Abstract
More than 15 human genetic diseases, including Huntington’s disease, result from the expansion of a trinucleotide repeat. The expansions are unstable in specific somatic tissues, which can lead to disease acceleration. Here we discuss the role of transcription elongation in tissue-selective trinucleotide repeat instability.
Keywords: Huntington's disease, R-loops, chromatin structure, somatic CAG/CTG instability, transcription dynamics, transcription elongation, trinucleotide repeat diseases
Trinucleotide repeats involved in diseases are dynamic mutations
Several neurological, neurodegenerative and neuromuscular diseases, including Huntington’s disease (HD), myotonic dystrophy 1 (DM1) and Friedreich ataxia (FRDA) are caused by abnormal expansion of a trinucleotide repeat (TNR) within genes otherwise unrelated.1,2 Various TNRs are involved in disease, though CAG/CTG repeats are most frequent. In addition, TNRs associated with diseases can lead to various disease mechanisms, ranging from loss-of-function to protein or RNA gain-of-function mechanisms, depending on the location of the repeat within the affected gene. Remarkably, TNRs involved in diseases are all located in transcribed regions. Moreover, TNRs associated with diseases are dynamic mutations within tissues and across generations, due to somatic and germline instability, respectively. TNR instability results in repeat size variation and, since the instability mechanism is biased toward further expansion in both the somatic tissues and the germline, the progression of the disease tends to accelerate over time in affected individuals and the severity of the disease tends to aggravate in successive generations. The degree of variation of the repeat tract is highly dependent upon tissue or cell-type. Interestingly, in most CAG/CTG diseases, which include HD, DM1 and several dominant spinocerebellar ataxias, repeat instability is elevated in the striatum and cortex, whereas it is minimal in the cerebellum, suggesting that tissue-specific factors and mechanisms drive instability. Several mechanisms have been involved in TNR instability, including DNA repair, replication and transcription.1-3 Yet, it is not clear how these mechanisms would lead to tissue-specific instability. Here we discuss advances in our understanding of the role of transcription in tissue-selective CAG/CTG instability.
Transcription and TNR instability: resolving the paradox
The models proposed to explain TNR instability rely upon the view that TNRs form stable DNA secondary structures and aberrant repair of those structures by overwhelmed DNA repair machineries results in TNR instability.1 Studies based on cell models suggest that transcription across TNRs induces stable DNA:RNA hybrids (R-loops), which would induce the formation of DNA secondary structures on the coding strand, followed by their error-prone repair. Using engineered cells, it has been shown that increasing transcriptional activity at transgenes with CAG/CTG repeats results in increased instability.4,5 Interestingly, bidirectional transcription across TNRs further increased TNR instability.5,6 Furthermore, treating such cells with RNaseH, which removes R-loops, improved the stability of CAG/CTG repeats.7 In addition, using in vitro transcription, it was found that CAG/CTG repeats form R-loops.8 Thus, in vitro and in cellula, transcription contributes to CAG/CTG instability. However, in the studies described above, transcription of expanded genes was under the control of artificial promoters and transgene expression levels were not physiological.
The in vivo role of transcription in TNR instability has actually remained unclear. Several studies, including our recent study, found no correlation between mRNA levels of TNR genes and repeat instability in tissues. Specifically, Huntingtin (HTT) mRNA of HD patients or rodent models was not increased in the striatum when compared with the cerebellum.9,10 Similarly, the level of somatic CTG instability did not correlate with the level of Dmpk transcript in tissues of DM1 mice.11
Moreover, in some TNR diseases, including DM1 and FRDA, expansion of TNRs induces a heterochromatinization process, which in FRDA leads to transcriptional silencing.12 Thus, in some instances, transcriptional repression is coincident with TNR instability.
Transcriptionally active chromatin correlates with increased CAG instability in HD mice
To explore the possible in vivo role of transcription in somatic CAG instability, in our recent study, we took advantage of the HD transgenic mouse lines R6/1 and R6/2.13,14 The two mice express a similar HD transgene consisting of human HTT exon-1 with an expanded CAG repeat, ≈1000 bp of the HTT promoter and 262 pb of HTT intron-1. The transgene is integrated at different genomic regions of R6/1 and R6/2 mice. Both lines show a comparable HD-like phenotype; however, the progression of the disease is much faster in R6/2 than in R6/1 mice. Interestingly, we found that CAG instability rates were increased in R6/2 tissues when compared with R6/1-matched tissues, which included striatum and cerebellum.9,15 The greater CAG instability of R6/2 mice relative to R6/1 mice was not simply the consequence of increased inherited repeat size, since R6/2 mice with inherited expansion of 100 and 160 repeats showed increased CAG instability when compared with R6/1 with 130 repeats, which suggested that the integration site of the transgene contributed to differential instability between R6/1 and R6/2 mice. In addition, transgene expression in the striatum and cerebellum was 2- to 3-fold increased in R6/2 mice relative to R6/1 mice.9 The level of transgene expression was in the same range as endogenous murine Htt expression. Transgene expression was slightly higher than murine Htt expression in R6/2 mice and slightly reduced compared with murine Htt expression in R6/1 mice, suggesting that transgene transcript levels are close to physiological levels in both R6/1 and R6/2 mice. Finally, chromatin experiments (ChIP) using antibodies to H3K9me2 and H3K4me3 revealed that chromatin at the HD transgene was in a more accessible state in R6/2 when compared with R6/1 matched tissues (e.g., striatum and cerebellum).9 Thus, our in vivo data suggested that integration site of the transgene influences chromatin structure and transcriptional activity, and as a consequence repeat instability.
However, despite increased CAG instability in striatum relative to cerebellum, we measured similar levels of transgene mRNA in the striatum and cerebellum of R6/1 or R6/2 mice. We also found similar amounts of H3K9me2 and H3K4me3 at the HD transgene in the striatum and cerebellum of R6/1 or R6/2 mice, indicating that the level of these histone marks does not correlate with tissue-selective CAG instability.
Tissue-selective recruitment of H3K36me3 and RNAP II at the HD locus
H3K4me3 marks more specifically transcription start sites (TSS) and is required for transcription initiation. Thus, the results described above suggest that steps involved in the initiation of HTT transcription are not contributing to tissue-selective repeat instability. Interestingly however, ChIP performed using an antibody to RNAP II revealed that RNAP II at the HD transgene strongly correlates with CAG instability levels in tissues of R6/1 and R6/2 mice.9 Specifically, RNAP II at the HD transgene was higher in striatum relative to cerebellum and higher in R6/2 tissues when compared with R6/1-matched tissues. Furthermore, the correlation between CAG instability and RNAP II level at the HD transgene was strong when using an antibody to elongating RNAP II. In addition, the level at the HD transgene of the histone mark H3K36me3, which is associated with transcription elongation, was strongly correlated with CAG instability in tissues of both R6/1 and R6/2 mice. Together, these data suggest that events associated with transcription elongation might be regulated in a tissue-dependent manner, contributing to tissue-selective CAG instability.
Given that similar levels of HD transcript were measured in the striatum and cerebellum of R6/1 mice or R6/2 mice, how could transcription elongation be involved in differential CAG instability between striatum and cerebellum? HTT mRNA processing might be regulated in a tissue-specific manner. Interestingly, previous studies showed that two mRNA species originate from the HTT gene, resulting from usage of two distinct polyadenylation signals.16,17 The relative abundance of the two species was tissue-specific, consistent with recent deep sequencing data of mRNA populations from multiple tissues showing that 86% of human genes display variants due to alternative polyadenylation.18 PolyA sites are essential for 3′-end maturation, stability and degradation of mRNAs. Thus, one might hypothesize that despite increased transcriptional activity in the striatum as compared with the cerebellum, HTT mRNA level could be similar in the two tissues, due to alternate polyadenylation site usage and increased HTT mRNA stability in the cerebellum. However, it is unlikely that this mechanism occurs in R6/1 and R6/2 mice, since the HD transgene that is expressed in these mice does not contain HTT natural polyadenylation sites. Other mechanisms involved in RNA processing, which are coupled to transcription elongation, might explain our results. Additionally, the dynamics of transcription of the HTT gene might be different between the two tissues. We are discussing below these possibilities.
Does RNAP II dynamics at proximal-promoters underlie tissue-selective CAG instability?
In higher eukaryotes, the transition from transcription initiation to productive elongation is a tightly regulated process, which together with transcription initiation regulation, controls transcription rate and dynamics of many genes.19,20 Promoter proximal pausing, e.g., pausing of RNAP II typically 30 to 50 nucleotides downstream of TSS, is a key feature of genes subject to control of productive elongation. RNAP II dynamics regulation at proximal promoters is complex and depends on several factors. In metazoans, RNAP II pausing is mediated by DRB sensitivity-inducing factor (DSIF), a heterodimer of SPT4 and SPT5, which promotes the recruitment of negative elongating factor (NELF). Pausing release requires positive transcription elongation factor-b (P-TEFb), a factor composed of CDK9 and cyclin T1. CDK9 phosphorylates RNAP II on Ser2 as well as SPT5, turning RNAP II into its elongating form and DSIF from a negative to a positive elongating factor. Recent studies showed that CDK7, which is part of TFIIH, is also implicated in RNAP II dynamics at promoter pausing sites.21 CDK7 activity is required for both pausing establishment, mediating recruitment of DSIF, and pausing release, activating CDK9. It was suggested that CDK7 antagonistic functions control RNAP II levels and pause duration at proximal-promoter regions. Our ChIP data showed that RNAP II at the HD transgene peaks downstream of the TSS, suggesting that HTT transcription is regulated through promoter-proximal pausing.9 RNAP II level was increased in striatum relative to cerebellum, suggesting that RNAP II dynamics at the HTT proximal-promoter region is different between the two tissues. The level and activity of factors involved in pausing establishment and release might be different between striatum and cerebellum. If true, one would expect that the transcription dynamics of genes regulated through promoter-proximal pausing is globally different between the two tissues.
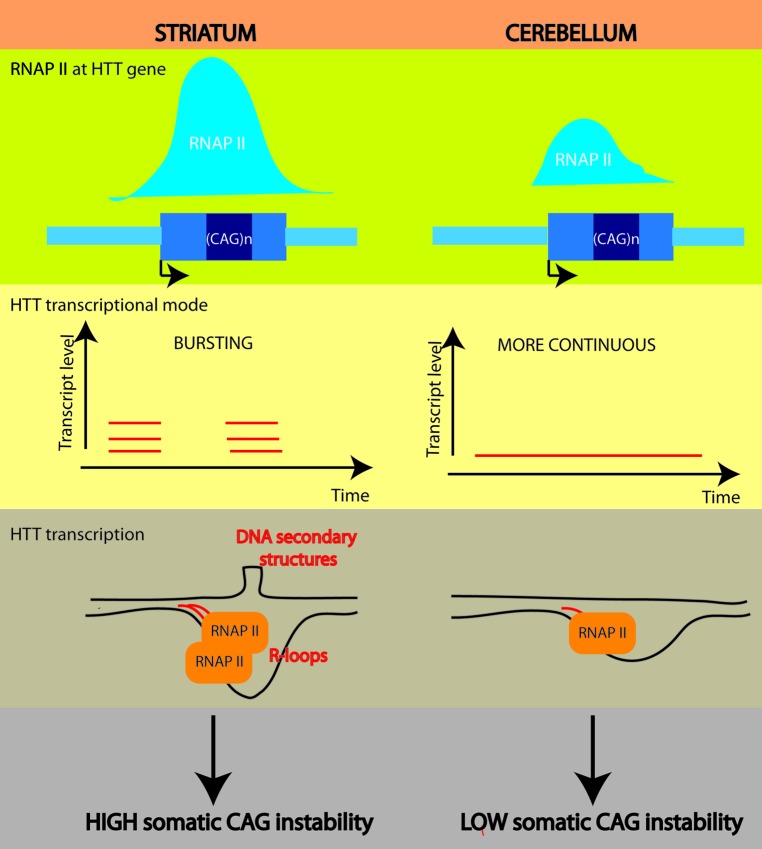
Then, how would RNAP II dynamics at the HD transgene modulate CAG instability? Comparable levels of mRNA transgene were measured in the striatum and cerebellum of R6 mice, despite different RNAP II levels at the HTT promoter-proximal pausing site.9 In a wide range of species, including mammals, gene transcription can occur through episodic bursting as opposed to constitutive expression, and gene expression is dependent upon burst size and frequency.22-25 A recent genome-wide study using time-lapse fluorescence microscopy and human cells showed that transcriptional bursting dominates throughout the genome.26 Although the mechanisms underlying transcriptional bursting are not yet clear, it has been proposed that RNAP II dynamics at promoter-proximal pausing sites controls burst size and frequency. Is HTT transcribed through a bursting mode and are bursting size and frequency different between striatum and cerebellum? The differential RNAP II level at HTT promoter proximal-region could reflect different bursting parameters between striatum and cerebellum. Using cell models, it was shown that transcription across CAG/CTG repeats induces R-loop formation, which promotes CAG/CTG instability.7 Thus, it is tempting to speculate that transcriptional bursting parameters might modulate the propensity for formation of R-loops in vivo, favoring more R-loop formation at the HD locus in the striatum relative to cerebellum (Fig. 1). Yet, whether R-loops form in vivo at the HD locus is unknown.
Figure 1. Model of transcription-dependent CAG instability in striatum and cerebellum. RNAP II level at the HTT gene promoter proximal region is increased in striatum when compared with cerebellum. This might be associated with a bursting transcriptional mode in the striatum and a more continuous transcriptional mode in the cerebellum. As a result, HTT transcription might lead to formation of stable DNA:RNA hybrids (R-loops) in the striatum, creating a window of opportunity for formation of DNA secondary structures, a prerequisite to CAG instability. The probability that this mechanism occurs in the cerebellum would be reduced.
Promoter-proximal pausing factors have been coupled to RNA processing. Specifically, interplay was evidenced between RNAP II, DSIF and capping enzyme.27,28 DSIF interacts with capping enzyme, facilitating capping. Moreover, capping enzyme promotes formation of R-loops.29 Since R-loops can negatively regulate the stability and processivity of elongating RNAP II, it has been suggested that capping enzyme-mediated R-loop formation modulates RNAP II dynamics at promoter-proximal pause sites, providing a kinetic “window of opportunity”19,29. The CAG repeat within the HTT gene is close to TSS (< 200 nucleotides). It is therefore possible that the proximity between promoter-proximal region and CAG repeats provides a particular environment favoring stable R-loop formation at the HD locus. Whether capping enzyme is involved in tissue-selective CAG instability in HD is an intriguing possibility.
Events associated with transcription elongation correlate with increased CAG instability in HD mouse tissues. We propose that tissue-dependent regulation of promoter proximal pausing at the HTT gene might underlie tissue-selective CAG instability in HD. Additional studies are required to specify the mechanisms involved in transcription-mediated CAG/CTG instability in HD. In HD, the striatum is both the tissue that preferentially degenerates and presents highest CAG instability. Interestingly, depleting SPT4 in yeast and mammalian cells specifically decreased transcription of CAG-expanded genes, thereby reducing toxicity of the gene products.30 Our results suggest that decreasing transcription elongation might also reduce toxicity, limiting repeat instability. Thus, targeting transcription elongation might represent an attractive therapeutic perspective.
Acknowledgments
We thank H. Puccio for support. This work was supported by funds from CNRS, INSERM, Strasbourg University and ANR (ANR-2011-JSV6–003–01 to KM)
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/transcription/article/25971
References
- 1.López Castel A, Cleary JD, Pearson CE. Repeat instability as the basis for human diseases and as a potential target for therapy. Nat Rev Mol Cell Biol. 2010;11:165–70. doi: 10.1038/nrm2854. [DOI] [PubMed] [Google Scholar]
- 2.McMurray CT. Mechanisms of trinucleotide repeat instability during human development. Nat Rev Genet. 2010;11:786–99. doi: 10.1038/nrg2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin Y, Hubert L, Jr., Wilson JH. Transcription destabilizes triplet repeats. Mol Carcinog. 2009;48:350–61. doi: 10.1002/mc.20488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin Y, Dion V, Wilson JH. Transcription promotes contraction of CAG repeat tracts in human cells. Nat Struct Mol Biol. 2006;13:179–80. doi: 10.1038/nsmb1042. [DOI] [PubMed] [Google Scholar]
- 5.Nakamori M, Pearson CE, Thornton CA. Bidirectional transcription stimulates expansion and contraction of expanded (CTG)*(CAG) repeats. Hum Mol Genet. 2011;20:580–8. doi: 10.1093/hmg/ddq501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin Y, Leng M, Wan M, Wilson JH. Convergent transcription through a long CAG tract destabilizes repeats and induces apoptosis. Mol Cell Biol. 2010;30:4435–51. doi: 10.1128/MCB.00332-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin Y, Dent SY, Wilson JH, Wells RD, Napierala M. R loops stimulate genetic instability of CTG.CAG repeats. Proc Natl Acad Sci U S A. 2010;107:692–7. doi: 10.1073/pnas.0909740107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reddy K, et al. Determinants of R-loop formation at convergent bidirectionally transcribed trinucleotide repeats. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goula AV, Stys A, Chan JP, Trottier Y, Festenstein R, Merienne K. Transcription elongation and tissue-specific somatic CAG instability. PLoS Genet. 2012;8:e1003051. doi: 10.1371/journal.pgen.1003051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dure LS, 4th, Landwehrmeyer GB, Golden J, McNeil SM, Ge P, Aizawa H, Huang Q, Ambrose CM, Duyao MP, Bird ED, et al. IT15 gene expression in fetal human brain. Brain Res. 1994;659:33–41. doi: 10.1016/0006-8993(94)90860-5. [DOI] [PubMed] [Google Scholar]
- 11.Lia AS, Seznec H, Hofmann-Radvanyi H, Radvanyi F, Duros C, Saquet C, Blanche M, Junien C, Gourdon G. Somatic instability of the CTG repeat in mice transgenic for the myotonic dystrophy region is age dependent but not correlated to the relative intertissue transcription levels and proliferative capacities. Hum Mol Genet. 1998;7:1285–91. doi: 10.1093/hmg/7.8.1285. [DOI] [PubMed] [Google Scholar]
- 12.Kumari D, Usdin K. Chromatin remodeling in the noncoding repeat expansion diseases. J Biol Chem. 2009;284:7413–7. doi: 10.1074/jbc.R800026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, Hetherington C, Lawton M, Trottier Y, Lehrach H, Davies SW, et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/S0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 14.Mangiarini L, Sathasivam K, Mahal A, Mott R, Seller M, Bates GP. Instability of highly expanded CAG repeats in mice transgenic for the Huntington’s disease mutation. Nat Genet. 1997;15:197–200. doi: 10.1038/ng0297-197. [DOI] [PubMed] [Google Scholar]
- 15.Goula AV, Berquist BR, Wilson DM, 3rd, Wheeler VC, Trottier Y, Merienne K. Stoichiometry of base excision repair proteins correlates with increased somatic CAG instability in striatum over cerebellum in Huntington’s disease transgenic mice. PLoS Genet. 2009;5:e1000749. doi: 10.1371/journal.pgen.1000749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin B, Rommens JM, Graham RK, Kalchman M, MacDonald H, Nasir J, Delaney A, Goldberg YP, Hayden MR. Differential 3′ polyadenylation of the Huntington disease gene results in two mRNA species with variable tissue expression. Hum Mol Genet. 1993;2:1541–5. doi: 10.1093/hmg/2.10.1541. [DOI] [PubMed] [Google Scholar]
- 17.Casanova E, Alonso-Llamazares A, Zamanillo D, Garate C, Calvo P, Chinchetru MA. Identification of a long huntingtin mRNA transcript in mouse brain. Brain Res. 1996;743:320–3. doi: 10.1016/S0006-8993(96)00701-9. [DOI] [PubMed] [Google Scholar]
- 18.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–6. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet. 2012;13:720–31. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol. 2006;7:557–67. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- 21.Larochelle S, Amat R, Glover-Cutter K, Sansó M, Zhang C, Allen JJ, Shokat KM, Bentley DL, Fisher RP. Cyclin-dependent kinase control of the initiation-to-elongation switch of RNA polymerase II. Nat Struct Mol Biol. 2012;19:1108–15. doi: 10.1038/nsmb.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zenklusen D, Larson DR, Singer RH. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat Struct Mol Biol. 2008;15:1263–71. doi: 10.1038/nsmb.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hebenstreit D. Are gene loops the cause of transcriptional noise? Trends Genet. 2013;29:333–8. doi: 10.1016/j.tig.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 24.Pedraza JM, Paulsson J. Effects of molecular memory and bursting on fluctuations in gene expression. Science. 2008;319:339–43. doi: 10.1126/science.1144331. [DOI] [PubMed] [Google Scholar]
- 25.Kaern M, Elston TC, Blake WJ, Collins JJ. Stochasticity in gene expression: from theories to phenotypes. Nat Rev Genet. 2005;6:451–64. doi: 10.1038/nrg1615. [DOI] [PubMed] [Google Scholar]
- 26.Dar RD, Razooky BS, Singh A, Trimeloni TV, McCollum JM, Cox CD, Simpson ML, Weinberger LS. Transcriptional burst frequency and burst size are equally modulated across the human genome. Proc Natl Acad Sci U S A. 2012;109:17454–9. doi: 10.1073/pnas.1213530109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wen Y, Shatkin AJ. Transcription elongation factor hSPT5 stimulates mRNA capping. Genes Dev. 1999;13:1774–9. doi: 10.1101/gad.13.14.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mandal SS, Chu C, Wada T, Handa H, Shatkin AJ, Reinberg D. Functional interactions of RNA-capping enzyme with factors that positively and negatively regulate promoter escape by RNA polymerase II. Proc Natl Acad Sci U S A. 2004;101:7572–7. doi: 10.1073/pnas.0401493101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaneko S, Chu C, Shatkin AJ, Manley JL. Human capping enzyme promotes formation of transcriptional R loops in vitro. Proc Natl Acad Sci U S A. 2007;104:17620–5. doi: 10.1073/pnas.0708866104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu CR, Chang CR, Chern Y, Wang TH, Hsieh WC, Shen WC, Chang CY, Chu IC, Deng N, Cohen SN, et al. Spt4 is selectively required for transcription of extended trinucleotide repeats. Cell. 2012;148:690–701. doi: 10.1016/j.cell.2011.12.032. [DOI] [PubMed] [Google Scholar]