Abstract
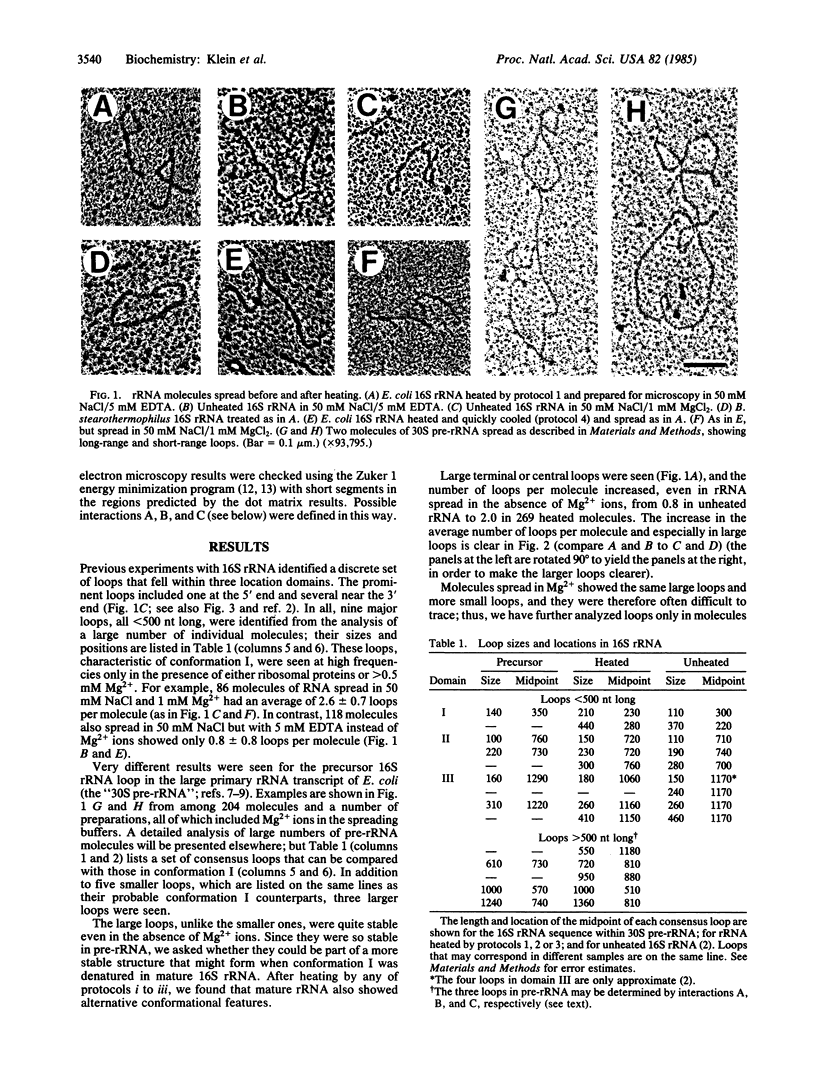
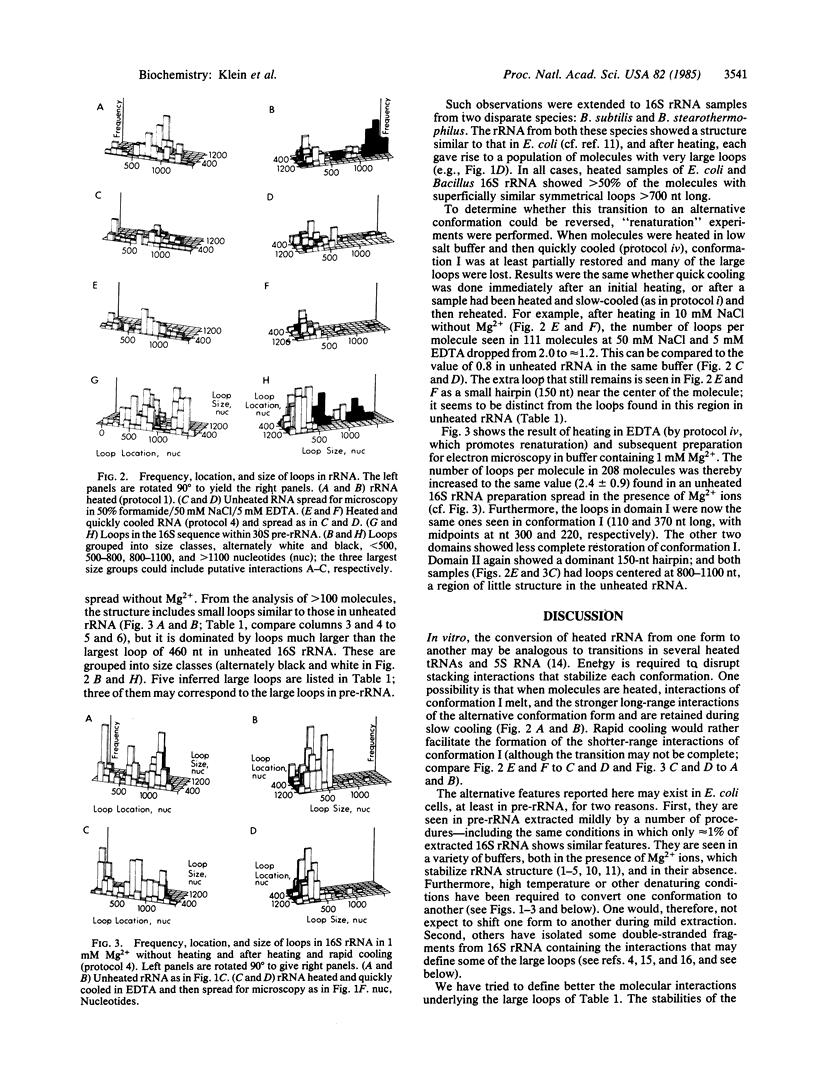
Partially denatured 16S rRNA from 30S ribosomes shows features of secondary structure in electron microscopy that correspond to the well accepted secondary structure model derived from chemical modification and phylogenetic data. However, a very different conformation is seen in precursor 16S rRNA sequences contained within 30S pre-rRNA transcripts: the major 5'-terminal loop is absent, and several additional quite stable large loops, symmetrically placed in the molecule, are present. Features of the alternative structure are also seen in mature 16S rRNA from Escherichia coli and from two Bacillus species when heated in certain buffers. Microscopy thus reveals specific features of alternative conformations and their relative stabilities, suggesting a possible transition during ribosome formation.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brimacombe R., Maly P., Zwieb C. The structure of ribosomal RNA and its organization relative to ribosomal protein. Prog Nucleic Acid Res Mol Biol. 1983;28:1–48. doi: 10.1016/s0079-6603(08)60081-1. [DOI] [PubMed] [Google Scholar]
- Carbon P., Ebel J. P., Ehresmann C. The sequence of the ribosomal 16S RNA from Proteus vulgaris. Sequence comparison with E. coli 16S RNA and its use in secondary model building. Nucleic Acids Res. 1981 May 25;9(10):2325–2333. doi: 10.1093/nar/9.10.2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech T. R., Tanner N. K., Tinoco I., Jr, Weir B. R., Zuker M., Perlman P. S. Secondary structure of the Tetrahymena ribosomal RNA intervening sequence: structural homology with fungal mitochondrial intervening sequences. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3903–3907. doi: 10.1073/pnas.80.13.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. T7 early RNAs and Escherichia coli ribosomal RNAs are cut from large precursor RNAs in vivo by ribonuclease 3. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3296–3300. doi: 10.1073/pnas.70.12.3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotz C., Brimacombe R. An experimentally-derived model for the secondary structure of the 16S ribosomal RNA from Escherichia coli. Nucleic Acids Res. 1980 Jun 11;8(11):2377–2395. doi: 10.1093/nar/8.11.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman W. E., Goldberg G., Bowman L. H., Steinmetz D., Schlessinger D. Mouse rDNA: sequences and evolutionary analysis of spacer and mature RNA regions. Mol Cell Biol. 1983 Aug;3(8):1488–1500. doi: 10.1128/mcb.3.8.1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson A. B., Good L., Simonetti J., Zuker M. Some simple computational methods to improve the folding of large RNAs. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):45–52. doi: 10.1093/nar/12.1part1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein B. K., Forman P., Shiomi Y., Schlessinger D. Electron microscopy of secondary structure in partially denatured Escherichia coli 16S rRNA and 30S subunits. Biochemistry. 1984 Aug 14;23(17):3927–3933. doi: 10.1021/bi00312a021. [DOI] [PubMed] [Google Scholar]
- Klein B. K., King T. C., Schlessinger D. Structure of partially denatured Escherichia coli 23 S ribosomal RNA determined by electron microscopy. J Mol Biol. 1983 Aug 25;168(4):809–830. doi: 10.1016/s0022-2836(83)80076-x. [DOI] [PubMed] [Google Scholar]
- Mangiarotti G., Turco E., Perlo C., Altruda F. Role of precursor 16S RNA in assembly of E. coli 30S ribosomes. Nature. 1975 Feb 13;253(5492):569–571. doi: 10.1038/253569a0. [DOI] [PubMed] [Google Scholar]
- Nikolaev N., Schlessinger D. Binding of ribosomal proteins to 30S preribosomal ribonucleic acid of Escherichia coli. Biochemistry. 1974 Oct 8;13(21):4272–4278. doi: 10.1021/bi00718a005. [DOI] [PubMed] [Google Scholar]
- Spirin A. S., Serdyuk I. N., Shpungin J. L., Vasiliev V. D. Quaternary structure of the ribosomal 30S subunit: model and its experimental testing. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4867–4871. doi: 10.1073/pnas.76.10.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiegler P., Carbon P., Zuker M., Ebel J. P., Ehresmann C. Structural organization of the 16S ribosomal RNA from E. coli. Topography and secondary structure. Nucleic Acids Res. 1981 May 11;9(9):2153–2172. doi: 10.1093/nar/9.9.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Gutell R., Gupta R., Noller H. F. Detailed analysis of the higher-order structure of 16S-like ribosomal ribonucleic acids. Microbiol Rev. 1983 Dec;47(4):621–669. doi: 10.1128/mr.47.4.621-669.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. A., Steitz J. A. Complementary sequences 1700 nucleotides apart form a ribonuclease III cleavage site in Escherichia coli ribosomal precursor RNA. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3593–3597. doi: 10.1073/pnas.75.8.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwieb C., Glotz C., Brimacombe R. Secondary structure comparisons between small subunit ribosomal RNA molecules from six different species. Nucleic Acids Res. 1981 Aug 11;9(15):3621–3640. doi: 10.1093/nar/9.15.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]