10−7 | 101 | 0.7820.010 | 0.2460.013 | 97.4 | 5.6 | 0.86 | |||
| 2.25 × 10−7 | 200 | 0.6160.007 | 0.4850.011 | 192 | 2.9 | 0.850.02 | |||
| 3.38 × 10−7 | 301 | 0.4720.011 | 0.7510.023 | 297 | 3.5 | 0.880.03 | |||
| 4.50 × 10−7 | 401 | 0.3660.006 | 1.0050.016 | 398 | 2.4 | 0.880.02 | |||
| 9.00 × 10−7 | 801 | 0.1180.006 | 2.1430.051 | 848 | 3.0 | 0.940.03 | |||
| 1.35 × 10−6 | 1202 | 0.0550.006 | 2.913 | 115047.8 | 4.2 | 0.850.04 | |||
CONCLUSIONS
In conclusion, we have developed a droplet microfluidic method for quantifying enzyme molecules by digital counting of droplets under single molecule conditions. The core engine to this method is an integrated microfluidic device designed and fabricated for high throughput uniform droplet generation, monolayer droplet collection, incubation, detection, and release. Compared with other droplet generation and trapping devices, our design offers the advantages of simple device fabrication, scalable droplet generation, facile droplet trapping, and rapid device regeneration. Using the device, single β-Gal molecules and fluorogenic substrate FDG were injected from two separate inlets to form uniform 20 μm droplets at frequency as high as 6.6 kHz. A capture well was designed to obtain stable close-packed droplet monolayers for high throughput in situ detection and analysis of droplets. A series of β-Gal solutions at different single molecule concentrations were analysed, and the fraction of fluorescent droplets increased as the β-Gal concentration increased. Fluorescence intensity distributions of empty and enzyme containing droplets at three single molecule concentrations, 0.1, 0.5, and 1 cpd, were found to match well with the value calculated from the Poisson distribution, proving the feasibility of detecting single enzyme molecules in emulsion droplets. The agreement of measured concentrations to input concentrations and the small standard deviations demonstrate the accuracy and precision of the digital detection method for the quantification of enzyme molecules. The advantages of using droplets for digital detection include small volume consumption, flexible scalability, high throughput, single-molecule sensitivity, and improved precision. For example, by performing a single molecule reaction in a single picoliter droplet, as compared to a 96-well plate well, one can reduce reagent consumption by 6-7 orders of magnitude. More importantly, hundreds of thousands of reactions can be easily performed in parallel with low cost. Even for one experiment with 200 000 reactions in 20 μm droplets, total reagent consumption is only 0.8 μl. The capability of highly parallel detection of single enzyme molecules in uniform picoliter droplets paves the way for microdroplet based digital detection of proteins. By integrating with single-molecule ELISA in droplets, a wide variety of protein molecules can be digitally detected at the single-molecule level by this method.
ACKNOWLEDGMENTS
We thank the National Basic Research Program of China (2010CB732402 and 2013CB933703), the National Science Foundation of China (91313302, 21205100, 21275122, and 21075104), National Instrumentation Program (2011YQ03012412), and the National Science Foundation for Distinguished Young Scholars of China (21325522) for their financial support.
References
- Leamon J. H., Link D. R., Egholm M., and Rothberg J. M., Nat. Methods 3(7), 541 (2006). 10.1038/nmeth0706-541 [DOI] [PubMed] [Google Scholar]
- Theberge A. B., Courtois F., Schaerli Y., Fischlechner M., Abell C., Hollfelder F., and Huck W. T., Angew. Chem., Int. Ed. 49(34), 5846 (2010). 10.1002/anie.200906653 [DOI] [PubMed] [Google Scholar]
- Joensson H. N. and Andersson Svahn H., Angew. Chem., Int. Ed. 51(49), 12176 (2012). 10.1002/anie.201200460 [DOI] [PubMed] [Google Scholar]
- Teh S. Y., Lin R., Hung L. H., and Lee A. P., Lab Chip 8(2), 198 (2008). 10.1039/b715524g [DOI] [PubMed] [Google Scholar]
- Zhu Z., Jenkins G., Zhang W., Zhang M., Guan Z., and Yang C. J., Anal. Bioanal. Chem. 403(8), 2127 (2012). 10.1007/s00216-012-5914-x [DOI] [PubMed] [Google Scholar]
- Vogelstein B. and Kinzler K. W., Proc. Natl. Acad. Sci. U.S.A. 96(16), 9236 (1999). 10.1073/pnas.96.16.9236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottesen E. A., Hong J. W., Quake S. R., and Leadbetter J. R., Science 314(5804), 1464 (2006). 10.1126/science.1131370 [DOI] [PubMed] [Google Scholar]
- Baker M., Nat. Methods 9(6), 541 (2012). 10.1038/nmeth.2027 [DOI] [PubMed] [Google Scholar]
- Baker M., Nat. Methods 7(5), 351 (2010). 10.1038/nmeth0510-351 [DOI] [Google Scholar]
- Leng X., Zhang W., Wang C., Cui L., and Yang C. J., Lab Chip 10(21), 2841 (2010). 10.1039/c0lc00145g [DOI] [PubMed] [Google Scholar]
- Hatch A. C., Fisher J. S., Tovar A. R., Hsieh A. T., Lin R., Pentoney S. L., Yang D. L., and Lee A. P., Lab Chip 11(22), 3838 (2011). 10.1039/c1lc20561g [DOI] [PubMed] [Google Scholar]
- Hindson B. J., Ness K. D., Masquelier D. A., Belgrader P., Heredia N. J., Makarewicz A. J., Bright I. J., Lucero M. Y., Hiddessen A. L., Legler T. C., Kitano T. K., Hodel M. R., Petersen J. F., Wyatt P. W., Steenblock E. R., Shah P. H., Bousse L. J., Troup C. B., Mellen J. C., Wittmann D. K., Erndt N. G., Cauley T. H., Koehler R. T., So A. P., Dube S., Rose K. A., Montesclaros L., Wang S., Stumbo D. P., Hodges S. P., Romine S., Milanovich F. P., White H. E., Regan J. F., Karlin-Neumann G. A., Hindson C. M., Saxonov S., and Colston B. W., Anal. Chem. 83(22), 8604 (2011). 10.1021/ac202028g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Jenkins G., Zou Y., Zhu Z., and Yang C. J., Anal. Chem. 84(8), 3599 (2012). 10.1021/ac2033084 [DOI] [PubMed] [Google Scholar]
- Chen W. W., Balaj L., Liau L. M., Samuels M. L., Kotsopoulos S. K., Maguire C. A., Loguidice L., Soto H., Garrett M., Zhu L. D., Sivaraman S., Chen C., Wong E. T., Carter B. S., Hochberg F. H., Breakefield X. O., and Skog J., Mol. Ther. Nucl. Acids 2, e109 (2013). 10.1038/mtna.2013.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressman D., Yan H., Traverso G., Kinzler K. W., and Vogelstein B., Proc. Natl. Acad. Sci. U.S.A. 100(15), 8817 (2003). 10.1073/pnas.1133470100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekin D., Skhiri Y., Baret J. C., Le Corre D., Mazutis L., Salem C. B., Millot F., El Harrak A., Hutchison J. B., Larson J. W., Link D. R., Laurent-Puig P., Griffiths A. D., and Taly V., Lab Chip 11(13), 2156 (2011). 10.1039/c1lc20128j [DOI] [PubMed] [Google Scholar]
- Shuga J., Zeng Y., Novak R., Lan Q., Tang X., Rothman N., Vermeulen R., Li L., Hubbard A., Zhang L., Mathies R. A., and Smith M. T., Nucl. Acids Res. 41(16), e159 (2013). 10.1093/nar/gkt613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agresti J. J., Antipov E., Abate A. R., Ahn K., Rowat A. C., Baret J. C., Marquez M., Klibanov A. M., Griffiths A. D., and Weitz D. A., Proc. Natl. Acad. Sci. U.S.A. 107(9), 4004 (2010). 10.1073/pnas.0910781107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotman B., Proc. Natl. Acad. Sci. U.S.A. 47, 1981 (1961). 10.1073/pnas.47.12.1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondelez Y., Tresset G., Tabata K. V., Arata H., Fujita H., Takeuchi S., and Noji H., Nat. Biotechnol. 23(3), 361 (2005). 10.1038/nbt1072 [DOI] [PubMed] [Google Scholar]
- Deiss F., LaFratta C. N., Symer M., Blicharz T. M., Sojic N., and Walt D. R., J. Am. Chem. Soc. 131(17), 6088 (2009). 10.1021/ja901876z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorris H. H. and Walt D. R., J. Am. Chem. Soc. 131(17), 6277 (2009). 10.1021/ja9008858 [DOI] [PubMed] [Google Scholar]
- Li Z., Hayman R. B., and Walt D. R., J. Am. Chem. Soc. 130(38), 12622 (2008). 10.1021/ja8053018 [DOI] [PubMed] [Google Scholar]
- Konry T. and Walt D. R., J. Am. Chem. Soc. 131(37), 13232 (2009). 10.1021/ja905125b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissin D. M., Kan C. W., Campbell T. G., Howes S. C., Fournier D. R., Song L., Piech T., Patel P. P., Chang L., Rivnak A. J., Ferrell E. P., Randall J. D., Provuncher G. K., Walt D. R., and Duffy D. C., Nat. Biotechnol. 28(6), 595 (2010). 10.1038/nbt.1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim J. U., Ranasinghe R. T., Smith C. A., Ibrahim S. M., Hollfelder F., Huck W. T., Klenerman D., and Abell C., ACS Nano 7(7), 5955 (2013). 10.1021/nn401661d [DOI] [PubMed] [Google Scholar]
- Ota S., Kitagawa H., and Takeuchi S., Anal. Chem. 84(15), 6346 (2012). 10.1021/ac301204v [DOI] [PubMed] [Google Scholar]
- Arayanarakool R., Shui L., Kengen S. W., van den Berg A., and Eijkel J. C., Lab Chip 13(10), 1955 (2013). 10.1039/c3lc41100a [DOI] [PubMed] [Google Scholar]
- Holtze C., Rowat A. C., Agresti J. J., Hutchison J. B., Angile F. E., Schmitz C. H., Koster S., Duan H., Humphry K. J., Scanga R. A., Johnson J. S., Pisignano D., and Weitz D. A., Lab Chip 8(10), 1632 (2008). 10.1039/b806706f [DOI] [PubMed] [Google Scholar]
- Baret J. C., Lab Chip 12(3), 422 (2012). 10.1039/c1lc20582j [DOI] [PubMed] [Google Scholar]
- Kiss M. M., Ortoleva-Donnelly L., Beer N. R., Warner J., Bailey C. G., Colston B. W., Rothberg J. M., Link D. R., and Leamon J. H., Anal. Chem. 80(23), 8975 (2008). 10.1021/ac801276c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baret J. C., Beck Y., Billas-Massobrio I., Moras D., and Griffiths A. D., Chem. Biol. 17(5), 528 (2010). 10.1016/j.chembiol.2010.04.010 [DOI] [PubMed] [Google Scholar]
- See supplementary material at http://dx.doi.org/10.1063/1.4866766 for (1) influence of fluorinated oil GH-135 and E2K0660 to the activity of β-Gal, (2) representative time traces of enzyme activity measured in microfluidic droplets under single-molecule condition, and (3) fluorescence intensity distribution for different β-Gal concentrations at single-single molecule concentrations.