Abstract
There are an estimated 285 million people with visual impairment worldwide, of whom 39 million are blind. The pathogenesis of many eye diseases remains poorly understood. The human eye is currently an emerging proteome that may provide key insight into the biological pathways of disease. We review proteomic investigations of the human eye and present a catalogue of 4842 non-redundant proteins identified in human eye tissues and biofluids to date. We highlight the need to identify new biomarkers for eye diseases using proteomics. Recent advances in proteomics now allow the identification of hundreds to thousands of proteins in tissues and fluids, characterization of various post-translational modifications, and simultaneous quantification of multiple proteins. To facilitate proteomic studies of the eye, the Human Eye Proteome Project (HEPP) was organized in September 2012. The HEPP is one of the most recent components of the Biology/Disease-driven Human Proteome Project (B/D-HPP) whose overarching goal is to support the broad application of state-of-the-art measurements of proteins and proteomes by life scientists studying the molecular mechanisms of biological processes and human disease. The large repertoire of investigative proteomic tools has great potential to transform vision science and enhance understanding of physiology and disease processes that affect sight.
Keywords: Biomarker, Cornea, Eye, Proteomics, Retina, Vision
1 Introduction
There are an estimated 285 million people with visual impairment worldwide, of whom 39 million are blind [1]. The main causes of blindness include age-related macular degeneration, glaucoma, corneal opacities, diabetic retinopathy, trachoma, and vitamin A deficiency. The global health cost of visual impairment and blindness was an estimated $3 trillion in 2010 [2]. The underlying pathophysiology of many blinding eye diseases remains poorly understood. The identification of individuals who are at high risk for visual impairment and blindness from eye diseases is problematic. Despite existing treatments such as anti-angiogenic drugs, topical medications, surgery, and laser photocoagulation, millions of patients with age-related macular degeneration, glaucoma, and diabetic retinopathy still progress to visual impairment and blindness.
The human eye is a complex organ that comprises the tear film, conjunctiva, cornea, aqueous humor, iris, ciliary body, lens, vitreous humor, retina, choroid, sclera, and optic nerve. The adjacent tissues, or ocular adnexae, consist of the lacrimal apparatus, eyelids, extraocular muscles, and orbit. There are certain structural qualities of the eye that make it unique in the human body, such as the transparency of the cornea, the focal abilities of the crystalline lens, and the light receptive qualities of the retina. Small disturbances in these delicate tissues can result in impaired vision and blindness.
Proteomics can provide new insights into the pathogenesis of eye disease by characterization of the diversity and quantity of proteins – including their isoforms and posttranslational modifications – in both health and natural aging versus those that occur with disease or upon treatment. There is also the possibility that the application of proteomics may lead to the discovery of biomarkers that identify individuals at risk for disease, monitor responses to treatment, and reveal biological pathways of disease. The goals of this review are: (1) to summarize the progress that has been made in proteomic investigations of the human eye, (2) to highlight the need for identification of new biomarkers for specific eye diseases, and (3) to show how the tremendous advances in proteomics can be used to gain insight into the pathogenesis of eye diseases. Finally, we present the new initiative, the Human Eye Proteome Project, that was organized at the recent Human Proteome Organization (HUPO) 11th World Congress in Boston, September 9-13, 2012 and recognized as an official initiative of HUPO by the Biology/Disease Human Proteome Project (B/D-HPP) [3] in December 2012. The overarching goal of the B/D-HPP is to support the broad application of state-of-the-art measurements of proteins and proteomes by life scientists studying the molecular mechanisms of biological processes and human disease [3].
2 Proteomes of human eye tissues and fluids
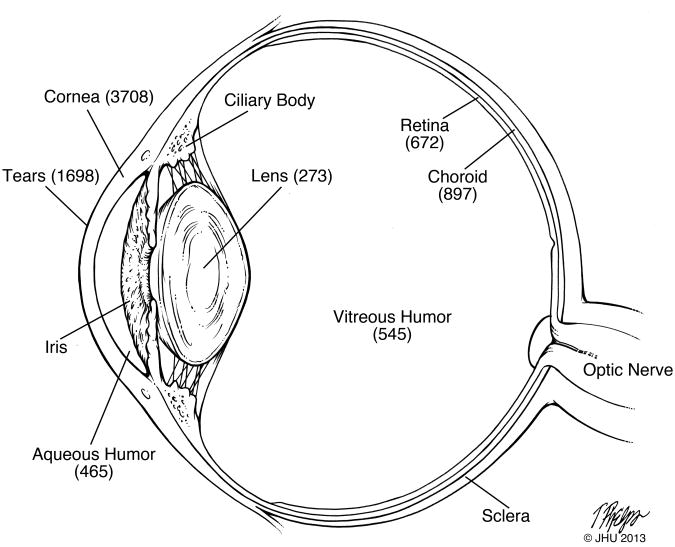
The proteomes of the different tissues of the human eye and its associated fluids have only been partially characterized. Most studies have been conducted with the more accessible compartments of the eye, i.e., tears, aqueous humor, and vitreous humor. In part this is due to the limited availability of samples and limited volume and quantity of different ocular tissues and biofluids. The number of non-redundant proteins identified in various tissues and fluids of the eye are shown in Figure 1.
Figure 1.
Schematic diagram of the human eye with the number of non-redundant proteins identified in various tissues and biofluids.
2.1 Tears
The tear film consists of an outer lipid layer, a middle aqueous layer, and an inner mucous layer that overlies the corneal and conjunctival epithelium. The human tear proteome has been investigated in normal subjects [4-8]. A recent study identified 1466 non-redundant proteins in tears, the largest number of tear proteins to date [7]. One obstacle to the proteomic analysis of tears is that highly abundant proteins such a lactoferrin, lysozyme, secretory IgA, and albumin mask the proteins that are present in lower abundance [6]. The method of tear collection can also influence the results, as the protein profile of tears can be quite different between closed eyes and open eyes [7] and between collection using Schirmer strips versus capillary tubes [6]. Post-translational modifications such as phosphorylation and glycosylation have been reported in tear proteins [4]. In a study of six healthy volunteers followed over seven days, no significant variability in tear protein profiles from day-to-day or between eyes of the same individual was found [8].
Dry eye syndrome, as defined at a recent expert group meeting, is “a multifactorial disease of the tears and ocular surface that results in symptoms of discomfort, visual disturbance, and tear instability with potential damage to the ocular surface. It is accompanied by increased osmolarity of the tear and inflammation of the ocular surface” [9]. Dry eye syndrome is common, with a reported age-adjusted prevalence of dry eye syndrome among men and women of about 4% and 8%, respectively, in the U.S. [10,11]. The tear proteome has been compared in patients with and without dry eye syndrome using a variety of techniques for tear collection, sample processing, and mass spectrometry instrumentation [12-18]. Three studies used isobaric tag for relative and absolute quantitation (iTRAQ) to compare the tear proteome in patients and controls [14,16,18]. In a discovery phase study using iTRAQ and LC-nano ESI-MS/MS (Q-Star XL, Applied Biosystems), Zhou and colleagues identified 91 non-redundant proteins in tears, of which α-enolase, α-1-acid glycoprotein 1, S100 A8 (calgranulin A), S100 A9 (calgranulin B), S100 A4, and S100 A5 (calgizzarin) were upregulated, and prolactin-inducible protein (PIP), lipocalin-1, lactoferrin, and lysozyme were downregulated in dry eye syndrome compared with controls [14]. Using iTRAQ and an Orbitrap mass spectrometer (LTQ-Orbitrap-XL, Thermo Scientific), Srinivasan and colleagues identified 386 proteins in tears, of which ezrin, apolipoprotein, Ig gamma-3 chain C region, vitamin D-binding protein, and peroxiridoxin were upregulated, and lysozyme, lipocalin-1, lactotransferrin, and IgA-alpha were downregulated in dry eye syndrome compared with controls [18]. While these two iTRAQ studies conducted by independent groups had comparable findings in the downregulated proteins, there was no overlap between the studies in upregulated proteins associated with dry eye syndrome. Many of the downregulated proteins, such as lysozyme, lactotransferrin, and lipocalin, are related to antibacterial protection of the ocular surface [18].
The human tear proteome has also been studied in a variety of other conditions, such as keratoconus [19-21], autoimmune thyroid eye disease [22], blepharitis [23], conjunctivochalasis [24], diabetic retinopathy [25], primary open angle glaucoma and pseudoexfoliative glaucoma [26], in contact lens wearers [27], and in patients on chronic use of topical glaucoma medications [28] and after ocular surface surgery [29].
2.2 Cornea
The number of non-redundant proteins described in the normal human cornea has rapidly increased from 138 proteins in 2005 [30] to 2070 proteins in 2011 [31] and 3248 proteins in 2012 [32], largely due to advances in mass spectrometry instrumentation. The main causes of corneal blindness worldwide are vitamin A deficiency (xerophthalmia), onchocerciasis, trachoma, and leprosy [33]; these conditions occur primarily in developing countries and have not been the subject of proteomic investigations.
Corneal diseases often manifest within one of the layers of the cornea. To account for these differences, Dyrlund and colleagues [32] separated the cornea into sub-proteomes consisting of epithelium and Bowman's layer, the stroma, and Descemet's membrane and endothelium. Some of the major corneal diseases that result in visual disability and blindness in developed countries include Fuchs endothelial corneal dystrophy and keratoconus. Fuchs' dystrophy is the third leading indication for corneal transplantation in the United States [34]. Fuchs' dystrophy is characterized by the progressive loss of corneal endothelial cells and thickening of Descemet's membrane. Loss of corneal endothelial cells can lead to corneal thickening, loss of transparency of the cornea, and impaired vision. The progression of Fuchs' dystrophy is associated with changes in the size, shape, and density of endothelial cells, increased deposition of collagen VIII in Descemet's membrane, and loss of Na+-K+-ATPase pump sites [35]. Recent proteomic investigations of human corneal endothelium by Jurkunas and colleagues have implicated transforming growth factor-beta-induced protein (TGFβIp), clusterin, and peroxiredoxins in Fuchs' dystrophy [36,37]. The pathogenesis of Fuchs' dystrophy appears to be related to oxidative stress and its effects on the endothelium [35].
Keratoconus is a condition of unknown cause in which the cornea assumes a conical shape as a result of non-inflammatory thinning of the corneal stroma. Keratoconus is one of the most common indications for corneal transplantation [38]. The incidence of keratoconus is about 2 per 100,000/year [38]. The proteome of the cornea has been studied in keratoconus using 2-dimensional gel electrophoresis and mass spectrometry [39-41]. One study suggested that additional biochemical methods such as immunohistochemistry may be helpful in distinguishing contaminating versus endogenous cytokeratins in the cornea proteome [42]. In addition to the more common conditions, rare corneal diseases that are under investigation include lattice and granular corneal dystrophies, linked with accumulations of transforming growth factor beta induced protein (TGFBIp). More than fifty different mutations in TGFBIp have been described [43,44], of which many are characterized by TGFBIp deposits in the cornea causing severe visual impairment. A recent study by Karring and colleagues describes the sub-proteomes of protein deposits isolated from patients suffering from either lattice or granular corneal dystrophy [45].
2.3 Aqueous humor
In studies of human aqueous, controls or presumably healthy subjects have been defined as adults undergoing elective cataract surgery [46-49] due to the ethical issues involved in collecting aqueous humor from the eyes of patients with no ocular disease. An aqueous sample of 100-150 μL can be obtained from the anterior chamber of the eye during surgery. Bennett and colleagues collected aqueous humor from ten patients during cataract surgery and reported a mean total protein concentration of 0.36 mg/mL (range 0.21-0.57 mg/mL) [49]. A total of 198 unique proteins were identified the aqueous of cataract patients, of which the predominant molecular functions were associated with binding (protein, metal ion, heparin, and DNA) and inhibition of proteolytic activity [49]. Another recent study that used an LTQ Orbitrap reported 676 unique proteins in aqueous humor, of which most proteins had catalytic, enzymatic, and structural properties. Most of the quantified proteins were in the concentration range of 0.1 to 2.5 ng/mL [48].
Major eye diseases that may be related, in part, to alterations in the aqueous humor include primary open angle glaucoma, uveitis, corneal graft rejection, and possibly Fuchs' dystrophy and keratoconus. Primary open angle glaucoma is the leading cause of irreversible blindness worldwide [50]. Elevated intraocular pressure is a risk factor for primary open angle glaucoma, as chronic elevated pressures may lead to loss of nerve fibers in the optic nerve, visual field loss, and blindness. The proteome of the human aqueous humor has been studied in patients with primary open angle glaucoma [51-56]. Saccá and colleagues compared the aqueous humor proteome in fourteen patients with glaucoma and eleven patients undergoing cataract surgery using antibody microarray. Thirteen proteins were significantly increased in glaucoma patients that were involved in inflammation (ELAM 1), cholesterol delivery (apolipoprotein B and E), muscle cell differentiation and function (myotrophin, myoblast determination protein 1, myogenin, vasodilator-stimulated phosphoprotein, ankyrin-2), stress response and removal of damaged proteins (heat shock 50 and 90 kDa proteins, ubiquitin fusion degradation 1-like), and signal transduction and neural development (phospholipase C β and γ) [56].
The aqueous proteome has been investigated in patients with acute corneal graft rejection [55], myopia [58], Fuchs' dystrophy [59], and primary congenital glaucoma [60]. Kim and colleagues compared aqueous humor in three patients with age-related macular degeneration and three controls with cataracts. Using a linear trap quadrupole mass spectrometer (LTQ-XL, Thermo Fisher), they identified 158 non-redundant proteins in aqueous. Seven aqueous biomarkers of sufficient abundance were compared in cases and controls using multiple reaction monitoring (MRM). Of these, four were significantly increased in the patients with age-related macular degeneration: pigment epithelium-derived factor, plasma protease C1 inhibitor, TGFBI, and ceruloplasmin [61].
2.4 Iris and ciliary body
The proteome of the human iris and ciliary body has not been well characterized.
2.5 Lens
The lens is a transparent and avascular tissue composed of epithelial and fiber cells. The lens fibers have the highest protein content in the body: >35% of their wet weight [62]. About 90% of the lens proteins are structural proteins known as crystallins in three distinct families: α-crystallin, β-crystallin, and γ-crystallin. As noted in a recent review by Kyselova, crystallins are extremely long-lived with virtually no protein turnover, thus providing an ideal opportunity for the study of post-translational modifications [62]. Cataract, the opacification of the lens, is a reversible cause of impaired vision and blindness – over 1.2 million cataract operations are performed in the U.S. alone at a cost of over $3.4 billion [63]. Risk factors for cataract include age, diabetes, smoking, and chronic ultraviolet light exposure [64], factors that could all potentially contribute to pathologic alterations of lens crystallins.
The proteins of the human lens have been studied using a variety of mass spectrometry approaches [65-75]. The most abundant post-translational modifications of crystallins with advancing age and cataracts are deamidation [68,72,74] and phosphorylation [65,75]. Huang and colleagues characterized the phosphoproteome of the human lens and identified 32 phosphoproteins and 73 phosphorylation sites [75]. Recent studies have also reported S-methylation [68], and glycation [68] of lens crystallins. Kyselova reviewed some of the older literature that described post-translational modifications in lens proteins [62].
2.6 Vitreous humor
In studies of the human vitreous, most studies have defined controls or presumably healthy eyes as patients undergoing vitrectomy for idiopathic macular hole due to the ethical issues involved in collecting vitreous from healthy human eyes. Several studies have compared the proteome of vitreous humor in patients with varying severity of diabetic retinopathy in comparison with patients with macular holes [76-85]. One group described 460 non-redundant proteins in human vitreous [81].
2.7 Retina
The retina consists of the neuroretina and retinal pigment epithelium. The two most common blinding retinal diseases are age-related macular degeneration and diabetic retinopathy. Age-related macular degeneration is the leading cause of visual loss among adults aged 65 or older in developed countries [86]. With the growing population of older adults, the prevalence of advanced age-related macular degeneration will increase by 50% to nearly 3 million in 2010 [87]. The global cost of visual impairment due to age-related macular degeneration alone is currently an estimated $343 billion, including $255 billion in direct health care costs [88]. Despite advances in treatment and prevention, age-related macular degeneration has no effective cure and remains the leading cause of irreversible blindness in older adults.
Crabb and colleagues described oxidative protein modifications such as cross-linked vitronectin and metalloproteinase inhibitor-3, carboxyethyl pyrrole protein adducts, and carboxymethyllysine, in drusen from eyes with age-related macular degeneration [89] and lipofuscin from retinal pigment epithelium in eye bank specimens [90]. Ferrington and colleagues used 2-dimensional gels and matrix-assisted laser desorption ionization (MALDI-TOF) mass spectrometry to study the proteome of the retinal pigment epithelium [91], central and peripheral retina [92], and mitochondria of the retinal pigment epithelium [93] in age-related macular degeneration. In a study of retinal pigment epithelial blebs, basigin and matrix metalloproteinase-14 were identified in blebs and were more common in age-related macular degeneration [94].
Two-dimensional gels and mass spectrometry were used to investigate the proteome of the retinal pigment epithelium in early diabetic retinopathy [95] and vitreous in diabetic macular edema [96]. Proteomic studies have also been conducted in idiopathic macular telangiectasia [97] (a rare condition), and with retinal pigment epithelial cells isolated from an autopsy eye [98].
2.8 Choroid
The proteome of the isolated choroid has not been well characterized. Crabb and colleagues described the proteome of the macular Bruch membrane/choroid complex in age-related macular degeneration [99]. In the region of the macula, Bruch membrane and choroid were dissected free from the overlying retina pigment epithelium and used for proteomic study. A total of 897 non-redundant proteins were identified, of which 56 proteins were elevated and 43 proteins were reduced in eyes with age-related macular degeneration compared with controls. A majority of the elevated proteins were involved in immune response and host defense, including complement proteins, α-defensins 1-3, protein S100s, crystallins, histones, and galectin-3 [99].
2.9 Optic nerve
In a proteomic analysis of the optic nerve head in normal eyes and eyes with primary open angle glaucoma, elevated peptidyl arginine deiminase 2, an enzyme that converts arginine to citrulline, was found in the optic nerve head of glaucoma eyes but not in normal eyes [100]. Further study showed that myelin basic protein was a major citrullinated protein in primary open angle glaucoma [100]. Two studies described the proteome of lamina cribrosa cells cultured from the optic nerve [101,102].
2.10 Sclera
The proteome of human sclera has not been well characterized. The sclera may be of interest for proteomic investigations of the trabecular meshwork and Schlemm's canal, which drains aqueous humor from the eye and influences intraocular pressure. Bhattacharya and colleagues have developed an enrichment strategy to study intracellular proteases in the trabecular meshwork proteome [103].
3 Compilation of the current human eye proteome
The supplementary data from the various papers described above were used to retrieve the corresponding protein accession numbers). The papers included in the compilation of the human eye proteome were those in which the authors applied proteomics and mass spectrometry methods to characterize the proteome of specific eye tissues and biofluids. The compilation did not include studies in which low abundance proteins, such as vascular endothelial growth factor, were described using immunological methods rather than mass spectrometry and proteomics. IPI accessions were converted to UniProt accessions either by lookup in the IPI [105] database or by blast searches (>90% sequence homology). All obsolete proteins, duplicate identifications, and contaminations (e.g. trypsin and non-human proteins) were then removed. For all protein lists with one or more non Swiss-Prot proteins (TrEMBL, NCBI, etc.), UniProt [106] and IPI [105] were used to create a FASTA file of sequences that were run through the CD-HIT Web Server clustering tool [106]. The tool was used to cluster highly similar proteins with sequence homology above 90% over the entire protein in order to ensure the removal of duplicate entries and isoform redundancy in protein lists from individual papers. Protein lists were imported into MS Data Miner (v.1.1.3) [107], and this platform was used to generate subproteome lists for tears, cornea, aqueous humor, lens, vitreous humor, retina, and choroid (Supplementary Table 1). In total, 4842 non-redundant proteins have been identified, 4080 of which are Swiss-Prot proteins. Some caveats deserve mention in regard to the compiled lists of proteins: (1) some reported isoforms have no accompanying peptide data, thus, it is not possible to determine whether a peptide amino acid sequence that is unique to the specific isoform was actually observed, (2) the unambiguous identification of proteins in older instrumentation is less certain, (3) older peptide databases contain some redundancy, (4) criteria used for protein identification varied widely among different studies (Supplementary Table 2), and (5) source of material included both surgical and postmortem samples, which could influence findings. Thus, the true list of proteins from each subproteome is likely to be smaller than the compiled lists. In the future, the redundancy of protein identifications and quality of reporting of peptide data from the eye could be improved with standards established by the Human Eye Proteome Project (section 6 below). Informed by the history of the Plasma Peptide Atlas, it is certain that a high overall false discovery rate would emerge from these data if a standardized analysis, like the TransProteomicPipeline (TPP), were applied to the raw spectra. As shown in Supplementary Table 2, many studies identified proteins based upon one or more peptides, and false discovery rates greater than 1% were commonly used. If highly stringent criteria such as those used to create the Plasma Peptide Atlas were applied, the number of identifications would certainly be expected to shrink from the provisional list of 4842 proteins shown in Supplementary Table 1. The Plasma Peptide Atlas has a stringent 1 percent false discovery rate at the protein level (corresponding to a 0.2 percent FDR at the peptide level). The resulting Plasma Peptide Atlas [108] of 1929 canonical proteins also excludes any ambiguous matches.
4 Plasma biomarkers for eye disease
As noted earlier, there is a need to identify biomarkers that can identify subjects at risk for disease and to monitor response to treatment. No discovery phase investigations of the plasma proteome have been undertaken for major blinding eye diseases such as age-related macular degeneration, primary open angle glaucoma, and diabetic retinopathy. In addition, the relationship between plasma proteins and proteins found in tears, aqueous, and vitreous remains poorly understood. The retina and choroid are highly vascularized and may shed proteins into the circulation. In addition, the ciliary body produces aqueous humor, which circulates through the anterior chamber of the eye, exits via the trabecular meshwork and Schlemm's canal, and enters the circulation through episcleral veins. Thus, plasma proteins may provide important clues to biological processes that occur in the eye.
In order to learn whether proteins detected in the current human eye proteome are present in plasma, we compared the former with the latest version of the Human Plasma Peptide Atlas. IPI accessions from the plasma list were converted to UniProt accessions and compared with the human eye proteome. There were 1317 proteins common to both eye and plasma, 3525 proteins unique to the eye, and 611 proteins unique to plasma (Supplementary Table 3).
4.1 Age-related macular degeneration
Age-related macular degeneration is a disorder of the macular area of the retina characterized by soft or confluent drusen, areas of increased pigment in the outer retina or choroid associated with drusen, and areas of depigmentation or hyperpigmentation of the retinal pigment epithelium. Late stages of macular degeneration can be either non-neovascular (dry, atrophic, or nonexudative) characterized by drusen and geographic atrophy, or neovascular, characterized by choroidal neovascularization and its sequelae [86]. The major epidemiological risk factors for age-related macular degeneration include increasing age, smoking, obesity, white race, and low intake of dietary antioxidants [100-111]. Genetic association analyses have identified variants in and around complement factor H and the ARMS2/HTRA1 region with increased susceptibility to age-related macular degeneration [112]. Although susceptibility genes play a role in over half of cases, many individuals carrying macular degeneration risk phenotypes never develop the disease, and only a fraction with diagnosed disease progress to advanced age-related macular degeneration with visual loss.
Age-related inflammatory processes, as reflected by elevated plasma C-reactive protein [113], and complement dysregulation appear to be involved in the etiology of disease, but understanding of the precise biological mechanisms is far from clear. In one case-control study, elevated plasma carboxyethylpyrrole was elevated in patients with age-related macular degeneration compared with controls [114].
4.2 Primary open angle glaucoma
Primary open angle glaucoma is a multifactorial, chronic degenerative optic neuropathy characterized by loss of retinal ganglion cells and their axons, optic nerve cupping, and visual field loss [115]. Several circulating biomarkers have been associated with primary open angle glaucoma, including antibodies against neuron specific enolase, antibodies to heat shock proteins, and brain-derived neurotrophic factor, as summarized in two recent reviews [116,117]. The diagnosis of primary open angle glaucoma is often difficult to make, thus, the accurate classification of cases and controls will be a challenge to proteomic studies of glaucoma.
4.3 Diabetic retinopathy
Diabetic retinopathy is the leading cause of blindness in working-aged persons in the United States [118]. Known risk factors, such as duration of diabetes, hemoglobin A1c levels, hypertension, hyperlipidemia, pregnancy, and renal disease, only explain a limited amount of variance in the risk of diabetic retinopathy [118]. The underlying pathogenesis of diabetic retinopathy is poorly understood [119]. The search for circulating biomarkers for diabetes and diabetic retinopathy in particular remains unsatisfactory. Elevated plasma levels of markers for inflammation and hemostasis are non-specific and have limited use as biomarkers for prediction of diabetic retinopathy [120]. An interesting approach for future studies may be to compare biomarkers of diabetic retinopathy with biomarker candidates for diabetic nephropathy and diabetic neuropathy.
5 Advances in proteomics and their potential clinical applications in ophthalmology and vision science
Major advances in mass spectrometry instrumentation, proteomic methodologies, and bioinformatics have dramatically transformed the field within the last few years. These advances are familiar to established investigators in proteomics and are summarized here to orient new investigators as to the potential transformative role of the proteomics revolution in ophthalmology and vision science.
Early proteomic studies of the eye used the traditional protein separation method of two-dimensional gel electrophoresis combined with iontrap or MALDI-TOF mass spectrometry. Higher resolving power and mass measurement accuracy have been facilitated with the advent of the Orbitrap mass spectrometer [121,122] and the TripleTOF (AB Sciex) with a scan rate of 100 Hz allowing for the identification of more proteins. New depletion columns allow the removal of the most highly abundant proteins from plasma and aqueous, thus allowing the identification of a greater number of proteins of lower abundance. With these new developments, it is now possible to observe thousands of proteins in tissues and body fluids, something that was not possible even five years ago.
The quantitation of proteins can be either relative, using label-free methods (spectral counts, extracted ion chromatograms, and SWATH), or stable-isotope labeling such as iTRAQ [123], stable-isotope labeling by amino acids in cell culture (SILAC) [124], or isotope-coded affinity tags (ICAT) [125], or absolute, using methods such as stable-isotope labeled synthetic peptides [126]. Selected reaction monitoring (SRM) (also synonymous with multiple reaction monitoring, or MRM) is coming into wide application for the quantitation of multiple proteins across large numbers of samples [127]. SRM utilizes the triple quadrupole (QQQ) mass spectrometer that has the capability to act as mass filters and can selectively monitor for specific analytes, thus allowing for rapid quantification of predetermined sets of proteins. A novel and innovative data-independent acquistion available with the TripleTOF instrument, SWATH, uses fragment ion spectral libraries for qualitative and quantitative probing of proteomic samples in a single LC-MS/MS injection [128].
Protein isoformps that arise from alternative splicing of mRNA or from single nucleotide polymorphisms (SNPs) are the subject of increasing interest [129-131]. Each isoform that results from alternative splicing can have different biological roles. Structure prediction modeling can be applied to infer structural and functional consequences of alternative splicing of proteins [132]. SNPs can presumably affect the function of the protein by altering its structural configuration. Improvements in chromatography and separation methods may help facilitate the detection of protein isoforms [131]. Bioinformatic tools and protein databases are also being expanded to facilitate the identification of protein isoforms. Post-translational modifications (PTMs) represent a challenging area of study, as many PTMs are labile to mass spectrometry analyses and a single protein can have multiple PTMs. PTMs such as phosphorylation, acetylation, and glycosylation are common. Specific methods have been developed to identify certain PTMs [133-136]. New mass spectrometry instrumentation, such as the Orbitrap Elite, is able to detect more PTMs. O-GlcNAcylation, a PTM that is being increasingly recognized as interacting with protein phosphorylation, is amenable to study using electron transfer dissociation for ionization of peptides.
Protein complexes are involved in the majority of cellular processes, and affinity purification-mass spectrometry and in vivo tagging with fluorescent probes can facilitate both the characterization of particular protein complexes and their localization in the cell [137,138]. In situ click chemistry utilizes synthetic ligands to facilitate protein capture [139] and will likely advance the study of protein complexes in different tissues.
Formalin-fixed paraffin-embedded tissues are routinely prepared by pathology laboratories and are available in archives. The use of such tissues for proteomic analyses was initially overlooked, as the assumption was made that tissue fixation would obviate their use. However, recent studies show that robust proteomic analyses can be conducted with formalin-fixed paraffin-embedded tissues, and that these fixed tissues are largely equivalent to frozen tissue [140-142]. In addition, analysis of archival specimens stored for a decade showed no evidence of deterioration in the proteome [140].
Other innovative developments that will facilitate the study of the ocular proteome include single molecule ELISAs that can detect serum proteins to the subfemtomolar level [143], and advances in imaging that allow measurements of proteins down to single-molecule sensitivity [144].
6 The Human Eye Proteome Project
The audacious goal of the Human Eye Proteome Project is to characterize the proteome of the human eye in health and disease in order to gain insight into the pathophysiology of eye diseases and to contribute to new preventive and therapeutic modalities for the prevention of visual disability and blindness. The Human Eye Proteome Project is an open initiative of HUPO that welcomes all interested scientists.
The provisional mission of the Human Eye Proteome Project is: (1) To develop tools for the identification and quantification of proteins in the human eye, and to apply these tools to identify disease pathways that will lead to new therapeutic interventions and biomarkers for the prevention and treatment of blinding eye diseases, (2) To build a network and community of scientists who will work together and facilitate common interests in the application of proteomics to eye diseases, (3) To characterize the proteome of the normal human eye during the entire life cycle, including protein variants and post-translational modifications, (4)To establish standard operating procedures for study of the human eye proteome, such as the processing, dissection, and storage of eye bank eyes, collection of tears, aqueous humor, and vitreous, collection of ocular tissues during surgery, and storage of samples, (5) To establish standards for the reporting of proteomic data in order to develop a high-confidence human eye proteome reference set in PeptideAtlas and to compare the eye proteome with the plasma proteome in the plasma PeptideAtlas, (6)To establish repositories of human samples to facilitate exchange and collaboration, (7) To collect eye proteome data and research protocols from members for open access, and (8) To raise funds for exchange of post-doctoral fellows, symposia at HUPO, and collaborative symposia with the Association for Research in Vision and Ophthalmology (ARVO). Further development of the Human Eye Proteome Project will take place in special sessions at the annual World HUPO meetings.
Supplementary Material
Acknowledgments
This work was supported by NIH Grants R01 AG027012, R01 HL111271, R01 EY012712, R21 HL112662, P50 HL084946, and NHLBI contract TAS::75 0872::TAS.
Abbreviations
- TGFβIp
transforming growth factor-beta-induced protein
Footnotes
The authors declare that they have no conflict of interest.
References
- 1.Pascolini D, Mariotti SP. Global estimates of visual impairment. Br J Ophthalmol. 2012;96:614–618. doi: 10.1136/bjophthalmol-2011-300539. [DOI] [PubMed] [Google Scholar]
- 2.Gordois A, Cutler H, Pezzullo L, Gordon K, Cruess A, Winyard S, Hamilton W, Chua K. An estimation of the worldwide economic and health burden of visual impairment. Glob Public Health. 2012;7:465–481. doi: 10.1080/17441692.2011.634815. [DOI] [PubMed] [Google Scholar]
- 3.Aebersold R, Bader GD, Edwards AM, van Eyk JE, Kussmann M, Qin J, Omenn GS. The biology/disease-driven human proteome project (B/D-HPP): enabling protein research for the life sciences community. J Proteome Res. 2013;12:23–27. doi: 10.1021/pr301151m. [DOI] [PubMed] [Google Scholar]
- 4.Li N, Wang N, Zheng J, Liu XM, Lever OW, Erickson PM, Li L. Characterization of human tear proteome using multiple proteomic analysis techniques. J Proteome Res. 2005;4:2051–2061. doi: 10.1021/pr0501970. [DOI] [PubMed] [Google Scholar]
- 5.De Souza GA, de Godoy LMF, Mann M. Identification of 491 proteins in the tear fluid proteome reveals a large number of proteases and protease inhibitors. Genome Biol. 2006;7:R72. doi: 10.1186/gb-2006-7-8-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Green-Church KB, Nichols KK, Kleinholz NM, Zhang L, Nichols JJ. Investigation of the human tear film proteome using multiple proteomic approaches. Mol Vis. 2008;14:456–470. [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou L, Zhao SZ, Koh SW, Chen L, Vaz C, Tanavde V, Li XR, Beuerman RW. In-depth analysis of the human tear proteome. J Proteomics. 2012;75:3877–3885. doi: 10.1016/j.jprot.2012.04.053. [DOI] [PubMed] [Google Scholar]
- 8.González N, Iloro I, Durán JA, Elortza F, Suárez T. Evaluation of inter-day and inter-individual variability of tear peptide/protein profiles by MALDI-TOF MS analyses. Mol Vis. 2012;18:1572–1582. [PMC free article] [PubMed] [Google Scholar]
- 9.The definition and classification of dry eye disease: report of the Definition and Classification Subcommittee of the International Dry Eye Workshop (2007) Ocul Surf. 2007;5:75–92. doi: 10.1016/s1542-0124(12)70081-2. [DOI] [PubMed] [Google Scholar]
- 10.Schaumberg DA, Dana R, Buring JE, Sullivan DA. Prevalence of dry eye disease among US men: estimates from the Physicians' Health Studies. Arch Ophthalmol. 2009;127:763–768. doi: 10.1001/archophthalmol.2009.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schaumberg DA, Sullivan DA, Buring JE, Dana MR. Prevalence of dry eye syndrome among US women. Am J Ophthalmol. 2003;136:318–326. doi: 10.1016/s0002-9394(03)00218-6. [DOI] [PubMed] [Google Scholar]
- 12.Grus FH, Podust VN, Bruns K, Lackner K, Fu S, Dalmasso EA, Wirthlin A, Pfeiffer N. SELDI-TOF-MS ProteinChip array profiling of tears from patients with dry eye. Invest Ophthalmol Vis Sci. 2005;46:863–876. doi: 10.1167/iovs.04-0448. [DOI] [PubMed] [Google Scholar]
- 13.Tomosugi N, Kitagawa K, Takahashi N, Sugai S, Ishikawa I. Diagnostic potential of tear proteomic patterns in Sjögren's syndrome. J Proteome Res. 2005;4:820–825. doi: 10.1021/pr0497576. [DOI] [PubMed] [Google Scholar]
- 14.Zhou L, Beuerman RW, Chan CM, Zhao SZ, Li XR, Yang H, Ton L, Liu S, Stern ME, Tan D. Identification of tear fluid biomarkers in dry eye syndrome using iTRAQ quantitative proteomics. J Proteome Res. 2009;8:4889–4905. doi: 10.1021/pr900686s. [DOI] [PubMed] [Google Scholar]
- 15.Versura P, Nanni P, Bavelloni A, Blalock WL, Piazzi M, Roda A, Campos EC. Tear proteomics in evaporative dry eye disease. Eye. 2010;24:1396–1402. doi: 10.1038/eye.2010.7. [DOI] [PubMed] [Google Scholar]
- 16.Tong L, Zhou L, Beuerman RW, Zhao SZ, Li XR. Association of tear proteins with Meibomian gland disease and dry eye symptoms. Br J Ophthalmol. 2011;95:848–852. doi: 10.1136/bjo.2010.185256. [DOI] [PubMed] [Google Scholar]
- 17.Cojocaru VM, Ciurtin C, Uyy E, Antohe F. Nano-LC mass spectrometry proteomic tear secretion analysis in patients with secondary Sjögren's syndrome. Digest J Nanomaterials Biostructures. 2011;6:491–498. [Google Scholar]
- 18.Srinivasan S, Thangavelu M, Zhang L, Green KB, Nichols KK. iTRAQ quantitative proteomics in the analysis of tears in dry eye patients. Invest Ophthalmol Vis Sci. 2012;53:5052–5059. doi: 10.1167/iovs.11-9022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lema I, Brea D, Rodríguez-González R, Díez-Feijoo E, Sobrino T. Proteomic analysis of the tear film in patients with keratoconus. Mol Vis. 2010;16:2055–2061. [PMC free article] [PubMed] [Google Scholar]
- 20.Pannebaker C, Chandler HL, Nichols JJ. Tear proteomics in keratoconus. Mol Vis. 2010;16:1949–1957. [PMC free article] [PubMed] [Google Scholar]
- 21.Acera A, Vecino E, Rodríguez-Agirretxe I, Aloria K, Arizmendi JM, Morales C, Durán JA. Changes in tear protein profile in keratoconus disease. Eye. 2011;25:1225–1233. doi: 10.1038/eye.2011.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okrojek R, Grus FH, Matheis N, Kahaly GJ. Proteomics in autoimmune thyroid eye disease. Horm Metab Res. 2009;41:465–470. doi: 10.1055/s-0029-1214413. [DOI] [PubMed] [Google Scholar]
- 23.Koo BS, Lee DY, Ha HS, Kim JC, Kim CW. Comparative analysis of the tear protein expression in blepharitis patients using two-dimensional electrophoresis. J Proteome Res. 2005;4:719–724. doi: 10.1021/pr0498133. [DOI] [PubMed] [Google Scholar]
- 24.Acera A, Suárez T, Rodríguez-Agirretxe I, Vecino E, Durán JA. Changes in tear protein profile in patients with conjunctivochalasis. Cornea. 2011;30:42–49. doi: 10.1097/ICO.0b013e3181dea7d7. [DOI] [PubMed] [Google Scholar]
- 25.Csősz É, Boross P, Csutak A, Berta A, Tóth F, Póliska S, Török Z, Tőzsér J. Quantitative analysis of proteins in the tear fluid of patients with diabetic retinopathy. J Proteomics. 2012;75:2196–2204. doi: 10.1016/j.jprot.2012.01.019. [DOI] [PubMed] [Google Scholar]
- 26.Pieragostino D, Bucci S, Agnifili L, Fasanella V, D'Aguanno S, Mastropasqua A, Ciancaglini M, Mastropasqua L, Di Ilio C, Sacchetta P, Urbani A, Del Boccio P. Differential protein expression in tears of patients with primary open angle and pseudoexfoliative glaucoma. Mol Biosyst. 2012;8:1017–1028. doi: 10.1039/c1mb05357d. [DOI] [PubMed] [Google Scholar]
- 27.Markoulli M, Papas E, Cole N, Holden B. Differential gel electrophoresis of the tear proteome. Optom Vis Sci. 2012;89:E875–883. doi: 10.1097/OPX.0b013e318255dc46. [DOI] [PubMed] [Google Scholar]
- 28.Wong TT, Zhou L, Li J, Tong L, Zhao SZ, Li XR, Yu SJ, Koh SK, Beuerman RW. Proteomic profiling of inflammatory signaling molecules in the tears of patients on chronic glaucoma medication. Invest Ophthalmol Vis Sci. 2011;52:7385–7391. doi: 10.1167/iovs.10-6532. [DOI] [PubMed] [Google Scholar]
- 29.Zhou L, Huang LQ, Beuerman RW, Grigg ME, Li SF, Chew FT, Ang L, Stern ME, Tan D. Proteomic analysis of human tears: defensin expression after ocular surface surgery. J Proteome Res. 2004;3:410–416. doi: 10.1021/pr034065n. [DOI] [PubMed] [Google Scholar]
- 30.Karring H, Thøgersen IB, Klintworth GK, Møller-Pedersen T, Enghild JJ. A dataset of human cornea proteins identified by peptide mass fingerprinting and tandem mass spectrometry. Mol Cell Proteomics. 2005;4:1406–1408. doi: 10.1074/mcp.D500003-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Galiacy SD, Froment C, Mouton-Barbosa E, Erraud A, Chaoui K, Desjardins L, Monsarrat B, Malecaze F, Burlet-Schiltz O. Deeper in the human cornea proteome using nanoLC-Orbitrap MS/MS: an improvement for future studies on cornea homeostasis and pathophysiology. J Proteomics. 2011;75:81–92. doi: 10.1016/j.jprot.2011.09.020. [DOI] [PubMed] [Google Scholar]
- 32.Dyrlund TF, Poulsen ET, Scavenius C, Nikolajsen CL, Thøgersen IB, Vorum H, Enghild JJ. Human cornea proteome: identification and quantification of the proteins of the three main layers including epithelium, stroma, and endothelium. J Proteome Res. 2012;11:4231–4239. doi: 10.1021/pr300358k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitcher JP, Srinivasan M, Upadhyay MP. Corneal blindness: a global perspective. Bull World Health Organ. 2011;79:214–221. [PMC free article] [PubMed] [Google Scholar]
- 34.Darlington JK, Adrean SD, Schwab IR. Trends of penetrating keratoplasty in the United States from 1980 to 2004. Ophthalmology. 2006;113:2171–2175. doi: 10.1016/j.ophtha.2006.06.034. [DOI] [PubMed] [Google Scholar]
- 35.Elhalis H, Azizi B, Jurkunas UV. Fuchs endothelial corneal dystrophy. Ocul Surf. 2010;8:173–184. doi: 10.1016/s1542-0124(12)70232-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jurkunas UV, Rawe I, Bitar MS, Zhu C, Harris DL, Colby K, Joyce NC. Decreased expression of peroxiredoxins in Fuchs' endothelial dystrophy. Invest Ophthalmol Vis Sci. 2008;49:2956–2963. doi: 10.1167/iovs.07-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jurkunas UV, Bitar M, Rawe I. Colocalization of increased transforming growth factor-β-induced protein (TGFBIp) and clusterin in Fuchs endothelial corneal dystrophy. Invest Ophthalmol Vis Sci. 2009;50:1129–1136. doi: 10.1167/iovs.08-2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kennedy RH, Bourne WM, Dyer JA. A 48-year clinical and epidemiologic study of keratoconus. Am J Ophthalmol. 1986;101:267–273. doi: 10.1016/0002-9394(86)90817-2. [DOI] [PubMed] [Google Scholar]
- 39.Nielsen K, Vorum H, Fagerholm P, Birkenkamp-Demtröder K, Honoré B, Ehlers N, Orntoft TF. Proteome profiling of corneal epithelium and identification of marker proteins for keratoconus, a pilot study. Exp Eye Res. 2006;82:201–209. doi: 10.1016/j.exer.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 40.Srivastava OP, Chandrasekaran D, Pfister RR. Molecular changes in selected epithelial proteins in human keratoconus corneas compared to normal corneas. Mol Vis. 2006;12:1615–1625. [PubMed] [Google Scholar]
- 41.Joseph R, Srivastava OP, Pfister RR. Differential epithelial and stromal protein profiles in keratoconus and normal human corneas. Exp Eye Res. 2011;92:282–298. doi: 10.1016/j.exer.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 42.Lyngholm M, Vorum H, Nielsen K, Ehlers N, Honoré B. Attempting to distinguish between endogenous and contaminating cytokeratins in a corneal proteomic study. BMC Ophthalmol. 2011;11:3. doi: 10.1186/1471-2415-11-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kannabiran C, Klintworth GK. TGFBI gene mutations in corneal dystrophies. Hum Mut. 2006;27:615–625. doi: 10.1002/humu.20334. [DOI] [PubMed] [Google Scholar]
- 44.Yang J, Han X, Huang D, Yu L, Zhu Y, Tong Y, Zhu B, Li C, Weng M, Ma X. Analysis of TGFBI gene mutations in Chinese patients with corneal dystrophies and review of the literature. Mol Vis. 2010;16:1186–1193. [PMC free article] [PubMed] [Google Scholar]
- 45.Karring H, Runager K, Thøgersen IB, Klintworth GK, Højrup P, Enghild JJ. Composition and proteolytic processing of corneal deposits associated with mutations in the TGFBI gene. Exp Eye Res. 2012;96:163–170. doi: 10.1016/j.exer.2011.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Richardson MR, Price MO, Price FW, Pardo JC, Grandlin JC, You J, Wang M, Yoder MC. Proteomic analysis of human aqueous humor using multidimensional protein identification technology. Mol Vis. 2009;15:2740–2750. [PMC free article] [PubMed] [Google Scholar]
- 47.Escoffier P, Paris L, Bodaghi B, Danis M, Mazier D, Marinach-Patrice C. Pooling aqueous humor samples: bias in 2D-LC-MS/MS strategy? J Proteome Res. 2010;9:789–797. doi: 10.1021/pr9006602. [DOI] [PubMed] [Google Scholar]
- 48.Chowdhury UR, Madden BJ, Charlesworth MC, Fautsch MP. Proteome analysis of human aqueous humor. Invest Ophthalmol Vis Sci. 2010;51:4921–4931. doi: 10.1167/iovs.10-5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bennett KL, Funk M, Tschernutter M, Breitwieser FP, Planyavsky M, Mohien CU, Müller A, Trajanoski Z, Colinge J, Superti-Furga G, Schmidt-Erfurth U. Proteomic analysis of human cataract aqueous humour: comparison of one-dimensional gel LCMS with two-dimensional LCMS of unlabelled and iTRAQ®-labelled specimens. J Proteomics. 2011;74:151–166. doi: 10.1016/j.jprot.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 50.Quigley HA, Broman AT. The number of people with glaucoma worldwide in 2010 and 2020. Br J Ophthalmol. 2006;90:262–267. doi: 10.1136/bjo.2005.081224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grus FH, Joachim SC, Sandmann S, Thiel U, Bruns K, Lackner KJ, Pfeiffer N. Transthyretin and complex protein pattern in aqueous humor of patients with primary open-angle glaucoma. Mol Vis. 2008;14:1437–1445. [PMC free article] [PubMed] [Google Scholar]
- 52.Duan X, Xue P, Wang N, Dong Z, Lu Q, Yang F. Proteomic analysis of aqueous humor from patients with primary open angle glaucoma. Mol Vis. 2010;16:2839–2846. [PMC free article] [PubMed] [Google Scholar]
- 53.Izzotti A, Longobardi M, Cartiglia C, Saccà SC. Proteome alterations in primary open angle glaucoma aqueous humor. J Proteome Res. 2010;9:4831–4838. doi: 10.1021/pr1005372. [DOI] [PubMed] [Google Scholar]
- 54.Duan X, Xue P, Wang N, Dong Z, Lu Q, Yang F. Proteomic analysis of aqueous humor from patients with primary open angle glaucoma. Mol Vis. 2010;16:2839–2846. [PMC free article] [PubMed] [Google Scholar]
- 55.Anshu A, Price MO, Richardson MR, Segu ZM, Lai X, Yoder MC, Price JW., Jr Alterations in the aqueous humor proteome in patients with a glaucoma shunt device. Mol Vis. 2011;17:1891–1900. [PMC free article] [PubMed] [Google Scholar]
- 56.Saccà SC, Centofanti M, Izzotti A. New proteins as vascular biomarkers in primary open angle glaucomatous aqueous humor. Invest Ophthalmol Vis Sci. 2012;53:4242–4253. doi: 10.1167/iovs.11-8902. [DOI] [PubMed] [Google Scholar]
- 57.Funding M, Vorum H, Honoré B, Nexø E, Ehlers N. Proteomic analysis of aqueous humour from patients with acute corneal rejection. Acta Ophthalmol Scand. 2005;83:31–39. doi: 10.1111/j.1600-0420.2005.00381.x. [DOI] [PubMed] [Google Scholar]
- 58.Duan X, Lu Q, Xue P, Zhang H, Dong Z, Yang F, Wang N. Proteomic analysis of aqueous humor from patients with myopia. Mol Vis. 2008;14:370–377. [PMC free article] [PubMed] [Google Scholar]
- 59.Richardson RW, Segu ZM, Price MO, Lai X, Witzmann FA, Mechref Y, Yoder MC, Price FW. Alterations in the aqueous humor proteome in patients with Fuchs endothelial corneal dystrophy. Mol Vis. 2010;16:2376–2383. [PMC free article] [PubMed] [Google Scholar]
- 60.Bouhenni RA, Al Shawan S, Morales J, Wakim BT, Chomyk AM, Alkuraya FS, Edward DP. Identification of differentially expressed proteins in the aqueous humor of primary congenital glaucoma. Exp Eye Res. 2011;92:67–75. doi: 10.1016/j.exer.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 61.Kim TW, Kang JW, Ahn J, Lee EK, Cho KC, Han BNR, Hong NY, Park J, Kim KP. Proteomic analysis of the aqueous humor in age-related macular degeneration (AMD) patients. J Proteome Res. 2012;11:4034–4043. doi: 10.1021/pr300080s. [DOI] [PubMed] [Google Scholar]
- 62.Kyselova Z. Mass spectrometry-based proteomics approaches applied in cataract research. Mass Spectrometry Rev. 2011;30:1173–1184. doi: 10.1002/mas.20317. [DOI] [PubMed] [Google Scholar]
- 63.West SK. Looking forward to 20/20: a focus on the epidemiology of eye diseases. Epidemiol Rev. 2000;22:64–70. doi: 10.1093/oxfordjournals.epirev.a018025. [DOI] [PubMed] [Google Scholar]
- 64.West S. Epidemiology of cataract, accomplishments over 25 years and future directions. Ophthalmic Epidemiol. 2007;14:173–178. doi: 10.1080/09286580701423151. [DOI] [PubMed] [Google Scholar]
- 65.MacCoss MJ, McDonald WH, Saraf A, Sadygov R, Clark JM, Tasto JJ, Gould KL, Wolters D, Washburn M, Weiss A, Clark JI, Yates JR., III hotgun identification of protein modifications from protein complexes and lens tissues. Proc Natl Acad Sci USA. 2002;99:7900–7905. doi: 10.1073/pnas.122231399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Srivastava OP, Srivastava K. Existence of deamidated αB-crystallin fragments in normal and cataractous human lenses. Mol Vis. 2003;9:110–118. [PubMed] [Google Scholar]
- 67.Harrington V, McCall S, Huynh S, Srivastava K, Srivastava OP. Crystallins in water soluble-high molecular weight protein fractions and water insoluble protein fractions in aging and cataractous human lenses. Mol Vis. 2003;10:476–489. [PubMed] [Google Scholar]
- 68.Wilmarth PA, Tanner S, Dasari S, Nagalla SR, Riviere MA, Bafna V, Pevzner PA, David LL. Age-related changes in human crystallins determined from comparative analysis of post-translational modifications in young and aged lens: does deamidation contribute to crystallin insolubility? J Proteome Res. 2006;5:2554–2566. doi: 10.1021/pr050473a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Harrington V, Srivastava OP, Kirk M. Proteomic analysis of water insoluble proteins from normal and cataractous human lenses. Mol Vis. 2007;13:1680–1694. [PubMed] [Google Scholar]
- 70.Zhang C, Liu P, Wang N, Li Y, Wang L. Comparison of two tandem mass spectrometry-based methods for analyzing the proteome of healthy human lens fibers. Mol Vis. 2007;13:1873–1877. [PubMed] [Google Scholar]
- 71.Srivastava K, Chaves JM, Srivastava OP, Kirk M. Multi-crystallin complexes exist in the water-soluble high molecular weight protein fractions of aging normal and cataractous human lenses. Exp Eye Res. 2008;87:356–366. doi: 10.1016/j.exer.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 72.Hains PG, Truscott RJW. Proteome analysis of human foetal, aged and advanced nuclear cataract lenses. Proteomics Clin Appl. 2008;2:1611–1619. doi: 10.1002/prca.200800085. [DOI] [PubMed] [Google Scholar]
- 73.Hains PG, Truscott RJW. Proteomic analysis of the oxidation of cysteine residues in human age-related nuclear cataract lenses. Biochim Biophys Acta. 2008;1784:1959–1964. doi: 10.1016/j.bbapap.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 74.Hains PG, Truscott RJW. Age-dependent deamidation of lifelong proteins in the human lens. Invest Ophthalmol Vis Sci. 2010;51:3107–3114. doi: 10.1167/iovs.09-4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang CH, Wang YT, Tsai CF, Chen YJ, Lee JS, Chiou SH. Phosphoproteomics characterization of novel phosphorylation sites of lens proteins from normal and cataractous human eye lenses. Mol Vis. 2011;17:186–198. [PMC free article] [PubMed] [Google Scholar]
- 76.Nakanishi T, Koyama R, Ikeda T, Shimizu A. Catalogue of soluble proteins in the human vitreous humor: comparison between diabetic retinopathy and macular hole. J Chromatog B. 2002;776:89–100. doi: 10.1016/s1570-0232(02)00078-8. [DOI] [PubMed] [Google Scholar]
- 77.Koyama R, Nakanishi T, Ikeda T, Shimizu A. Catalogue of soluble proteins in human vitreous humor by one-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electrospray ionization mass spectrometry including seven angiogenesis-regulating factors. J Chromatog B. 2003;792:5–21. doi: 10.1016/s1570-0232(03)00133-8. [DOI] [PubMed] [Google Scholar]
- 78.Yamane K, Minamoto A, Yamashita H, Takamura H, Miyamoto-Myoken Y, Yoshizato K, Nabetani T, Tsugita A, Mishima HK. Proteome analysis of human vitreous proteins. Mol Cell Proteomics. 2003;2:1177–1187. doi: 10.1074/mcp.M300038-MCP200. [DOI] [PubMed] [Google Scholar]
- 79.Ouchi M, West K, Crabb JW, Kinoshita S, Kamei M. Proteomic analysis of vitreous from diabetic macular edema. Exp Eye Res. 2005;81:176–182. doi: 10.1016/j.exer.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 80.Kim SJ, Kim S, Park J, Lee HK, Park KS, Yu HG, Kim Y. Differential expression of vitreous proteins in proliferative diabetic retinopathy. Curr Eye Res. 2006;31:231–240. doi: 10.1080/02713680600557030. [DOI] [PubMed] [Google Scholar]
- 81.Kim T, Kim SJ, Kim K, Kang UB, Lee C, Park KS, Yu HG, Kim Y. Profiling of vitreous proteomes from proliferative diabetic retinopathy and nondiabetic patients. Proteomics. 2007;7:4203–4215. doi: 10.1002/pmic.200700745. [DOI] [PubMed] [Google Scholar]
- 82.García-Ramírez M, Canals F, Hernández C, Colomé N, Ferrer C, Carrasco E, García-Arumí J, Simó R. Proteomic analysis of human vitreous fluid by fluorescence-based difference gel electrophoresis (DIGE): a new strategy for identifying potential candidates in the pathogenesis of proliferative diabetic retinopathy. Diabetologia. 2007;50:1294–1303. doi: 10.1007/s00125-007-0627-y. [DOI] [PubMed] [Google Scholar]
- 83.Gao BB, Chen X, Timothy N, Aiello LP, Feener EP. Characterization of the vitreous proteome in diabetes without diabetic retinopathy and diabetes with proliferative diabetic retinopathy. J Proteome Res. 2008;7:2516–2525. doi: 10.1021/pr800112g. [DOI] [PubMed] [Google Scholar]
- 84.Shitama T, Hayashi H, Noge S, Uchio E, Oshima K, Haniu H, Takemori N, Komori N, Matsumoto H. Proteome profiling of vitreoretinal diseases by cluster analysis. Proteomics Clin Appl. 2008;2:1265–1280. doi: 10.1002/prca.200800017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang H, Feng L, Hu JW, Xie CL, Wang F. Characterisation of the vitreous proteome in proliferative diabetic retinopathy. Proteome Sci. 2012;10:15. doi: 10.1186/1477-5956-10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jager RD, Meiler WF, Miller JW. Age-related macular degeneration. N Engl J Med. 2008;358:2606–2617. doi: 10.1056/NEJMra0801537. [DOI] [PubMed] [Google Scholar]
- 87.Friedman DS, O'Colmain BJ, Muñoz B, Tomany SC, McCarty C, de Jong PTVM, Nemesure B, Mitchell P, Kempen J, Congdon N. Prevalence of age-related macular degeneration in the United States. Arch Ophthalmol. 2004;122:564–572. doi: 10.1001/archopht.122.4.564. [DOI] [PubMed] [Google Scholar]
- 88.AMD Alliance International, The Global Economic Cost of Visual Impairment. Canberra: AMD Alliance International; 2010. [Accessed 11/08/2011]. Available at: http://www.amdalliance.org/cost-of-blindness.html. [Google Scholar]
- 89.Crabb JW, Miyagi M, Gu X, Shadrach K, West KA, Sakaguchi H, Kamei M, Hasan A, Yan L, Rayborn ME, Salomon RG, Hollyfield JG. Drusen proteome analysis: an approach to the etiology of age-related macular degeneration. Proc Natl Acad Sci. 2002;99:14682–14687. doi: 10.1073/pnas.222551899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ng KP, Gugiu B, Renganathan K, Davies MW, Gu X, Crabb JS, Kim SR, Rózanowska MB, Bonilha VL, Rayborn ME, Salomon RG, Sparrow JR, Boulton ME, Hollyfield JG, Crabb JW. Retinal pigment epithelium lipofuscin proteomics. Mol Cell Proteomics. 2008;7:1397–1405. doi: 10.1074/mcp.M700525-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nordgaard CL, Berg KM, Kapphahn RJ, Reilly C, Feng X, Olsen TW, Ferrington DA. Proteomics of the retinal pigment epithelium reveals altered protein expression at progressive stages of age-related macular degeneration. Invest Ophthalmol Vis Sci. 2006;47:815–822. doi: 10.1167/iovs.05-0976. [DOI] [PubMed] [Google Scholar]
- 92.Ethen CM, Reilly C, Feng X, Olsen TW, Ferrington DA. The proteome of central and peripheral retina with progression of age-related macular degeneration. Invest Ophthalmol Vis Sci. 2006;47:2280–2290. doi: 10.1167/iovs.05-1395. [DOI] [PubMed] [Google Scholar]
- 93.Nordgaard CL, Karunadharma PP, Feng X, Olsen TW, Ferrington DA. Mitochondrial proteomics of the retinal pigment epithelium at progressive stages of age-related macular degeneration. Invest Ophthalmol Vis Sci. 2008;49:2848–2855. doi: 10.1167/iovs.07-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Alcazar O, Hawkridge AM, Collier TS, Cousins SW, Bhattacharya SK, Muddiman DC, Marin-Castano ME. Proteomics characterization of cell membrane blebs in human retinal pigment epithelium cells. Mol Cell Proteomics. 2009;8:2201–2211. doi: 10.1074/mcp.M900203-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Decanini A, Karunadhrama PR, Nordgaard CL, Feng X, Olsen TW, Ferrington DA. Human retinal pigment epithelium proteome changes in early diabetes. Diabetologia. 2008;51:1051–1061. doi: 10.1007/s00125-008-0991-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hernández C, García-Ramírez M, Colomé N, Villarroel M, Corraliza L, García-Pacual L, Casado J, Canals F, Simó R. New pathogenic candidates for diabetic macular edema detected by proteomic analysis. Diabetes Care. 2010;33:e92. doi: 10.2337/dc10-0232. [DOI] [PubMed] [Google Scholar]
- 97.Len ACL, Powner MB, Zhu L, Hageman GS, Song X, Fruttiger M, Gillies MC. Pilot application of iTRAQ to the retinal disease macular telangiectasia. J Proteome Res. 2012;11:537–553. doi: 10.1021/pr200889t. [DOI] [PubMed] [Google Scholar]
- 98.Hathout Y, Flippin J, Fan C, Liu P, Csaky K. Metabolic labeling of human primary retinal pigment epithelial cells for accurate comparative proteomics. J Proteome Res. 2005;4:620–627. doi: 10.1021/pr049749p. [DOI] [PubMed] [Google Scholar]
- 99.Yuan X, Gu X, Crabb JS, Yue X, Shadrach K, Hollyfield JG, Crabb JW. Quantitative proteomics: comparison of the macular Bruch membrane/choroid complex from age-related macular degeneration and normal eyes. Mol Cell Proteomics. 2010;9:1031–1046. doi: 10.1074/mcp.M900523-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bhattacharya SK, Crabb JS, Bonilha VL, Gu X, Takahara H, Crabb JW. Proteomics implicates peptidyl arginine deiminase 2 and optic nerve citrullination in glaucoma pathogenesis. Invest Ophthalmol Vis Sci. 2006;47:2508–2514. doi: 10.1167/iovs.05-1499. [DOI] [PubMed] [Google Scholar]
- 101.Rogers RS, Dharsee M, Ackloo S, Sivak JM, Flanagan JG. Proteomics analyses of human optic nerve head astrocytes following biomechanical strain. Mol Cell Proteomics. 2012;11:1–17. doi: 10.1074/mcp.M111.012302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rogers R, Dharsee M, Ackloo S, Flanagan JG. Proteomics analyses of activated human optic nerve head lamina cribrosa cells following biomechanical strain. Invest Ophthalmol Vis Sci. 2012;53:3806–3816. doi: 10.1167/iovs.11-8480. [DOI] [PubMed] [Google Scholar]
- 103.Picciani R, Junk AK, Bhattacharya SK. Technical brief: a novel strategy for enrichment of trabecular meshwork protease proteome. Mol Vis. 2008;14:871–877. [PMC free article] [PubMed] [Google Scholar]
- 104.Kersey PJ, Duarte J, Williams A, Karavidopoulou Y, Birney E, Apweiler R. The International Protein Index: an integrated database for proteomics experiments. Proteomics. 2004;4:1995–1998. doi: 10.1002/pmic.200300721. [DOI] [PubMed] [Google Scholar]
- 105.The UniProt Consortium Reorganizing the protein space at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2012;40:D71–D75. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Huang Y, Niu B, Gao Y, Fu L, Li W. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics. 2010;26:680–682. doi: 10.1093/bioinformatics/btq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dyrlund TF, Poulsen ET, Scavenius C, Sanggaard KW, Enghild JJ. MS Data Miners: a web-based software tool to analyze, compare, and share mass spectrometry protein identifications. Proteomics. 2012;12:2792–2796. doi: 10.1002/pmic.201200109. [DOI] [PubMed] [Google Scholar]
- 108.Farrah T, Deutsch EW, Omenn GS, Campbell DS, Sun Z, Bletz JA, Mallick P, Katz JE, Malmström J, Ossola R, Watts JD, Lin B, Zhang H, Moritz RL, Aebersold R. A high-confidence human plasma proteome reference set with estimated concentrations in PeptideAtlas. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.006353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Smith W, Assink J, Klein R, Mitchell P, Klaver CC, Klein BE, Hofman A, Jensen S, Wang JJ, De Jong PT. Risk factors for age-related macular degeneration: pooled findings from three continents. Ophthalmology. 2001;108:697–704. doi: 10.1016/s0161-6420(00)00580-7. [DOI] [PubMed] [Google Scholar]
- 110.Tomany SC, Wang JJ, Van Leeuwen R, Klein R, Mitchell P, Vingerling JR, Klein BE, Smith W, De Jong PT. Risk factors for incident age-related macular degeneration: pooled findings from three continents. Ophthalmology. 2004;111:1280–1287. doi: 10.1016/j.ophtha.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 111.Semba RD. Handbook of Nutrition and Ophthalmology. Totowa NJ: Humana Press; 2007. [Google Scholar]
- 112.DeAngelis MM, Silveira AC, Carr EA, Kim IK. Genetics of age-related macular degeneration: current concepts, future directions. Sem Ophthalmol. 2011;26:77–93. doi: 10.3109/08820538.2011.577129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hong T, Tan AG, Mitchell P, Wang JJ. A review and meta-analysis of the association between C-reactive protein and age-related macular degeneration. Surv Ophthalmol. 2011;56:184–194. doi: 10.1016/j.survophthal.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 114.Gu J, Pauer GJT, Yu X, Narendra U, Sturgill GM, Bena J, Gu X, Peachey NS, Salomon RG, Hagstrom SA, Crabb JW. Assessing susceptibility to age-related macular degeneration with proteomic and genomic biomarkers. Mol Cell Proteomics. 2009;8:1338–1349. doi: 10.1074/mcp.M800453-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kwon YH, Fingert JH, Kuehn MH, Alward WLM. Primary open-angle glaucoma. N Engl J Med. 2009;360:1113–1124. doi: 10.1056/NEJMra0804630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Knepper PA, Samples JR, Yue BYJT. Biomarkers of primary open-angle glaucoma. Expert Rev Ophthalmol. 5:731–742. doi: 10.1586/EOP.10.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kokotas H, Kroupis C, Chiras D, Grigoriadou M, Lamnissou K, Petersen MB, Kitsos G. Biomarkers in primary open angle glaucoma. Clin Chem Lab Med. 2012 Jun 29;:1–13. doi: 10.1515/cclm-2012-0048. [DOI] [PubMed] [Google Scholar]
- 118.Mohamed Q, Gillies MC, Wong TY. Management of diabetic retinopathy: a systematic review. JAMA. 2007;298:902–916. doi: 10.1001/jama.298.8.902. [DOI] [PubMed] [Google Scholar]
- 119.Frank RN. Diabetic retinopathy. N Engl J Med. 2004;350:48–58. doi: 10.1056/NEJMra021678. [DOI] [PubMed] [Google Scholar]
- 120.Nguyen TT, Alibrahim E, Islam FMA, Klein R, Klein BEK, Cotch MF, Shea S, Wong TY. Inflammatory, hemostatic, and other novel biomarkers for diabetic retinopathy: the Multi-Ethnic Study of Atherosclerosis. Diabetes Care. 2009;32:1704–1709. doi: 10.2337/dc09-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hu Q, Noll RJ, Makarov A, Hardman M, Graham Cooks R. TheOrbitrap: a new mass spectrometer. J Mass Spectrom. 2005;40:430–443. doi: 10.1002/jms.856. [DOI] [PubMed] [Google Scholar]
- 122.Michalski A, Damoc E, Lange O, Denisov E, Nolting D, Müller M, Viner R, Schwartz J, Remes P, Belford M, Dunyach JJ, Cox J, Horning S, Mann M, Makarov A. Ultra high resolution linear ion trap Orbitrap mass spectrometer (Orbitrap Elite) facilitates top down LC MS/MS and versatile peptide fragmentation modes. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.O111.013698. Epub 2011 Dec 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayashta S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 124.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 125.Smolka MB, Zhou H, Purkayastha S, Aebersold R. Optimization of the isotope-coded affinity tag-labeling procedure for quantitative proteome analysis. Anal Biochem. 2001;297:25–31. doi: 10.1006/abio.2001.5318. [DOI] [PubMed] [Google Scholar]
- 126.Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc Natl Acad Sci USA. 2003;100:6940–6945. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Picotti P, Aebersold R. Selected reaction monitoring-based proteomics, workflows, potential, pitfalls, and future directions. Nat Methods. 2012;9:555–566. doi: 10.1038/nmeth.2015. [DOI] [PubMed] [Google Scholar]
- 128.Gille LC, Navarro P, Tate S, Röst H, Selevsek N, Reiter L, Bonner R, Aebersold R. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteomic analysis. Mol Cell Proteomics. 2012;11:1–17. doi: 10.1074/mcp.0111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Tress ML, Bodenmiller B, Aebersold R, Valencia A. Proteomics studies confirm the presence of alternative isoforms on a large scale. Genome Biol. 2008;9:R162. doi: 10.1186/gb-2008-9-11-r162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Su ZD, Sun L, Yu DX, Li RX, Li HX, Yu ZJ, Sheng QH, Lin X, Zeng R, Wu JR. Quantitative detection of single amino acid polymorphisms by targeted proteomics. J Mol Cell Biol. 2011;3:309–315. doi: 10.1093/jmcb/mjr024. [DOI] [PubMed] [Google Scholar]
- 131.Stastna M, Van Eyk JE. Analysis of protein isoforms: can we do it better? Proteomics J. 2012 doi: 10.1002/pmic.201200161. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Menon R, Roy A, Mukherjee S, Belkin S, Zhang Y, Omenn GS. Functional implications of structural predictions for alternative splice proteins expressed in Her2/neu-induced breast cancers. J Proteome Res. 2011;10:5503–5511. doi: 10.1021/pr200772w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Palmisano G, Thingholm TE. Strategies for quantitation of phosphoproteomic data. Expert Rev Proteomics. 2010;7:439–456. doi: 10.1586/epr.10.19. [DOI] [PubMed] [Google Scholar]
- 134.Vertegaal ACO. Uncovering ubiquitin and ubiquitin-like signaling networks. Chem Rev. 2011;111:7923–7940. doi: 10.1021/cr200187e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Paruchuri VD, Zachara NE. Defining the heart and cardiovascular O-GlcNAcome: a review of approaches and methods. Circ Cardiovasc Genet. 2011;4:710. doi: 10.1161/CIRCGENETICS.110.957779. [DOI] [PubMed] [Google Scholar]
- 136.De Ceuleneer M, Van Steendam K, Dhaenens M, Deforce D. In vivo relevance of citrullinated proteins and the challenges in their detection. Proteomics. 2012;12:752–760. doi: 10.1002/pmic.201100478. [DOI] [PubMed] [Google Scholar]
- 137.Cristea IM, Williams R, Chait BT, Rout MP. Fluorescent proteins as proteomic probes. Mol Cell Proteomics. 2005;4:1933–1941. doi: 10.1074/mcp.M500227-MCP200. [DOI] [PubMed] [Google Scholar]
- 138.Sardiu ME, Washburn MP. Building protein-protein interaction networks with proteomics and informatics tools. J Biol Chem. 2011;286:23645–23651. doi: 10.1074/jbc.R110.174052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Millward SW, Agnew HD, Lai B, Lee SS, Lim J, Nag A, Pitram S, Rohde R, Heath JR. In situ click chemistry: from small molecule discovery to synthetic antibodies. Integr Biol (Cambridge) 2012 Jul 26; doi: 10.1039/c2ib20110k. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sprung RW, Jr, Brock JWC, Li M, Washington MK, Slebos RJC, Liebler DC. Equivalence of protein inventories obtained from formalin-fixed paraffin-embedded and frozen tissue in multidimensional liquid chromatography-tandem mass spectrometry shotgun proteomic analysis. Mol Cell Proteomics. 2009;8:1988–1998. doi: 10.1074/mcp.M800518-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Scicchitano MS, Dalmas DA, Boyce RW, Thomas HC, Frazier KS. Protein extraction of formalin-fixed, paraffin-embedded tissue enables robust proteomic profiles by mass spectrometry. J Histochem Cytochem. 2009;57:849–860. doi: 10.1369/jhc.2009.953497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sprung RW, Martinez MA, Carpenter KL, Ham AJL, Washington MK, Arteaga CL, Sanders ME, Liebler DC. Precision of multiple reaction monitoring mass spectrometry analysis of formalin-fixed, paraffin-embedded tissue. J Proteome Res. 11:3498–3505. doi: 10.1021/pr300130t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Rissin DM, Kan CW, Campbell TG, Howes SC, Fournier DR, Song L, Piech T, Patel PP, Chang L, Rivnak AJ, Ferrell EP, Randall JD, Provuncher GK, Walt DR, Duffy DC. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nature Biotechnol. 2010;28:595–599. doi: 10.1038/nbt.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bancaud A, Huet S, Rabut G, Ellenberg J. Fluorescence perturbation techniques to study mobility and molecular dynamics of proteins in live cells: FRAP, photoactivation, photoconversion, and FLIP. Cold Spring Harb Protoc. 2010 doi: 10.1101/pdb.top90. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.